Abstract
The voltage-dependent gating of the colicin channel involves a substantial structural rearrangement that results in the transfer of about 35% of the 200 residues in its pore-forming domain across the membrane. This transfer appears to represent an unusual type of protein translocation that does not depend on a large, multimeric, protein pore. To investigate the ability of this system to transport arbitrary proteins, we made use of a pair of strongly interacting proteins, either of which could serve as a translocated cargo or as a probe to detect the other. Here we show that both an 86-residue and a 134-residue hydrophilic protein inserted into the translocated segment of colicin A are themselves translocated and are functional on the trans side of the bilayer. The disparate features of these proteins suggest that the colicin channel has a general protein translocation mechanism.
The translocation of proteins across membranes typically requires aqueous pores made by large protein complexes, as found in mitochondria and endoplasmic reticulum (1, 2). Certain protein toxins that transport a lethal domain to the cytoplasm, such as pertussis and anthrax, likewise, do so via large, highly conductive, multimeric pores (3, 4). However, a few protein toxins appear to be able to translocate protein domains without forming such structures. The pore-forming colicins, a subfamily of bactericidal proteins whose lethal action is the formation of low-conductance channels in the inner membrane of targeted cells, have been shown to translocate a substantial fraction of their pore-forming domain in association with opening and closing (5, 6), although such translocation, per se, seems unrelated to their toxicity and does not appear to depend on a large multimeric pore (for a review see ref. 7). The observation that localized mutations in this transported segment, including insertions of up to 12 residues, do not block its movement (8) suggested that this system might be recruited to translocate suitably attached exogenous proteins.
In its water-soluble form, the pore-forming domain forms a bundle of 10 α-helices (9–12), which undergo substantial rearrangement upon membrane binding and channel formation. In colicin Ia, a region corresponding to helices 2, 3, 4, and 5 of the soluble structure crosses the bilayer in association with gating (6). Colicin A, used here, is thought to support a similar translocation function, although the exact extent of the translocated domain may differ somewhat from colicin Ia (13).
If colicin is biotinylated at any residue in its normally translocated segment, the attached biotin is carried along to the trans side of the membrane when the channel opens. If avidin is present on the trans side, it can bind to the biotin and prevent the channel from closing (14). We reasoned that this effect on channel closing could be used to test whether a candidate protein could be translocated, if there exists a soluble protein that can bind to it with high affinity. The pair of interacting proteins we used is based on another colicin, colicin E2, a non-pore-forming bacteriocin that is secreted as a heterodimer bound to its immunity protein, ImmE2 (15). (In vivo, the immunity protein serves to inactivate the DNase activity of the C-terminal domain of colicin E2 and thus protects the cell that produces it from its lethal action.) The 14-kDa ImmE2 binds to the C-terminal domain of colicin E2 (CTE2) with an affinity comparable to that of the avidin/biotin complex (16–18), and thus this pair of proteins can serve as another bipartite molecular tool for mapping and manipulating macromolecules. We constructed chimeric versions of colicin A containing either ImmE2 or an enzymatically inactive form of CTE2 inserted between helices 3 and 4 of colicin A, a region of colicin A that is, by analogy to colicin Ia, translocated in association with channel gating (Fig. 1). We then used the complementary protein to probe for the presence of the correctly folded insert.
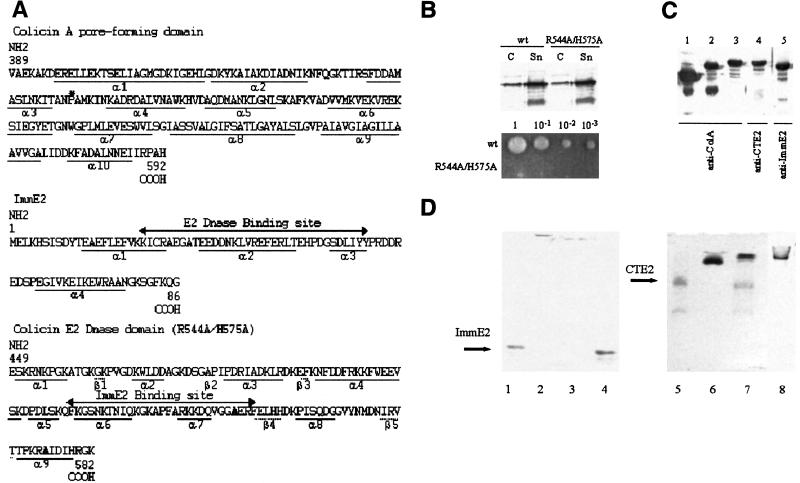
Figure 1.
