Abstract
The posttranslational γ-carboxylation of glutamate residues in secreted proteins to γ-carboxyglutamate is carried out by the vitamin K-dependent enzyme γ-glutamyl carboxylase. γ-Carboxylation has long been thought to be a biochemical specialization of vertebrates, essential for blood clotting. Recently, a γ-carboxylase was shown to be expressed in Drosophila, although its function remains undefined in this organism. We have characterized both cDNA and genomic clones for the γ-glutamyl carboxylase from the marine mollusc, Conus, the only nonvertebrate organism for which γ-carboxyglutamate-containing proteins have been biochemically and physiologically characterized. The predicted amino acid sequence has a high degree of sequence similarity to the Drosophila and vertebrate enzymes. Although γ-carboxylases are highly conserved, the Conus and mammalian enzymes have divergent substrate specificity. There are striking parallels in the gene organization of Conus and human γ-carboxylases. Of the 10 Conus introns identified, 8 are in precisely the same position as the corresponding introns in the human enzyme. This remarkable conservation of intron/exon boundaries reveals that an intron-rich γ-carboxylase was present early in the evolution of the animal phyla; although specialized adaptations in mammals and molluscs that require this extracellular modification have been identified, the ancestral function(s) and wider biological roles of γ-carboxylation still need to be defined. The data raise the possibility that most introns in the genes of both mammals and molluscs antedate the divergence of these phyla.
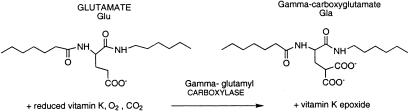
The vitamin K-dependent posttranslational modification of glutamate to γ-carboxyglutamate (Gla) is a striking biochemical feature of the vertebrate blood-clotting cascade (1, 2). This modification carried out by an integral endoplasmic reticulum membrane protein (3), γ-glutamyl carboxylase, results in the carboxylation of specific glutamate residues in vitamin K-dependent proteins to Gla in the presence of carbon dioxide, oxygen, and reduced vitamin K. In the process, vitamin K is converted to vitamin K epoxide, which is subsequently converted to vitamin K by vitamin K epoxide reductase and used in the carboxylation reaction.
Several clotting factors, as well as proteins that regulate the blood-clotting process, require this modification to function properly. Inhibitors that prevent the vitamin K-dependent carboxylation of Glu to Gla are in wide use therapeutically (4). Thus, not only was γ-carboxylation first discovered in the mammalian blood-clotting system (5–8), but this posttranslational modification continues to be intensively studied primarily by blood-clotting specialists.
Subsequent to its initial discovery, Gla was identified in other extracellular mammalian proteins, including the bone Gla protein and the bone matrix protein (9), implicating γ-carboxylation in bone metabolism. Other vertebrate Gla-containing proteins have been identified; among these are Gas6, a ligand for a tyrosine kinase receptor (10), believed to play a role in cell growth and differentiation (11), as well as several proline-rich polypeptides of undefined function (12). Such results strongly suggest additional physiological roles in mammals for this posttranslational modification of extracellular proteins.
In 1984 (13), Gla was shown to be present in a highly specialized invertebrate system, venom peptides of the marine cone snails (Conus). It was found that a neuroactive Conus peptide, conantokin-G, which is a 17-aa peptide ligand produced in the venom of the fish-hunting cone snail, Conus geographus, contained five residues of Gla. This peptide targets a subtype of the glutamate receptor, the N-methyl-d-aspartate receptor (14). A variety of other Conus venom peptides, some in the conantokin family related to conantokin-G, but others with completely unrelated amino acid sequences, also proved to have Gla residues (15–19). These discoveries provided initial evidence suggesting that the role of this posttranslational modification in blood clotting might be only a small part of the total biological picture. Not only was Gla present in invertebrates (which was totally unexpected), but it was found in several entirely different structural and functional contexts. Biochemical experiments established that the posttranslational conversion of glutamate to Gla in Conus peptides had the same general requirements as had been established for mammalian γ-carboxylation, i.e., reduced vitamin K, carbon dioxide, molecular oxygen, and a recognition signal sequence (20, 21).
A recent discovery that strongly reinforces the proposal for a broader biological role for γ-carboxylation is the identification of a γ-glutamyl carboxylase in the fruit fly, Drosophila melanogaster (22, 23). However, no substrates for γ-carboxylation have yet been identified in Drosophila, and therefore the physiological function of the posttranslational modification in this organism is unknown.
In this report, cDNA clones encoding γ-glutamyl carboxylase from Conus are identified and characterized, as well as Conus γ-carboxylase gene clones. Together, the data provide compelling evidence that a single ancestral gene gave rise to all γ-carboxylases found in the three phylogenetic systems (mammalian, Conus, and Drosophila). The results support the hypothesis that γ-carboxylation is a phylogenetically widespread posttranslational modification of ancient evolutionary origins, whose “ancestral” physiological roles remain undefined.
