Abstract
A challenge in understanding the mechanism of protein function in biology is to establish the correlation between functional form in the intracellular environment and high-resolution structures obtained with in vitro techniques. Here we present a strategy to probe conformational changes of proteins inside cells. Our method involves: (i) engineering binding proteins to different conformations of a target protein, and (ii) using them to sense changes in the surface property of the target in cells. We probed ligand-induced conformational changes of the estrogen receptor α (ERα) ligand-binding domain (LBD). By using yeast two-hybrid techniques, we first performed combinatorial library screening of “monobodies” (small antibody mimics using the scaffold of a fibronectin type III domain) for clones that bind to ERα and then characterized their interactions with ERα in the nucleus, the native environment of ERα, in the presence of various ligands. A library using a highly flexible loop yielded monobodies that specifically recognize a particular ligand complex of ERα, and the pattern of monobody specificity was consistent with the structural differences found in known crystal structures of ERα-LBD. A more restrained loop library yielded clones that bind both agonist- and antagonist-bound ERα. Furthermore, we found that a deletion of the ERα F domain that is C-terminally adjacent to the LBD increased the crossreactivity of monobodies to the apo-ERα-LBD, suggesting a dynamic nature of the ERα-LBD conformation and a role of the F domain in restraining the LBD in an inactive conformation.
Many biological processes are regulated by proteins. Regulatory proteins undergo conformational changes to alter the surface properties that in turn modulate their interactions with partners and/or alter their catalytic efficiency. Thus, it is essential to detect conformational changes of proteins to understand the molecular mechanism underlying their functions. It is generally accepted that the “molecular crowding” within the cellular environment can significantly affect ligand binding, catalysis, stability, and folding of macromolecules (1). Thus, the structures and relative populations of “active” and “inactive” conformations of a protein might be quite different from those determined by using in vitro biophysical methods. In vivo techniques such as fluorescence resonance energy transfer (FRET) (2) and enzyme fragment complementation (3) have been successfully used to detect gross conformational changes of proteins (e.g., folding, large structural rearrangement, and subunit association). The goal of this study has been to establish an in vivo technique to detect more subtle conformational changes of proteins (e.g., secondary structure rearrangement) that nevertheless lead to significant changes in surface property.
An alternative approach to directly obtaining geometric information (e.g., FRET) is to characterize the binding of conformation-specific probes to a target of interest. A classic example is conformation-specific antibodies used by Anfinsen and colleagues to characterize in vitro refolding (4). It is conceivable that one can introduce conformation-specific probes, such as antibodies, inside cells and detect their respective binding to a target to probe changes in the surface property of the target. To implement this strategy, one must first obtain conformation-specific probes and establish detection methods for probe binding. However, antibodies and their fragments usually require the formation of disulfide bonds for proper folding and thus do not always function in the reducing environment inside cells. Short peptides may also be used, but they tend to be rapidly degraded in cells because of their low resistance to proteolysis.
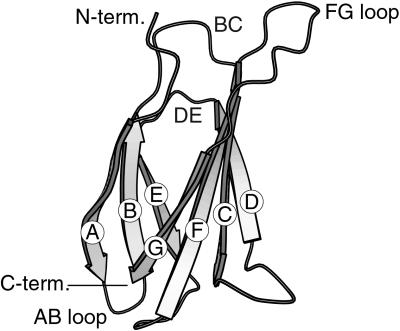
We have developed small antibody mimics, termed “monobodies,” using a small β-sheet protein scaffold, the tenth fibronectin type III domain from human fibronectin (FNfn10; Fig. 1) (5). We have shown that monobodies with a novel binding function can be engineered by screening phage-display libraries of FNfn10 in which loop regions are diversified. FNfn10 is highly stable, although it does not contain disulfide bonds or metal-binding sites (5–7). These physical properties allow monobodies to function regardless of the redox potential of the environment, making them well suited as probes used inside cells.
Figure 1.
Schematic drawing of the structure of FNfn10. β-Strands are labeled as A–G, and the loops used for monobody library construction are also labeled.
We have chosen the yeast two-hybrid technique as the method to both screen monobody libraries and detect binding of a monobody to a target in living cells. The yeast two-hybrid system has been established as a standard method to identify and characterize protein interactions in the nucleus of yeast cells (8, 9). Furthermore, R. Brent and colleagues pioneered the use of the yeast two-hybrid system for combinatorial library screening of peptide aptamers (10, 11).