(A) Sequence of the pore-forming domain of colicin A (pfColA), ImmE2, and CTE2. The asterisk in the pfColA sequence is the insertion location. The bold letters in the CTE2 sequence are the mutated amino acids R544A/H575A. (B) In vivo activity of colicin E2 (R544A/H575A). Five hours after induction with mitomycin C, equal amounts of cells (C) and supernatants (Sn) of medium were subjected to SDS/PAGE and proteins were detected by immunoblotting with anti-colicin E2 Ab. The wt and mutated colicin E2 were secreted at the same level. Undiluted wt and mutated colicin E2 (1 μl of supernatant and indicated serial dilutions) were spotted onto a lawn of sensitive cells. The zone of inhibition on the lawn appears as a clear hole on the photo. Note that the mutated R544A/H575H colicin E2 is totally devoid of toxicity. (C) Expression of colicin A, ColA-CTE2, and ColA-ImmE2 proteins. Bacteria producing colicin A, ColA-CTE2, or ColA-ImmE2 were grown in LB medium at 37°C to OD600 = 0.8 and induced for 30 min with 300 ng/ml mitomycin C. Then total cell extracts producing colicin A (lane 1), ColA-ImmE2 (lanes 2 and 5), and ColA-CTE2 (lanes 3 and 4) were loaded onto SDS/PAGE and analyzed by immunoblotting with the indicated Ab. ColA-ImmE2 and ColA-CTE2 were detected with Abs directed against colicin A or ImmE2 and colicin E2, respectively. (D) Protein binding in solution shown by nondenaturing gels. The gel on the left (lanes 1–4) was run − to + at pH 8.8, and the gel on the right (lanes 5- 8) was run + to − at pH 5.0. Lanes 1–4 contain 2 μg of each ImmE2. In addition, lane 2 contains 6 μg of CTE2, lane 3 contains 20 μg of ColA-CTE2, and lane 4 contains 6 μg of colicin Ia. Notice that CTE2 and ColA-CTE2 bind to ImmE2 and prevent it from entering the gel, whereas the basic protein colicin Ia does not. Lanes 5–8 contain 1.2 μg each of CTE2. In addition, lane 6 contains 3 μg of ImmE2, lane 7 contains 5 μg of colicin A, and lane 8 contains 3 μg of ColA-ImmE2. Notice that ColA-ImmE2 and ImmE2 bind to CTE2 and shift its position in the gel, whereas wt colicin A does not.
Materials and Methods
Construction of the Hybrid Proteins.
To eliminate the lethal enzymatic DNase activity of wild-type (wt) colicin E2, the double point mutation R544A/H575A of colicin E2 was introduced into pBRE2 (19) by using the (Stratagene) Quick Change system (Fig. 1 A and B). DNA fragments extending from codons 389 to 458 and from 459 to 592 of the colicin A gene were amplified by PCR (PCR 1 and 2, respectively) using pLR1 (20) as template. A DNA fragment extending from codons 449 to 582 of the colicin E2 gene and a DNA fragment containing the Imm2 gene were amplified by PCR (PCR 3 and 4, respectively) using pBRE2 as template. The 5′-primers used for PCR 3 and 4 also encoded codons 451–458 of the colicin A gene. The 3′-primers used for PCR 3 and 4 also encoded codons 459–466 of the colicin A gene. Thus, PCR 3 contained codons 451–458 of the colicin A gene, codons 449–582 of the mutated colicin E2 gene, and codons 459–592 of the colicin A gene. PCR 4 contained codons 451–458 of the colicin A gene, codons 1–86 of Imm2 gene, and codons 459–592 of the colicin A gene. PCR fragments 1, 2, and 3 were assembled by the overlap method as the ColA-CTE2 gene. PCR fragments 1, 2, and 4 were assembled by the overlap method as the ColA-ImmE2 gene. These genes were inserted between the AvrII and XhoI sites of pLR1 and the entire construct was sequenced.