Methods
Characterization of γ-Glutamyl Carboxylase cDNAs.
Conus carboxylase partial cDNA was synthesized through reverse transcription–PCR by using primers corresponding to regions conserved between mammals and Drosophila (amino acids 395–405 and 465–470 in the human sequence). By using venom duct poly(A)+RNA (24) and primers derived from the partial cDNA sequence, we obtained the 3′ end of the carboxylase gene using the methods of 3′ RACE (25). The cDNA was cloned, and the sequence was determined. 5′RACE experiments were carried out by using a GIBCO/BRL kit, according to instructions provided by the vendor. The 5′ RACE products were cloned and the sequence was determined. The sequences from the 5′ and 3′ RACE experiments were assembled to obtain the complete sequence of Conus γ-glutamyl carboxylase. The complete cDNA sequence from Conus textile and 3′ sequences from C. omaria, C. episcopatus, and C. imperialis have been submitted to GenBank (accession nos. AY044904, AF448233, AF448234, and AF448235). Analysis of amino acid homology between human (26), Drosophila (23), and Conus were carried out with GAP and PILEUP VERSION 4.0 (Genetics Computer Group, Madison, WI). Hydrophobicity analysis and transmembrane segment analysis were done by using a computer algorithm (TOP-PRED) with a window of 21 amino acids (27).
Isolation of Genomic Clone of Conus γ-Carboxylase.
A partial genomic clone of C. textile carboxylase (gCGx.1, 1.4 kb in size) was obtained by PCR amplification of genomic DNA by using oligonucleotide primers corresponding to amino acids 433–441 and 504–511 of the Conus sequence (Fig. 1). A genomic library of Conus genomic DNA constructed in Lambda FIX II (Stratagene) was probed with gCGx.1 DNA. Four positive clones were isolated from a screen of 500,000 plaques. One of the clones containing an 18-kb insert was sequenced.
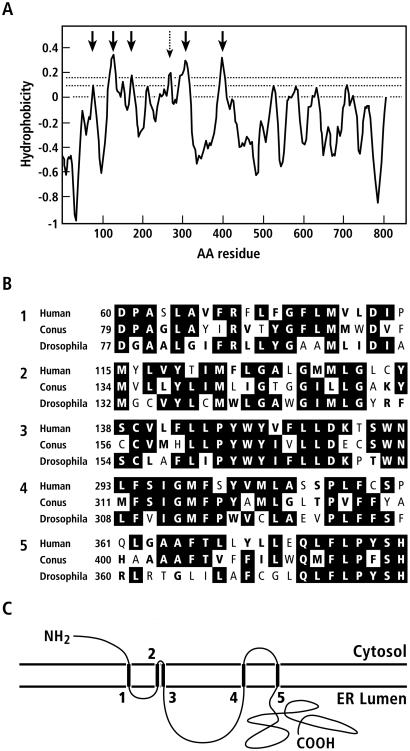
Figure 1.
Amino acid sequence alignment of human, C. textile, and Drosophila carboxylases.
γ-Carboxylation Reaction.
Partially purified enzyme was isolated according to Stanley et al. (20), and the γ-carboxylation reaction was carried out according to methods described therein.
Results
Identification and Characterization of cDNA Clones Encoding C. textile Venom Duct γ-Carboxylase.
C. textile venom duct mRNA was used to obtain cDNA clones that had sequence homology to regions highly conserved between mammalian and (the recently elucidated) Drosophila γ-carboxylases. The predicted amino acid sequence of the ORF was reconstructed from the C. textile cDNAs, as described under Methods (Fig. 1).
cDNA clones from venom duct mRNA derived from three other Conus species (Conus omaria, C. episcopatus, and C. imperialis) also were identified and partially sequenced. All three were analyzed in parallel by using 3′ RACE. Different 3′ RACE primers were used, and cDNAs were prepared by a variety of techniques with both strands of the cDNA clones sequenced. The sequences at the 3′ end for all three Conus species are homologous. For C. textile, C. omaria and C. episcopatus, there are only a few nucleotide changes that lead to mostly conservative amino acid substitutions. These are all species that belong to the same mollusc-hunting clade of Conus. Conus imperialis is a more divergent worm-hunting species, belonging to a different clade (28). As might be expected, there is greater divergence in the overlapping sequences determined by the 3′ RACE experiment. The ORF of C. textile, C. episcopatus, and C. omaria is 811 aa compared with 799 aa for C. imperialis.
Comparison of C. textile, Drosophila, and Human Sequences.
The predicted translation product of the putative γ-carboxylase from C. textile cDNA is compared with the mammalian and Drosophila carboxylase sequences as shown in Fig. 1. The high degree of homology with the previously characterized carboxylases is consistent with the four Conus cDNA sequences encoding Conus γ-glutamyl carboxylase.