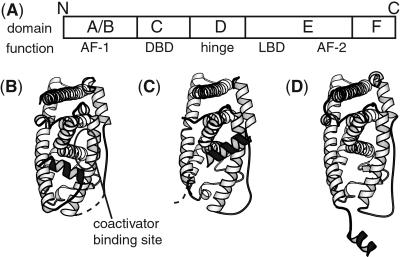
In this study, we probed ligand-induced conformational changes of the human estrogen receptor α (ERα) in the cell nucleus. ERα is a ligand-inducible transcription factor and a member of the nuclear receptor (NR) superfamily (12). ERα is a therapeutic target of, and a clinical marker for, estrogen-responsive breast tumor (13). A diverse group of ligands exist, including antiestrogens that are in clinical use, which modulate ER transcriptional activation and the physiological response of the hormone 17β-estradiol (E2) (14). ERα contains a domain structure typical of nuclear receptors (Fig. 2A) (12). The crystal structures of the DNA-binding domain (C domain) and the ligand-binding domain (LBD; also the E domain) of ERα have been determined (15–18), whereas the other domains (A/B, D, and F) are considered highly flexible. ERα agonists and antagonists bind exclusively to the LBD. The x-ray crystal structures of ERα LBD complexed with several ligands (16–18) revealed conformational changes induced by different ligands (Fig. 2 B–D). In a model based on these and other nuclear receptor crystal structures, the ligand-dependent transactivation function of NRs is accomplished through a conformational change of the LBD that alters its surface property, which in turn induces the interaction between NRs and coactivator proteins (19). However, there is a paucity of structural information of ERα in solution. Thus, probing surface property changes with conformation-specific probes may provide new insights into the solution structure and dynamics of ERα and its fragments and into mechanistic roles of the flexible regions. Also, the ability to rapidly characterize ERα surface property would be useful as a preliminary screening tool for estrogen- and antiestrogen-like molecules.
Figure 2.
(A) Nuclear receptor domain structure. AF-1, ligand-independent activation function; DBD, DNA-binding domain; AF-2, ligand-dependent activation function. (B–D) Schematic drawings of the crystal structures of ERα-LBD illustrating ligand-induced conformational changes. B and C are from ref. 16, and D is from ref. 18. Helix 12 is highlighted in black. In B, a LXXLL peptide is bound to the coactivator-binding site, but the peptide is omitted for clarity. In D, an aberrant intermolecular disulfide bond forced helix 12 to an extended conformation.
The combination of ERα and the yeast two-hybrid system is particularly suitable for the purpose of this study. Mammalian steroid receptors expressed in yeast function as hormone-dependent transcriptional activators (20, 21). ERα LBD conformation can be manipulated by a simple addition of ligands. Because yeast does not have endogenous NRs, one can study a single NR without complications arising from the coexistence of other NRs and NR-associated factors that are present in mammalian cells. The yeast two-hybrid method detects protein interactions in the nucleus, the native cellular compartment of NRs, and it has been used to discover proteins that interact with ERα in a ligand-dependent manner (22–26). Furthermore, McDonnell and colleagues have discovered a series of linear peptides that detect conformational changes of ERα by using the phage display method (27, 28), demonstrating that ERα conformation can be monitored by using small conformation-specific probes.
Here we successfully engineered a series of monobodies that recognize a specific ligand complex of ERα-LBD. Screening of two libraries with different degrees of loop flexibility yielded monobodies with distinct loop sequences and binding specificity. We then used these monobodies to probe the conformational changes of ERα in living cells. We found that the flexible F domain may have an important role in the conformational changes of the LBD.
Materials and Methods
Materials.
E2 and 4-hydroxy tamoxifen (OHT) were purchased from Sigma; diethylstilbestrol, estriol, and progesterone were from Steraloids (Newport, RI); ICI182,780 was purchased from Tocris Neuramin (Bristol, U.K.); and raloxifene is a product of Lilly Research Laboratories (Indianapolis). An anti-ERα (F domain) antibody, HC-20, was purchased from Santa Cruz Biotechnology, and anti-LexA antibody was kindly provided by E. Golemis. Secondary antibodies were purchased from Pierce. The ERα cDNA was kindly provided by the late A. Notides. The steroid receptor coactivator-1 (SRC-1) cDNA (29) was a generous gift from B. W. O'Malley.
Strains and Media.
Yeast strains EGY48, MATα his3 trp1 ura3 leu2∷6LexAop-LEU2, and RFY206, MATa his3Δ200 leu2–3 lys2Δ201 trp1Δ∷hisG ura3–52, have been described (30, 31) and were purchased from Origene (Rockville, MD). Yeast was grown in yeast extract/peptone/dextrose media or yeast complete dropout media following instructions from Invitrogen.
Construction of Yeast Two-Hybrid Vectors and Monobody Libraries.
The synthetic gene for FNfn10 (5) was subcloned in the plasmid pYESTrp2 (Invitrogen) so that FNfn10 is fused C terminal to the B42 activation domain (pYT45). The cDNA encoding residues 570–780 of SRC-1 (29) was cloned in pYESTrp2 (pYTSRC570–780). Plasmids encoding LexA-fusion proteins were constructed by subcloning an appropriate PCR fragment in the plasmid pEG202 (Origene): pEGERα297–595, the E domain of ERα (ERα-EF) (residues 297–595); pEGERα297–554, the E domain of ERα (ERα-E) (residues 297–554). Libraries and specific mutations were made by using Kunkel mutagenesis (32). The NNK or NNS sequences (N denotes a mixture of A, T, G, and C; K denotes a mixture of G and T; and S, a mixture of C and G) were used to randomize a codon. The FG7 library (2 × 106 independent clones) contained seven diversified residues in the FG loop [residues 78–84 of FNfn10 (numbering according to ref. 5)]. The AB7 library (2 × 105 independent clones) had an insertion of seven diversified residues between residues 15 and 16.