Protein Manipulation.
ColA-ImmE2 and ColA-CTE2 were expressed and purified (Fig. 1C) as previously described for mutated colicin A (20). Purified ColA-ImmE2 was biotinylated at Cys-23 of ImmE2 with N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (HPDP-biotin; Pierce) and purified on a monomeric avidin column (Pierce) as described for biotinylated colicin Ia (6). Binding in solution of ImmE2 to CTE2 and to ColA-CTE2 was verified on nondenaturing 15% polyacrylamide gels run at pH 8.8 and stained with Coomassie blue (Fig. 1D). Binding in solution of CTE2 to ImmE2 and to ColA-ImmE2 was verified on nondenaturing 12% polyacrylamide gels run at pH 5.0 (run from + to −).
Bilayer Experiments.
Artificial planar membranes were formed at room temperature from asolectin (type II, Sigma) from which nonpolar lipids had been removed. Lipid (1% in hexane) was layered on top of buffer solutions on either side of a Teflon partition containing an 80- to 100-μm hole pretreated with squalene dissolved in petroleum ether. After evaporation of the solvents, membranes were formed by raising the buffer level over the hole to a final volume of ≈1 ml, while monitoring the membrane capacitance (21). The transmembrane voltage was clamped and currents were monitored as previously described (22).
Results
Translocation of ImmE2.
We constructed the hybrid proteins ColA-ImmE2 (colicin A in which ImmE2 was inserted between residues 458 and 459), and ColA-CTE2 [colicin A in which CTE2 (R544A/H575A) was inserted at the same site] (Fig. 1). Both exhibit toxicity against sensitive cells, and, like wt colicin A, both form voltage-gated channels in planar lipid bilayers. Channels containing either insert exhibit somewhat altered closing kinetics compared to wt channels, but the insertions were fundamentally benign with respect to channel behavior. For both the hybrids and for wt, kinetics of closing are complex, with several rate constants that depend on the pH, voltage, and the history of the channel in the membrane. As seen in Fig. 2, channels open at positive voltage and close at negative voltage. In Fig. 2A, channels containing the ImmE2 insert close on a timescale of tens of seconds at −60 mV before the addition of CTE2 to the trans side of the bilayer. After exposure to trans CTE2, open channels are locked in the open state, and cannot be closed even at −150 mV because the inserted immunity protein has been translocated across the membrane and is able to bind CTE2, which prevents closing (much as trans avidin does to channels biotinylated in this region). If excess ImmE2 is then added to the trans side, the channels that are locked open do not close promptly, even though the concentration of CTE2 has been reduced to near zero because of formation of the soluble ImmE2/CTE2 complex (not shown). Channels that open subsequently close normally. We see no reversal of binding after 500 sec, which (assuming that we would detect a 5% drop in conductance) allows us to place an upper limit on the koff of ≈10−4/sec, consistent with the reported value of 10−6/sec for the closely related colicin complex E9/ImmE9 but not for that of the mismatched E9/ImmE2 (koff = 0.73/sec)(23). Trans CTE2 has no effect on colA-ImmE2 channels in the closed state because the ImmE2 insert is on the cis side when the channel is closed.