A hydropathy plot of the C. textile carboxylase sequence is shown in Fig. 2A. The hydrophobicity profile obtained is very similar to that of the Drosophila and human enzymes. The mammalian γ-carboxylase has been shown to be an endoplasmic reticulum membrane protein (3). In vitro translation studies using reporter-tagged carboxylase were used by Tie et al. (29) to assess the availability of sites for N-glycosylation. Based on these studies, a model in which the enzyme has five transmembrane (TM) segments with the N terminus in the cytoplasm and C terminus in the lumen was proposed. The sequence alignments from Fig. 1 suggest the five putative transmembrane domains for Conus, Drosophila, and human enzymes shown in Fig. 2 B and C. The degree of sequence similarity between the three enzymes in the transmembrane domains is strongly consistent with a conserved topology around the endoplasmic reticulum (ER) membrane.
Figure 2.
(A) Hydropathy plot of Conus carboxylase aa sequence. The upper and lower cutoffs for tentative transmembrane segments are shown. The dotted arrow indicates a predicted transmembrane domain not evident in the Tie analysis (29). (B) Comparison of amino acid sequences of proposed Conus transmembrane domains derived from A to the corresponding human and Drosophila sequences. (C) Proposed topology of γ-glutamyl carboxylase originally proposed for the human enzyme by Stafford and coworkers (29).
Immediately following TM5 is a remarkably conserved sequence (amino acids 383–405 in the human enzyme; refs. 23 and 30). At every amino acid position, at least two and in most cases all three enzymes have identical amino acids.
Human: FLTQGYNNWTNGLYGYSWDMMVH
Conus: FITKGNNSWTQGLYGYSWDMMVH
Drosophila: FITKGYNNWTNGLYGYSWDMMVH
A mutation in residue 395 (Leu to Arg, L in bold above) in the human enzyme (31) results in a clinical syndrome characterized by a general deficiency of blood coagulation that can be treated by an infusion of high doses of vitamin K. Site-directed mutagenesis studies suggest that the propeptide binding site may in part be in the vicinity of residues 234, 406, and 513 (32). Thus, this conserved region, predicted to be in the ER lumen, may be part of the substrate binding site.
Cysteine residues have been determined to be essential for carboxylation (33). Dowd et al. (34) proposed the participation of two cysteine residues in the catalytic event. By using the amino acid alignment in Fig. 1, the only cysteines conserved in human, Drosophila, and Conus carboxylases are Cys-99, -139 and -450 (coordinates are for the human carboxylase). Results from mutational studies and chemical modification of Cys-99 and -450 suggest that these residues are necessary for both epoxidation and carboxylation by the carboxylase (35).
Map of Conus Carboxylase Gene.
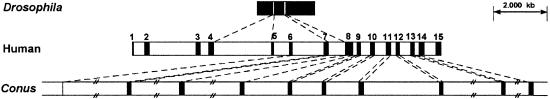
From a library of C. textile genomic DNA in bacteriophage λ, we identified and partially sequenced a genomic clone of Conus γ-glutamyl carboxylase. The genomic map of the γ-glutamyl carboxylase gene has been determined in human (36), in rat (37), and in Drosophila (22, 23). Whereas the mammalian gene has 15 exons, Drosophila has only 3. Fig. 3 shows a partial genomic map of the Conus γ-glutamyl carboxylase gene; intron positions are compared with both the human and Drosophila genes. Of the 10 Conus introns identified, the positions of 8 of them are identical to those found in the human gene. Two human introns have been split into two each in the Conus genomic sequence. The Conus introns are longer than their corresponding human counterparts. Table 1 shows the nucleic acid and amino acid sequences at the exon-exon junctions of human, Drosophila, and Conus carboxylases—the positions of introns are precisely conserved. The introns have traditional spliceosomal intron 5′-GT donor and 3′-AG acceptor sites for splicing.
Figure 3.
Partial genomic map of Conus γ-glutamyl carboxylase compared with human and Drosophila carboxylases. Exons (numbered in human) are shown as black boxes. //, incomplete Conus intron sequence.
Table 1.