A yeast expression vector for a glutathione S-transferase (GST)-monobody fusion protein was constructed as follows. The XbaI-KpnI fragment of the modified pYEX4T-1 vector that encodes the CUP promotor and GST gene, kindly provided by E. M. Phizicky (33), was cloned between the XbaI and KpnI sites of YEplac181 (34) to make pGSTleu. Then the gene for a monobody was cloned between the NcoI and BamHI sites of pGSTleu after introducing NcoI and BamHI sites at the 5′ and 3′ ends of the monobody gene, respectively, by using PCR.
Library Screening.
The yeast strain RFY206 harboring pEGERα297–595 and a LacZ reporter plasmid, pSH18–34 (Origene), was mated with EGY48 containing a monobody library (31). Diploid cells that contain an ERα-binding monobody were selected by using the LEU+ phenotype on minimal dropout media (Gal Raf Leu− His− Ura− Trp−). A series of library screening was performed in the presence of different ERα ligands (E2, estriol, and OHT; 1 μM). Colonies grown after 3 days of incubation were further tested for galactose dependence of the LEU+ phenotype and β-galactosidase activity. The plasmids coding for a monobody were recovered from yeast clones following instructions from Origene, and the amino acid sequences of monobodies were deduced by DNA sequencing.
β-Galactosidase Assay.
The yeast RFY206 was first transformed with pEGERα297–595 (or pEGERα297–554) and pSH18–34 and subsequently with a derivative of the pYT45 plasmid encoding a particular monobody (or pYTSRC570–780). Yeast cells were grown overnight at 30°C in the YC Glc His− Ura− Trp− media. The cells were pelleted and resuspended in YC Gal/Raf/His/Ura/Trp media containing a ligand at a final cell density of 0.2 OD660 in a total volume of 175 μl in the wells of a deep 96-well plate. Ligands used were E2, ICI182,780, OHT, raloxifene, progesterone, estriol, diethylstilbestrol, and genistein. The ligand concentration was 1 μM except for genistein (10 μM). After incubating for 6 h at 30°C with shaking, 175 μl of β-galactosidase assay buffer [60 mM Na2HPO4/40 mM NaH2PO4/10 mM KCl/1 mM MgSO4/0.27% β-mercaptoethanol/14 mM SDS/4 mg/ml of 2-nitrophenyl-β-d-galactoside/50% Y-PER (Pierce)] was added to the culture and incubated at 30°C, then the reaction was stopped by adding 150 μl of 1 M Na2CO3. After centrifugation, OD420 was measured to determine β-galactosidase activity. For Western blotting analysis, yeast cells were pelleted and then suspended in 5 μl of Y-PER (Pierce) per milligram of cells, then 1 mM PMSF and 540 μg/ml of leupeptine were added, and the samples were incubated at room temperature for 20 min with gentle agitation. The suspension was spun down, and the supernatant and suspension were examined by Western blotting.
Results
Selection of Monobodies That Interact with ERα.
We used the ERα E and F domains (ERα-EF) as the target. We included the F domain so that the bait protein is closer to the full-length ER than just the LBD. The F domain is ≈45 residues long and is believed to be highly flexible. Potential roles of this domain in the ligand-dependent transcription activation have been reported (35, 36). None of the published crystal structures of ER-LBD include the F domain. We prepared two combinatorial libraries of monobodies by diversifying seven residues in the FG loop (“FG7”) and by inserting seven diversified residues in the AB loop (“AB7”), respectively (Fig. 1). The FG loop is the most flexible among FNfn10 loops, whereas the AB loop is a tight turn (6). Mutations in these loops have minor effects on the overall structure and stability of FNfn10 (V. Batori, A.K., and S.K., unpublished results).
We performed yeast two-hybrid screening of the monobody libraries for clones that interact with ERα-EF in the presence of a ligand. We used E2, estriol, and OHT in the screening of the FG7 library. The first two are agonists, whereas OHT functions as an antagonist in the breast. Because of their tissue-specific functions, chemicals such as OHT and raloxifene are classified as selective estrogen receptor modulators (37). We obtained multiple positive clones from each screening (Table 1). Monobodies that have been selected in the presence of an agonist (E2 and E3) contain motifs similar to LXXLL that are the consensus of the NR boxes of coactivators (22). Interestingly, we found a significant number of LXXML sequences among these clones. Because of the degeneracy of the codons, Leu is expected to appear three times as often as Met at a given diversified position in the library, suggesting that Met in the LXXML sequence is preferred over Leu. In addition, many of the clones contain an amino acid with a carboxyl or amino side chain at the third position of the LXXLL-like motifs. These motifs resemble the LLEML sequence within helix 12 of ERα and -β. In the ERα/OHT crystal structure, the LLEML segment of helix 12 occupies the coactivator-binding site (Fig. 2C) (16). The sequence similarity of the isolated monobodies to the coactivator motif strongly suggests that these monobodies directly bind to ERα. In contrast, monobodies identified from screening of the FG7 library in the presence of OHT contain an amino acid sequence that is distinctly different from the LXXLL motif. These sequences do not show obvious homology to those of linear peptides selected for binding to the ERα/OHT complex by Norris et al. (28).