Comparison of exon/exon junctions among human, Drosophila, and Conus carboxylases
| Exon junction | Amino acid sequences |
|---|---|
| 6 | L S K L L S E |
| Human 237 | CTC TTC AGT CCC TTC AA | A CTG CTG TTG TCT GAG GAG |
| Conus 255 | GTC TTT TAC CCG TTT AC | G TTC CTG ATG ACA GAA GAC |
| V F Y P F T F L M T E D | |
| 7 | R L V W V C |
| Drosophila 312 | CGA CTG TTT GTA ATA G | GC ATG TTT CCC TGG GTC TGT |
| L S V | |
| Human 297 | CAG CTT TTC AGC ATT G | GT ATG TTC TCC TAC GTC ATG |
| Conus 315 | CAG ATG TTC AGC ATA G | GA ATG TTT CCG TAT GCC ATG |
| Q M F S I G M F P Y A M | |
| 7A | |
| Conus 369 | CAG GCC AAA CCA GAA CTG | GCC AGC ACC CCT GAG CAT |
| Q A K P E L A S T P E H | |
| 8 | L Q Y N |
| Human 380 | TCT CAT TTT CTC AAC CAG | GGC TAT AAC AAC TGG ACA |
| Conus 419 | TCT CAT TTT ATC ACA AAG | GGC AAC AAC AGC TGG ACC |
| S H F I T K G N N S W T | |
| 9 | Y N G V F T Q R |
| Human 424 | GGC TAC CTT AAC CCT GGG | GTA TTT ACA CAG AGT CGG |
| Conus 463 | GGG TTC CTG GAC CCG CAG | GCA TGG AGC AAG TCA CAT |
| G F L D P Q A W S K S H | |
| 10 | D F D R |
| Human 475 | GAC CGC TTC CAG CAG AG | G ATT TTT GAC CCT CGT GTG |
| Conus 514 | CAT CGC TTC CAG CAA CG | G ATC GTG AAC CCC AAT GTG |
| H R F Q Q R I V N P N V | |
| 11 | I H |
| Human 532 | ATT GCA GAT TTC CCT G | GA CTG CAC TTG GAG AAT TTT |
| Conus 574 | CTG GCT GAC TTT CCT G | GT TTG TAC CTG GAG AAC TTT |
| L A D F P G L Y L E N F | |
| 12 | E E K M Q L A E Y |
| Human 575 | GAG GGA GAA AAA ATG CAG | TTG CCT GCT GGT GAG TAC |
| Conus 623 | GAT GGC CAA GAG TCA TTG | ATT CCC ACA GGG GTG TTC |
| D G Q E S L I P T G V F | |
| 13 | V E E T G L P |
| Human 625 | GTG GAG AAT GGA AGT G | AA ACA GGG CCT CTA CCC CCA |
| Conus 669 | GCC CTC AAC GGC TCC C | TG GAT GCT CCA GTT CCA GAC |
| A L N G S L D A P V P D | |
| 13A | |
| Conus 723 | CAT TAT ATG TCT ATG TAT | CGT GGA CTG CAG CTG ATA |
| H Y M S M Y R G L Q L I |
Substrate Recognition by Conus γ-Glutamyl Carboxylase and the Fidelity of Modification.
As is the case for mammalian carboxylases, the Conus carboxylase is microsomal and has an absolute requirement for reduced vitamin K for activity. A large variety of Gla-containing peptides have been isolated from different Conus species. In contrast to the mammalian Gla-containing Conus peptides, the sequence contexts of modification in these peptides can be very different from each other; examples of three divergent Gla-containing peptides are shown in Table 2. Precursors of Gla-containing Conus peptides are translated from mRNA, transported to the ER, and subsequently undergo posttranslational modification and proteolysis to form the mature Gla-containing peptide. In the case of mammalian Gla-containing peptides, N-terminal propeptide sequences of γ-carboxylated peptides contain recognition sequences (γ-carboxylation recognition signals, γ-CRSs) that mark potential substrates for γ-carboxylation. Similarly, γ-carboxylation of substrates using partially purified Conus carboxylase requires propeptides containing a putative γ-CRS N-terminal to the mature peptide region to be modified.
Table 2.
Comparison of Gla-containing Conus and human peptides and γ-CRS-containing regions
| Peptide (ref.) | Source | Modified peptide sequence | Corresponding γ-CRS-containing propeptide |
|---|---|---|---|
| Conantokin-G (13) | C. geographus | GEγγLQγNQγLIRγKSNGK | GKDRLTQMKRILKQRGNKAR |
| tx5a (16) | C. textile | γCCγDGW+CCT†AAO | PLSSLRDNLKRTIRTRLNIR |
| Spasmodic (19) | C. textile | GCNNSCQγHSDCγSHCICTFRGCGAVN | DNRRNLQSKWKPVSLYMSRR |
| h-FIX (47) | Human (Factor IX) | YNSGKLγγFVQGNLγRγCMγγKCSFγγARγVFγNTγKTTγFW | TVFLDHENANKILNRRKR |
Note that Gla in Conus peptides are not in homologous loci. Residues known to be functionally important in the human γ-CRS sequence are underlined; note that these are not conserved in the Conus propeptides. γ = Gla (γ-carboxypglutamate); W+ = 6-bromotryptophan; T
= glycosylated threonine; O = hydroxyproline.
Experiments to evaluate the effects of Conus γ-CRSs on the apparent Km with the pentapeptide FLEEL as the modification target are shown in Table 3. The Km for FLEEL decreased by 60- to 460-fold when an N-terminal Conus γ-CRS was present. A similar increase in affinity (121-fold) was observed when an authentic Conus peptide substrate region, conantokin-G, was used (see Table 3). These effects on affinity are considerably greater than those observed when a mammalian γ-CRS, that of the blood-clotting Factor IX, is attached to FLEEL (Km decreased by only 4-fold). We previously examined the carboxylation of a high-affinity mammalian substrate by both the bovine and Conus carboxylase (20). The propeptide-containing substrate composed of amino acids 18–41 of Factor IX (38) is a high-affinity substrate for the bovine enzyme. Under our experimental conditions, the Km for this substrate for bovine enzyme was determined to be ≈1 μM, whereas no activity was observed for the Conus enzyme. In analogous experiments (−20 to −1 ConG). ConG*, a high-affinity substrate for the Conus enzyme, was not carboxylated by the bovine enzyme. A comparison of the amino acid sequences of the γ-CRS-containing regions of three Gla-containing Conus peptides and Factor IX, the mammalian blood coagulation factor (Table 2), shows that the different Conus γ-CRSs do not bear any obvious sequence homology either to each other or to the mammalian γ-CRSs.
Table 3.
Affinity of Conus γ-glutamyl carboxylases for various substrates
| Peptide | Apparent Km, μM | Decrease in Km conferred by γ-CRS |
|---|---|---|
| FLEEL | 280 | |
| (−20 to −1 ConG).FLEEL | 4 | ×70 |
| (−20 to −1 tx5a).FLEEL | 0.6 | ×467 |
| (−20 to −1 spasmodic).FLEEL | 4.7 | ×60 |
| (−18 to −1 FIX).FLEEL | 73 | ×4 |
| ConG* | 3,400 | |
| (−20 to −1 ConG).ConG* | 28 | ×121 |
(−20 to −1 X).Y indicates a substrate in which the amino acid sequences −20 to −1 of the propeptide of Gla-containing conopeptide X is covalently linked to a Glu-containing substrate Y. Y is either the pentapeptide FLEEL or uncarboxylated conantokin-G, ConG
.
Discussion
γ-Carboxylase cDNA clones from venom ducts of the four species of Conus were analyzed. The γ-carboxylases from all four were closely similar in sequence and, furthermore, had extensive sequence homology with both the Drosophila (31% sequence identity, 43% sequence similarity) and the mammalian enzymes (43% sequence identity, 55% sequence similarity) to the human enzyme. As is demonstrated above, the degree of sequence identity is much higher in selected regions, including the transmembrane domains and a putative substrate binding pocket, predicted to be in the ER lumen after the last transmembrane domain.
The general topology of the Conus, Drosophila, and human enzymes is clearly similar. The data are consistent with, and provide support for, the proposal of Stafford and coworkers (29) regarding the topology of the mammalian enzyme: they identified five putative transmembrane domains in the mammalian enzyme that are predicted to be membrane spanning in the invertebrate enzymes as well (Fig. 2C). The high degree of sequence similarity throughout is consistent with the hypothesis that all three enzymes (Conus, Drosophila, and mammalian) have a common evolutionary origin.
Residues implicated in catalysis, such as Cys-99 and -450 (human coordinates), which have been identified as being essential for both epoxidation and carboxylation, are also conserved in Conus. Cys-99 and -450 form part of the active site of the enzyme and have been proposed to be in the proximity of both the propeptide binding site and the sites for modification (35). Thus, substrates may well interact with the widely separated amino (39) and carboxyl terminus (40) of the carboxylase protein. The lower degree of conservation in the carboxylase molecule toward the carboxyl terminus suggests that specific recognition interactions between the carboxylase and its polypeptide substrates may be mediated by this region of the molecule.
We also have presented data demonstrating that the Conus enzyme has high affinity for Conus propeptide regions known to contain γ-CRSs. In contrast, a propeptide region with a γ-CRS from a mammalian substrate gave a much smaller boost in affinity to the Conus enzyme. Thus, despite their striking sequence similarities, γ-carboxylases have clearly diverged in the different animal phyla with respect to substrate recognition, with each enzyme presumably having a preference for the spectrum of substrates found in that phylogenetic system. This provides an opportunity to systematically identify regions of the enzyme required for high-affinity substrate recognition, and ultimately, to identify physiologically relevant substrates in each phylum.