Table 1.
The amino acid sequences of the diversified loop of selected monobodies
| Ligand* | Name | Sequence |
|---|---|---|
| FG loop library (FG7) | ||
| – | Wild-type | RGDSPAS |
| E2 | E2#3 | ALVEMLR |
| E2 | E2#4 | RLLWNSL |
| E2 | E2#5 | RVLMTLL |
| E2 | E2#7 | GLRRLLR |
| E2 | E2#8 | GLRQMLG |
| E2 | E2#9 | RVLHSLL |
| E2 | E2#10 | RVRDLLM |
| E2 | E2#11 | RVMDMLL |
| E2 | E2#23 | LRLMLAG |
| E3 | E3#2 | GIAELLR |
| E3 | E3#6 | RILLNMLT† |
| OHT | OHT#31 | GGWLWCVT† |
| OHT | OHT#32 | TWVVRRV |
| OHT | OHT#33 | TWVRPNQ |
| AB loop library (AB7)‡ | ||
| E2 | AB7-A1 | WTWVLRE |
| E2 | AB7-B1 | WVLITRS |
Ligand used for library screening.
These clones contain an additional S to T mutation at residue 85. Sequences homologous to the LXXLL motif are highlighted in bold.
Seven randomized residues were inserted in the AB loop library, as opposed to replacing residues in the FG loop library.
We obtained two clones from the AB7 library that interact with the ERα-EF/E2 complex (Table 1). Their sequences contain the WVL sequence and are distinctly different from the LXXLL motif. We examined whether we obtain no LXXLL-like sequences from the AB7 library because the library did not contain such sequences. Direct DNA sequencing of the library revealed no obvious bias at the diversified positions. The interaction between ERα-EF and a monobody containing the LRLMLAG sequence (the same as E2#23) in the AB loop was below the detection limit. These results indicate that structural restraints of the AB loop prevent an LXXLL-like sequence from taking on a conformation capable of binding to the ERα–E2 complex.
Discrimination of ERα Conformations in Living Cells by Using Conformation-Specific Monobodies.
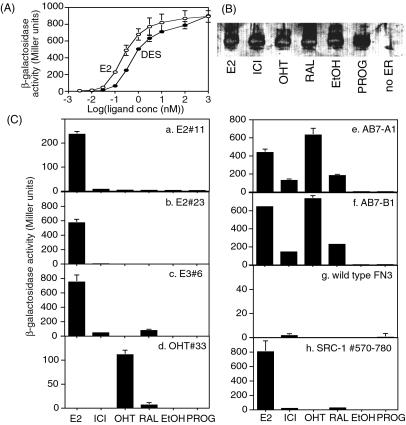
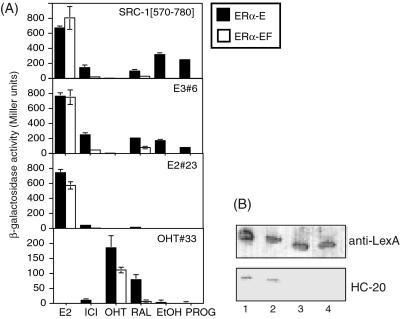
We then examined in vivo the interactions of the monobodies with different ERα-EF/ligand complexes by using β-galactosidase assays. It has been shown that β-galactosidase activity correlates modestly with the interaction affinity of different bait–prey pairs of the yeast two-hybrid system (38). In our assays, we use a single bait–prey pair for a series of ligands. Furthermore, we used a short incubation period (6 h) after the addition of a ligand and the initiation of monobody production to minimize the effect of different ligands on the expression level and degradation of the LexA-ER fusion protein. We confirmed that yeast samples prepared in the presence and absence of ligands contained similar levels of ERα-EF protein (Fig. 3B) and of monobodies (data not shown). β-Galactosidase activity of yeast cells containing LexA-ERα-EF and B42-SRC-1 exhibited a dose–response curve that is consistent with ligand binding of ERα-LBD (Fig. 3A). These results demonstrate that our procedure can quantify the degree of bait–prey interaction. Our results are consistent with a previous study on ER–coactivator interactions by using a similar yeast two-hybrid system (39).
Figure 3.
(A) Ligand dose dependence of the interaction between ERα-EF and SRC-1 determined by yeast two-hybrid β-galactosidase assays. Western blotting (B) showing similar amounts of LexA-ERa-EF in the presence of different ligands. (C) In vivo binding specificity of ERα-binding monobodies, as tested by using β-galactosidase assays. Binding specificity toward agonist, antagonist, and selective estrogen receptor modulators is shown. ICI, ICI182,780; RAL, raloxifene; PROG, progesterone; EtOH, no added ligand.