γ-Carboxylation clearly has been adapted by different animal phyla for specialized physiological purposes (in molluscs, for Conus venom peptides; in vertebrates, for the blood clotting cascade). At the present time, these are the only physiological phenomena where biochemical mechanisms for the role of Gla are reasonably well understood. The conservation of γ-carboxylase sequences in three phyla indicates that the ancestral gene function(s) must have been strongly selected to be retained as animal phyla diverged. The nature of these ancestral functions of γ-carboxylation is unknown. One possibility, based on the recent finding that Drosophila γ-carboxylase is highly expressed in later-stage embryos (23), is that this posttranslational modification of extracellular proteins plays some important, potentially conserved role(s) in development.
The gene organization of the Conus enzyme provides remarkable molecular evidence that the γ-carboxylation systems of three different animal phyla had a common origin. Although the γ-carboxylase genes differ greatly in size (the Drosophila gene is ≈2.1 kb with 2 introns, the human gene is ≈13 kb with 14 introns (36), and the Conus gene is even larger), there is a striking and unexpected conservation of intron/exon boundary locations. Within the Conus genomic region analyzed so far, all of the intron-exon junctions in the mammalian gene were found to be precisely conserved. We believe that this is the greatest correspondence of intron positions documented so far between homologous genes from different animal phyla. The single Drosophila intron in this interval is also at precisely the same locus (results shown in Fig. 3).
Although the positions of introns in the various carboxylase genes are conserved, the size of the introns is not conserved. Of the 10 Conus introns that have been identified, 4 have been fully sequenced. These 4 introns correspond to human introns 8–11 (average size, 292 nucleotides; range, 162–434). The four Conus introns have an average length of 1,650 nt (range, 909–2,835). All four introns are missing in Drosophila. Thus, the size distribution of these introns in the three different phyla does not overlap.
The unexpected conclusion that emerges from our study of the Conus γ-carboxylase gene is that all eight introns in the human gene, corresponding to the Conus genomic interval analyzed, are evolutionarily ancient, older than the Cambrian explosion (≈540 million years ago) when the molluscs and chordates are first detected in the fossil record. This finding raises the intriguing question of whether most introns in other human genes have a similarly ancient lineage. Our results suggest that Drosophila (and perhaps, other insects) may not be the appropriate invertebrate standard for evaluating whether vertebrate introns are likely to be relatively recent or more ancient than the Cambrian explosion.
We address the very different number of introns found in the same genomic interval for the Drosophila gene. Formally, there are two possibilities. Drosophila may have diverged from humans and Conus earlier, before 8 of the 10 introns conserved between Conus and humans appeared in their common ancestor. The second formal possibility is that ancestral introns originally present in the Drosophila lineage were subsequently lost. We believe that the second explanation is compelling from the available evidence.
The location of the 2 introns of Drosophila corresponds precisely to 2 of the 14 human introns. Furthermore, the data in Table 4 are consistent with very different pressures on ancestral introns in the three phyla. In Conus, the long period of divergence since its common ancestor with Drosophila and humans has led to the persistence of introns that are fairly long, almost all being >1 kb in size. Where the data are complete, the human introns are significantly smaller (between 3- and 14-fold shorter than the corresponding Conus intron). In the Drosophila γ-carboxylase, the two introns that persist are very much smaller (58 and 72 nt) than any γ-carboxylase intron in the two other phyla, and, in fact, most introns are absent. These results suggest that a different evolutionary history for each γ-carboxylase gene has led to the striking differences observed in the size distribution of introns in Conus, humans, and Drosophila. With Drosophila, the pressure for reduction led to the disappearance of most introns, and the minimal size of the two that remain.
Table 4.
Size comparison of corresponding introns
| Source of gene | Number of nucleotides in | ||||
|---|---|---|---|---|---|
| intron number (human)
| |||||
| 8 | 9 | 10 | 11 | Avg | |
| Conus textile | 1,640 | 909 | 1,215 | 2,835 | 1,650 |
| Homo sapiens | 161 | 371 | 434 | 201 | 292 |
| Drosophila melanogaster | 0 | 0 | 0 | 0 | 0 |
These findings may lend a new flavor to the “introns early-introns late” debate that has been raging in the literature regarding the origins of genes (41–46). The γ-carboxylase work suggests that the three animal phyla investigated were subject to very different pressures in the course of their separate evolutionary histories, resulting in a characteristic size spectrum of introns for each of the three genes. Clearly, the same factors could have had even more dramatic effects on introns of other taxa. The exon theory of genes provides an elegant rationale for the original presence of introns. The variance documented above in the sets of introns from three animal groups lends credence to the possibility that the evolutionary histories of some lineages might lead to a total loss of introns. In their homologous γ-carboxylase genes, Conus and humans retain most ancestral introns, albeit with strikingly different size distributions. Drosophila has lost >80%, with a severe reduction in size of the two that remain. Given these results, it seems less unreasonable to propose that some time in their long evolutionary history, eubacteria/archaebacteria could have undergone a much more accelerated loss, to the point where no spliceosomal introns are found in present-day genomes.
Scheme 1.