We tested in vivo interaction between monobody clones and ERα-EF in the presence of different ERα ligands (Fig. 3C). In general, monobodies selected from the FG7 library for an ERα-EF/agonist (E2 and estriol) complex interacted with ERα-EF in the presence of E2 but not in the presence of OHT or other antagonists. The binding specificity of these clones is similar to that of the NR-box fragment of the coactivator, SRC-1, suggesting that these clones recognize a surface of ER-LBD that is used for coactivator binding. The clone, E3#6, showed weak but significant interaction with the ERα-EF/raloxifene complex (Fig. 3C). In an analogous manner, monobodies selected from the FG7 library for the ERα-EF/OHT complex were specific to the same complex. In addition, the affinity of the selected monobodies to an unrelated protein (the pBait control protein; Origene) was below the detection limit of our assay (data not shown).
Monobodies selected from the AB7 library showed a very different interaction profile (Fig. 3C). They interacted strongly with the E2 and OHT complexes of ERα-EF, and they also showed weaker but significant interaction with the ICI182,780 and raloxifene complexes. Norris et al. found a linear peptide (“αII”) that binds E2, OHT, and raloxifene ERα complexes (28). Our AB7-A1 and -B1 clones, however, show no sequence similarities with αII.
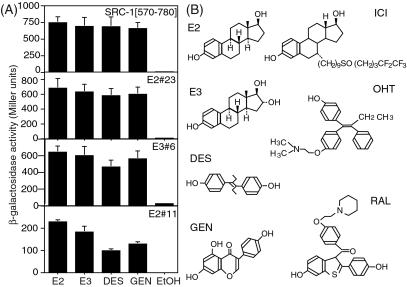
We then tested the effects of different agonists on the interactions between ERα-EF and monobodies (Fig. 4). We found that the clone, E2#11, showed clearly different reactivity to different agonist complexes of ERα-EF, whereas E2#23 and the NR-box fragment of coactivator SRC-1 bind equally well to these agonist complexes. Paige et al. have shown that these agonists induce distinct conformations in full-length ERα and ERβ that are detectable by using in vitro binding assays of ER-binding peptides (27). Taken together, these results demonstrate that one can isolate monobodies that are specific to different conformations of ERα-EF–ligand complexes, and that one can use such monobodies to detect conformational differences of ERα-EF complexes in the nucleus induced by various ligands, even smaller changes induced by different agonists.
Figure 4.
(A) In vivo binding specificity of monobodies to different ERα-EF/agonist complexes determined by β-galactosidase assays. E3, estriol; DES, diethylstilbestrol; GEN, genistein; EtOH, no added ligand. (B) The chemical structures of ligands used in this study.
Roles of the F Domain in the Conformational Dynamics of the ERα LBD.
We then tested whether the F domain (residues 551–595) affects the interactions of monobodies with the LBD (the E domain) of ERα. We compared β-galactosidase activity of cells containing a LexA-ERα-E domain fusion protein and a monobody-activation domain fusion protein with that of cells containing LexA-ERα-EF and the same monobody-activation domain fusion protein (Fig. 5). We confirmed that the expression levels of ERα-E and -EF bait proteins were similar, and that the cells containing the ERα-EF fusion protein do not have breakdown products similar to the ERα-E fusion protein (Fig. 5B). In the presence of E2, the deletion of the F domain had little effect on the interactions of E2#23, E3#6, and SRC-1 with the ERα fragments, strongly suggesting that the F domain does not constitute the binding site for these monobodies. In contrast, the deletion of the F domain resulted in a significant increase (more than 100-fold in β-galactosidase activity) in the binding of E3#6 and SRC-1 to apo-ERα. In a somewhat analogous manner, OHT#33 interactions were similar with ERα-E and -EF in the presence of OHT, whereas the interaction of this monobody with the ERα-E/raloxifene complex was significantly greater than that with the ERα-EF/raloxifene complex. In contrast to the data with monobodies that bind to ERα/agonist complexes, the deletion did not increase the interaction of OHT#33 and apo-ERα. Likewise, the deletion did not significantly affect the interactions of AB7-A1 with any of the ERα–ligand complexes tested (data not shown).
Figure 5.
(A) Effects of the F domain on the binding of ERα to SRC-1 and monobodies. Quantitative β-galactosidase assays were performed for yeast two-hybrid strains containing a monobody (or SRC-1)-activation domain fusion and either the ERα-EF- or E domain-DNA-binding domain fusion proteins. Experiments were performed in the same manner as in Fig. 3. (B) Western blotting of yeast cells containing LexA–ERα-EF (lanes 1 and 2) or LexA–ERα-E (lanes 3 and 4) probed with an anti-LexA antibody (Top) or anti-ERα-F domain antibody (Bottom). Yeast cells were grown in the presence (lanes 1 and 3) and absence (lanes 2 and 4) of E2. Note that these proteins are expressed at a similar level, and lanes 1 and 2 do not contain degradation products similar to LexA–ERα-E (lanes 2 and 4). Abbreviations are the same as in Fig. 3.
Potential Applications of Monobodies.