Acknowledgments
We thank Dr. Jean Rivier and Dr. Robert Schackmann for the synthesis of the peptides used for this study. This research was supported by National Institutes of Health Grant GM48677 and research funds from Cognetix, Inc.
Footnotes
References
- 1.Suttie J W. Annu Rev Biochem. 1985;54:459–477. doi: 10.1146/annurev.bi.54.070185.002331. [DOI] [PubMed] [Google Scholar]
- 2.Vermeer C. Biochem J. 1990;266:625–636. doi: 10.1042/bj2660625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helgeland L. Biochim Biophys Acta. 1977;499:181–193. doi: 10.1016/0304-4165(77)90001-0. [DOI] [PubMed] [Google Scholar]
- 4.Vermeer C, Schurgers L J. Blood Stasis Thromb. 2000;14:339–353. doi: 10.1016/s0889-8588(05)70137-4. [DOI] [PubMed] [Google Scholar]
- 5.Magnusson S, Sottrup-Jensen L, Petersen T E, Morris H R, Dell A. Mol Cell Biol. 1974;13:497–4985. [Google Scholar]
- 6.Nelsestuen G L, Zytokovicz T H, Howard J B. J Biol Chem. 1974;249:6347–6350. [PubMed] [Google Scholar]
- 7.Stenflo J, Fernlund P, Egan W, Roepstorff P. Proc Natl Acad Sci USA. 1974;71:2730–2733. doi: 10.1073/pnas.71.7.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stenflo J. J Biol Chem. 1974;249:5527–5535. [PubMed] [Google Scholar]
- 9.Price P A, Williamson M K. J Biol Chem. 1985;260:14971–14975. [PubMed] [Google Scholar]
- 10.Manfioletti G, Brancolini C, Avanzi G, Schneider C. Mol Cell Biol. 1993;13:4976–4985. doi: 10.1128/mcb.13.8.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crosier K E, Crosier P S. Pathology. 1997;29:131–135. doi: 10.1080/00313029700169744. [DOI] [PubMed] [Google Scholar]
- 12.Kulman J D, Harris J E, Haldeman B A, Davie E W. Proc Natl Acad Sci USA. 1997;94:9058–9062. doi: 10.1073/pnas.94.17.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McIntosh J M, Olivera B M, Cruz L J, Gray W R. J Biol Chem. 1984;259:14343–14346. [PubMed] [Google Scholar]
- 14.Olivera B M, Rivier J, Clark C, Ramilo C A, Corpuz G P, Abogadie F C, Mena E E, Woodward S R, Hillyard D R, Cruz L J. Science. 1990;249:257–263. doi: 10.1126/science.2165278. [DOI] [PubMed] [Google Scholar]
- 15.Haack J A, Rivier J, Parks T N, Mena E E, Cruz L J, Olivera B M. J Biol Chem. 1990;265:6025–6029. [PubMed] [Google Scholar]
- 16.Walker C, Steel D, Jacobsen R B, Lirazan M B, Cruz L J, Hooper D, Shetty R, DelaCruz R C, Nielsen J S, Zhou L, et al. J Biol Chem. 1999;274:30664–30671. doi: 10.1074/jbc.274.43.30664. [DOI] [PubMed] [Google Scholar]
- 17.Hooper D, Lirazan M B, Schoenfeld R, Cook B, Cruz L J, Olivera B M, Bandyopadhyay P. In: Peptides for the New Millennium: Proceedings of the Sixteenth American Peptide Symposium. Fields G B, Tam J P, Barany G, editors. Dordrecht, The Netherlands: Kluwer Academic; 2000. pp. 727–729. [Google Scholar]
- 18.Rigby A C, Lucas-Meunier E, Kalume D E, Czerwiec E, Hambe B, Dahlqvist I, Fossier P, Baux G, Roepstorff P, Baleja J D, et al. Proc Natl Acad Sci USA. 1999;96:5758–5763. doi: 10.1073/pnas.96.10.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lirazan M B, Hooper D, Corpuz G P, Ramilo C A, Bandyopadhyay P, Cruz L J, Olivera B M. Biochemistry. 2000;39:1583–1588. doi: 10.1021/bi9923712. [DOI] [PubMed] [Google Scholar]
- 20.Stanley T B, Stafford D W, Olivera B M, Bandyopadhyay P K. FEBS Lett. 1997;407:85–88. doi: 10.1016/s0014-5793(97)00299-8. [DOI] [PubMed] [Google Scholar]
- 21.Bandyopadhyay P K, Colledge C J, Walker C S, Zhou L-M, Hillyard D R, Olivera B M. J Biol Chem. 1998;273:5447–5450. doi: 10.1074/jbc.273.10.5447. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Yang C-T, Jin D, Stafford D W. J Biol Chem. 2000;275:18291–18296. doi: 10.1074/jbc.M001790200. [DOI] [PubMed] [Google Scholar]