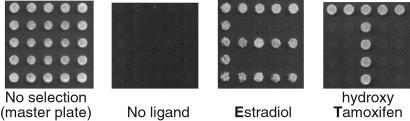
We now possess a collection of yeast strains that detect changes in the surface property of different ERα–ligand complexes. They could be used for discovering and classifying ER ligands. Fig. 6 demonstrates the concept of this application. We also tested whether a monobody could modulate the interaction between ER and the natural coactivator, SRC-1. We used the yeast two-hybrid system that monitored the interaction between ERα-EF and SRC-1 and coexpressed the monobody E2#23 under the control of a separate promotor. β-Galactosidase activity in the presence of E2 decreased by ≈30% when the monobody was expressed, whereas coexpression of the wild-type FNfn10 did not alter the level of the marker enzyme activity (data not shown). This inhibitory effect was reduced when the expression level of the SRC-1 activation domain fusion was increased (data not shown). These results suggest that the monobody binds to the coactivator-binding site of ERα in a competitive manner against SRC-1. The inhibitory effect of the monobody was modest, likely because the monobody is not expressed at a high level, and its binding affinity is not very high. Nevertheless, these results suggest that it may be possible to develop monobody-based inhibitors of NRs.
Figure 6.
Demonstration of the concept of using a monobody collection as chemical sensor. Yeast cells containing E2-, OHT-, and (E2 or OHT)-dependent monobodies were strategically placed on 5 × 5 grids (“No selection”). These cells were stamped on growth selection plates (Leu−) containing E2, OHT, or no ligand. White circles are yeast cells grown on a media plate.
Discussion
In this work, we have shown that one can obtain monobodies that are specific to a particular ERα–ligand complex, and that one can probe relatively small conformational changes among ERα–ligand complexes in living cells by using such monobodies. It should be noted that in the crystal structures, E2 and diethylstilbestrol are completely buried in the interior of the LBD (16, 17), and estriol and genistein are also likely to be completely buried because of their similar structures (Fig. 4B). Thus, it is highly likely that the differences in the surface property among ERα–agonist complexes detected by monobodies are because of different conformations of the ERα polypeptide, rather than steric interference of monobody–ER interactions by ligands. It is also highly likely that AB7-A1 and -B1 recognize surfaces of ERα (Fig. 3C), because E2 and OHT have drastically different structures. Obviously, the difference between the interaction of E3#6 with apo-ERα-EF and that with apo-ERα-E (Fig. 5) is due to a conformational difference between these two fragments. Alternatively, the binding surfaces for a monobody may include surfaces of a bound ligand, which may be the case for the ERα complexes with an antagonist containing bulky moieties. Also, direct steric hindrance by a bound ligand can abolish monobody binding. Thus a loss of monobody binding indicates a change in the surface property but not necessarily a change in the protein conformation itself. Taken together, our results demonstrate that our strategy can probe small but significant conformational changes of a protein in living cells.
The ability to detect conformational changes of proteins in the native environment should bridge the gap that currently exists between high-resolution structural information obtained from in vitro techniques and functional information from cell biology studies. The use of engineered probes, such as monobodies, allows discrimination of a wider variety of conformations than those that are responsible for interactions of the target protein with other natural proteins. In addition to probing ligand-induced conformational changes, our approach can detect effects of mutations, e.g., the deletion of the F domain. We have shown that a collection of yeast two-hybrid cells containing ERα and an appropriate monobody could be used for screening of estrogen- and drug-like molecules. Monobodies that interact with an ER–antagonist complex should be useful in classifying ER ligands and in predicting their in vivo action.
Our strategy should be applicable to other members of the nuclear receptor superfamily and possibly to other classes of proteins, with minor adjustments. In the present study, we used a yeast two-hybrid system as the means to detect interactions of monobodies with a target in living cells. The yeast two-hybrid system, as well as its mammalian counterpart, detects interactions in the nucleus and thus is ideally suited for applications to nuclear receptors. However, alternative methods may be better suited for probing conformational changes of proteins that are naturally located outside the nucleus. Potential methods include FRET, the split ubiquitin system (40), and dihydroforate reductase reconstitution (3). Indeed, Raquet et al. reported the use of the split-ubiquitin system to detect conformational differences of a protein in living cells (41). Our strategy using conformation-specific monobodies could readily be adapted to these systems. The surface property changes of ERα-E and -EF as discriminated by our monobody collection generally agree with the conformational differences of ERα- and ERβ-E domains found in a series of crystal structures. Thus, our results support that these crystal structures represent relevant conformations of ER in cells. However, the interaction profile of AB7-A1 and -B1 (Fig. 3C) suggests the formation of a common epitope in the E2 and OHT complexes of ERα. It will be interesting to identify the location of this epitope by mutagenesis of ERα or cocrystallization. The distinct profiles of interaction specificity among the clones from two different libraries (Fig. 3C) demonstrate the utility of libraries with different degrees of loop flexibility for expanding the conformational space achievable by peptides. The monobody scaffold offers a unique advantage because of the presence of multiple loops with different degrees of flexibility.