- 23.Walker C S, Shetty R P, Clark K, Kazuko S G, Letsou A, Olivera B M, Bandyopadhyay P K. J Biol Chem. 2001;276:7769–7774. doi: 10.1074/jbc.M009576200. [DOI] [PubMed] [Google Scholar]
- 24.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 25.Frohman M A. The Polymerase Chain Reaction. Boston: Birkhauser; 1994. pp. 14–37. [Google Scholar]
- 26.Wu S-M, Cheung W-F, Frazier D, Stafford D W. Science. 1991;254:1634–1636. doi: 10.1126/science.1749935. [DOI] [PubMed] [Google Scholar]
- 27.von Heijne G. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 28.Espiritu D J D, Watkins M, Dia-Monje V, Cartier G E, Cruz L J, Olivera B M. Toxicon. 2001;39:1899–1916. doi: 10.1016/s0041-0101(01)00175-1. [DOI] [PubMed] [Google Scholar]
- 29.Tie J, Wu S-M, Jin D, Nicchitta C V, Stafford D W. Blood. 2000;96:973–978. [PubMed] [Google Scholar]
- 30.Begley G S, Furie B C, Czerwiec E, Taylor K L, Furie G L, Bronstein L, Stenflo J, Furie B. J Biol Chem. 2000;275:36245–36249. doi: 10.1074/jbc.M003944200. [DOI] [PubMed] [Google Scholar]
- 31.Brenner B, Sanchez-Vega B, Wu S, Lanir N, Stafford D W, Solera J. Blood. 1998;92:4554–4559. [PubMed] [Google Scholar]
- 32.Sugita S, Ichtchenko K, Khvotchev M, Sudhof T C. J Biol Chem. 1998;273:32715–32724. doi: 10.1074/jbc.273.49.32715. [DOI] [PubMed] [Google Scholar]
- 33.Canfield L M. Biochem Biophys Res Commun. 1987;148:184–191. doi: 10.1016/0006-291x(87)91093-x. [DOI] [PubMed] [Google Scholar]
- 34.Dowd P, Hershline R, Ham S W, Naganathan S. Science. 1995;269:1684–1691. doi: 10.1126/science.7569894. [DOI] [PubMed] [Google Scholar]
- 35.Pudota B N, Miyagi M, Hallgren K W, West K A, Crabb J W, Misono K S, Berkner K L. Proc Natl Acad Sci USA. 2000;97:13033–13038. doi: 10.1073/pnas.97.24.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu S M, Stafford D W, Frazier L D, High K A, Chu K, Sanchez-Vega B, Solera J. Blood. 1997;89:4058–4062. [PubMed] [Google Scholar]
- 37.Romero E E, Deo R, Velazquez-Estades L J, Roth D A. Biochem Biophys Res Commun. 1998;248:783–788. doi: 10.1006/bbrc.1998.8987. [DOI] [PubMed] [Google Scholar]
- 38.Wu S-M, Morris D P, Stafford D W. Proc Natl Acad Sci USA. 1991;88:2236–2240. doi: 10.1073/pnas.88.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada M, Kuliopulos A, Nelson N P, Roth D A, Furie B, Furie B C, Walsh C T. Biochemistry. 1995;34:481–489. doi: 10.1021/bi00002a012. [DOI] [PubMed] [Google Scholar]
- 40.Wu S-M, Mutueumarana V P, Geromanos S, Stafford D W. J Biol Chem. 1997;272:11718–11722. doi: 10.1074/jbc.272.18.11718. [DOI] [PubMed] [Google Scholar]
- 41.Gilbert W. Cold Spring Harbor Symp Quant Biol. 1987;52:901–905. doi: 10.1101/sqb.1987.052.01.098. [DOI] [PubMed] [Google Scholar]
- 42.Gilbert W, DeSouza S J, Long M. Proc Natl Acad Sci USA. 1997;94:7698–7703. doi: 10.1073/pnas.94.15.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Logsdon J M J. Curr Opin Genet Dev. 1998;8:637–648. doi: 10.1016/s0959-437x(98)80031-2. [DOI] [PubMed] [Google Scholar]
- 44.Cavalier-Smith T. Trends Genet. 1991;7:145–148. [PubMed] [Google Scholar]
- 45.Darnell J E, Doolittle W F. Proc Natl Acad Sci USA. 1986;83:1271–1275. doi: 10.1073/pnas.83.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer J D, Logsdon J M J. Curr Opin Genet Dev. 1991;1:470–477. doi: 10.1016/s0959-437x(05)80194-7. [DOI] [PubMed] [Google Scholar]
- 47.Kurachi K, Davie E W. Proc Natl Acad Sci USA. 1982;79:6461–6464. doi: 10.1073/pnas.79.21.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]