We found a dramatic increase in the interactions of the monobody E3#6 and ERα on the deletion of the F domain (Fig. 5). A similar effect was observed between SRC-1 and ERα. These results may be interpreted as a dynamic conformational equilibrium in which ERα-E, in particular helix 12 (Fig. 1), is in equilibrium among multiple conformations, and the presence of the F domain shifts this equilibrium away from the “active” conformation. A number of mutations at residues 536 and 537, which are located in the loop connecting helices 11 and 12, resulted in a constitutively active phenotype (42–45), suggesting that these mutations can shift the conformational equilibrium within the LBD. A series of ERβ LBD crystal structures also suggest the dynamic nature of helix 12. In the genistein complex (16), helix 12 is in a position similar to that found in the ERβ-antagonist structure, as opposed to the “agonist” conformation that is expected from the agonistic activity of genistein. In the structure of ERβ bound to an antagonist, ICI164,384, the electron density for the entire helix 12, is missing, suggesting a conformational disorder (46). Furthermore, an NMR study of the LBD of peroxisome proliferator-activated receptor γ revealed that the apo-LBD, particularly ligand- and cofactor-binding regions, is in a dynamic conformational ensemble (47). Because the F domain of ERα is quite large (≈45 residues) and is directly linked to helix 12, it is plausible that the F domain can affect the balance of the conformational ensemble of the E domain even if the F domain is largely unstructured. These results suggest that fusing a foreign peptide as a conformational probe (e.g., a fluorescent protein for FRET) after the E or F domain might disturb the conformational equilibrium of the LBD. It should be noted that the observed effect of the F domain deletion may be mediated through a change in association of ERα with other macromolecules such as heat shock proteins. These results demonstrate that our approach can reveal conformational dynamics of a target protein in living cells and thus provide useful information complementary to static information obtained from x-ray crystal structure.
We have also shown that monobodies could be used as modulators of biological functions. Although the inhibitory activity of our first-generation monobody was modest, the binding affinity and specificity of monobodies could be improved by introducing additional mutations in adjacent loops (e.g., BC and DE; Fig. 1) and performing further rounds of selection with a higher degree of stringency. Our previous results demonstrated that the monobody scaffold could accommodate many mutations in multiple loops (5). Peptide aptamers based on a single loop and antibody fragments (“intrabodies”) have been shown to be effective inhibitors of intracellular processes (48, 49). We speculate that monobodies with potent inhibitory activity could also be developed.
Acknowledgments
We thank Drs. M. Dumont, E. Grayhack, M. Muyan, E. M. Phizicky, I. Serebriiskii, F. Sherman, and M. R. Yudt and V. Batori for helpful discussions; and the late Dr. A. C. Notides and Dr. E. M. Phizicky (University of Rochester), Dr. E. Golemis (Fox Chase Cancer Center, Philadelphia), and Dr. B. W. O'Malley (Baylor College of Medicine, Houston) for reagents. This work was supported in part by National Institutes of Health Grant R29-GM55042 and U.S. Army Grant DMAD17-97-1-7295 (to S.K.).
Abbreviations
- E2
17β-estradiol
- ERα
estrogen receptor α
- ERα-E
the E domain of ERα
- ERα-EF
the E and F domains of ERα
- FRET
fluorescence resonance energy transfer
- LBD
ligand-binding domain
- NR
nuclear receptor
- OHT
4-hydroxy tamoxifen
- FNfn10
the tenth fibronectin type III domain from human fibronectin
- SRC-1
steroid receptor coactivator-1
References
- 1.Minton A P. Curr Opin Struct Biol. 2000;10:34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 2.Truong K, Ikura M. Curr Opin Struct Biol. 2001;11:573–578. doi: 10.1016/s0959-440x(00)00249-9. [DOI] [PubMed] [Google Scholar]
- 3.Remy I, Wilson I A, Michnick S W. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- 4.Sachs D H, Schechter A N, Eastlake A, Anfinsen C B. Proc Natl Acad Sci USA. 1972;69:3790–3794. doi: 10.1073/pnas.69.12.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koide A, Bailey C W, Huang X, Koide S. J Mol Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 6.Main A L, Harvey T S, Baron M, Boyd J, Campbell I D. Cell. 1992;71:671–678. doi: 10.1016/0092-8674(92)90600-h. [DOI] [PubMed] [Google Scholar]
- 7.Plaxco K W, Spitzfaden C, Campbell I D, Dobson C M. Proc Natl Acad Sci USA. 1996;93:10703–10706. doi: 10.1073/pnas.93.20.10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 9.Uetz P, Hughes R E. Curr Opin Microbiol. 2000;3:303–308. doi: 10.1016/s1369-5274(00)00094-1. [DOI] [PubMed] [Google Scholar]
- 10.Colas P, Brent R. Trends Biotechnol. 1998;16:355–363. doi: 10.1016/s0167-7799(98)01225-6. [DOI] [PubMed] [Google Scholar]
- 11.Mendelsohn A R, Brent R. Curr Opin Biotechnol. 1994;5:482–486. doi: 10.1016/0958-1669(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 12.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan V C, Jeng M H, Jiang S Y, Yingling J, Stella A L. Semin Oncol. 1992;19:299–307. [PubMed] [Google Scholar]
- 14.Anstead G M, Carlson K E, Katzenellenbogen J A. Steroids. 1997;62:268–303. doi: 10.1016/s0039-128x(96)00242-5. [DOI] [PubMed] [Google Scholar]
- 15.Schwabe J W R, Chapman L, Finch J T, Rhodes D. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 16.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 17.Brzozowski A M, Pike A C, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J A, Carlquist M. Nature (London) 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 18.Tanenbaum D M, Wang Y, Williams S P, Sigler P B. Proc Natl Acad Sci USA. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pike A C, Brzozowski A M, Hubbard R E. J Steroid Biochem Mol Biol. 2000;74:261–268. doi: 10.1016/s0960-0760(00)00102-3. [DOI] [PubMed] [Google Scholar]
- 20.Metzger D, White J H, Chambon P. Nature (London) 1988;334:31–36. doi: 10.1038/334031a0. [DOI] [PubMed] [Google Scholar]
- 21.Schena M, Yamamoto K R. Science. 1988;241:965–967. doi: 10.1126/science.3043665. [DOI] [PubMed] [Google Scholar]
- 22.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 23.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong H, Darimont B D, Ma H, Yang L, Yamamoto K R, Stallcup M R. J Biol Chem. 1999;274:3496–3502. doi: 10.1074/jbc.274.6.3496. [DOI] [PubMed] [Google Scholar]
- 25.Montano M M, Ekena K, Delage-Mourroux R, Chang W, Martini P, Katzenellenbogen B S. Proc Natl Acad Sci USA. 1999;96:6947–6952. doi: 10.1073/pnas.96.12.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 27.Paige L A, Christensen D J, Gron H, Norris J D, Gottlin E B, Padilla K M, Chang C Y, Ballas L M, Hamilton P T, McDonnell D P, et al. Proc Natl Acad Sci USA. 1999;96:3999–4004. doi: 10.1073/pnas.96.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norris J D, Paige L A, Christensen D J, Chang C Y, Huacani M R, Fan D, Hamilton P T, Fowlkes D M, McDonnell D P. Science. 1999;285:744–746. doi: 10.1126/science.285.5428.744. [DOI] [PubMed] [Google Scholar]
- 29.Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 30.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 31.Finley R L, Jr, Brent R. Proc Natl Acad Sci USA. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 33.Martzen M R, McCraith S M, Spinelli S L, Torres F M, Fields S, Grayhack E J, Phizicky E M. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- 34.Gietz R D, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 35.Nichols M, Rientjes J M, Logie C, Stewart A F. Mol Endocrinol. 1997;11:950–961. doi: 10.1210/mend.11.7.9944. [DOI] [PubMed] [Google Scholar]
- 36.Montano M M, Muller V, Trobaugh A, Katzenellenbogen B S. Mol Endocrinol. 1995;9:814–825. doi: 10.1210/mend.9.7.7476965. [DOI] [PubMed] [Google Scholar]
- 37.Jordan V C. J Natl Cancer Inst. 1998;90:967–971. doi: 10.1093/jnci/90.13.967. [DOI] [PubMed] [Google Scholar]
- 38.Estojak J, Brent R, Golemis E A. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishikawa J, Saito K, Goto J, Dakeyama F, Matsuo M, Nishihara T. Toxicol Appl Pharmacol. 1999;154:76–83. doi: 10.1006/taap.1998.8557. [DOI] [PubMed] [Google Scholar]
- 40.Johnsson N, Varshavsky A. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raquet X, Eckert J H, Muller S, Johnsson N. J Mol Biol. 2001;305:927–938. doi: 10.1006/jmbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- 42.Weis K E, Ekena K, Thomas J A, Lazennec G, Katzenellenbogen B S. Mol Endocrinol. 1996;10:1388–1398. doi: 10.1210/mend.10.11.8923465. [DOI] [PubMed] [Google Scholar]
- 43.White R, Sjoberg M, Kalkhoven E, Parker M G. EMBO J. 1997;16:1427–1435. doi: 10.1093/emboj/16.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Q X, Borg A, Wolf D M, Oesterreich S, Fuqua S A. Cancer Res. 1997;57:1244–1249. [PubMed] [Google Scholar]
- 45.Eng F C, Lee H S, Ferrara J, Willson T M, White J H. Mol Cell Biol. 1997;17:4644–4653. doi: 10.1128/mcb.17.8.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pike A C, Brzozowski A M, Walton J, Hubbard R E, Thorsell A, Li Y, Gustafsson J, Carlquist M. Structure (London) 2001;9:145–153. doi: 10.1016/s0969-2126(01)00568-8. [DOI] [PubMed] [Google Scholar]
- 47.Johnson B A, Wilson E M, Li Y, Moller D E, Smith R G, Zhou G. J Mol Biol. 2000;298:187–194. doi: 10.1006/jmbi.2000.3636. [DOI] [PubMed] [Google Scholar]
- 48.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Nature (London) 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 49.Richardson J H, Marasco W A. Trends Biotechnol. 1995;13:306–310. doi: 10.1016/S0167-7799(00)88970-2. [DOI] [PubMed] [Google Scholar]