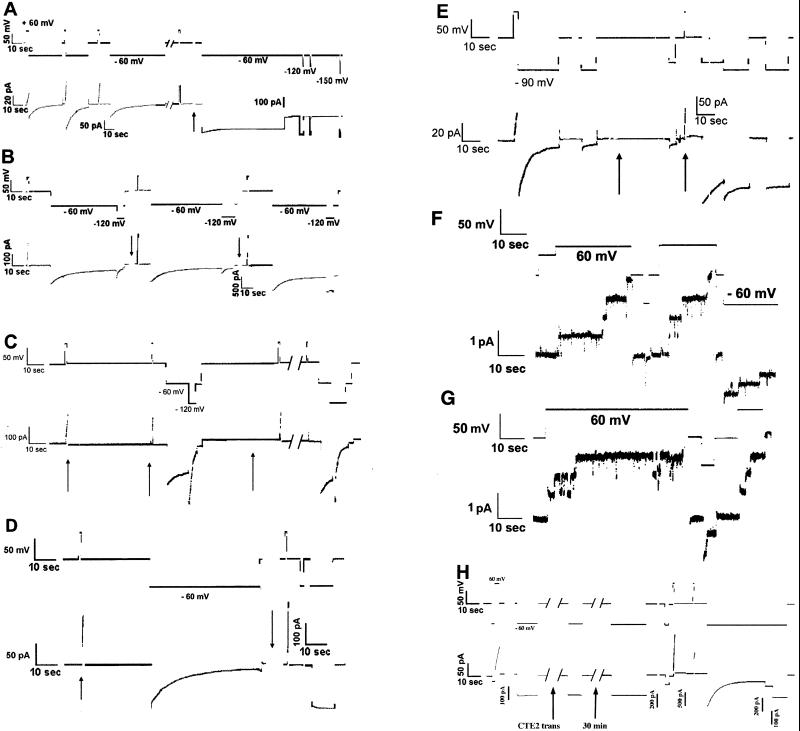
Figure 2.
Current (lower)/voltage (upper) traces from planar lipid bilayer experiments. At the beginning of each trace, both the current and the voltage are 0. The solution on both sides of the membrane is 0.1 M KCl/20 mM malate/5 mM CaCl2/1 mM EDTA, pH 5, unless otherwise noted. Independent experiments verified that ImmE2, CTE2, avidin, and streptavidin did not cause membrane conductance under our conditions. Channel-forming colicin was added to the solution on one side of the membrane (defined as the cis side) before the section of record shown. (A) ColA-ImmE2 effect of trans CTE2. ColA-ImmE2 (0.5 μg) was added to the chamber just before the trace shown. At the beginning of the trace, a series of pulses to + and −60 mV opened and closed a population of channels. A few seconds after the first break (of 60 sec), ≈700 pS of conductance was turned on with a brief +60-mV pulse. During the second 60-sec break (arrow), 3 μg of CTE2 was added to the trans side. A large fraction of the conductance failed to turn off at −60 mV after exposure to trans CTE2. (Note the gain change.) Short pulses to more negative potentials (−120 and −150 mV) also were ineffective. (Notice that a few percent of the conductance fails to close during the 1-min pulse to −60 mV before the first break. This behavior is typical of colicin A and represents a deep, open state that can be closed only at large negative voltages. Entry into the deep, open state is slow under these conditions, and it is easily distinguished from the locked-open state engendered by any of the trans-binding proteins studied here.) (B) ColA-ImmE2 block of trans-CTE2 effect by trans ImmE2. The hybrid (1 μg) was added to the chamber, and, after opening and closing 3 nS of conductance, 6 μg of CTE2 and 1.5× molar excess of ImmE2 were added to the trans solution (first arrow). Channels continued to gate normally. At the second arrow, 5 μg more CTE2 was added to the trans compartment, an amount sufficient to create ≈2 μg of free CTE2. Channels were subsequently opened with a brief pulse to +60 mV and then exposed to a pulse of −60 mV. A substantial fraction of the conductance failed to close, even when the membrane was stepped to −120 mV. Note the gain change. (C) Prebinding of CTE2 to ColA-ImmE2. Before adding it to the chamber, ColA-ImmE2 (0.5 μg) was incubated with a 3× molar excess of CTE2. A brief +60-mV pulse at the first arrow turned on ≈2 nS of conductance. Another, short pulse at the second arrow showed that the channels remained open during the 1-min delay at 0 mV, after which the channels were closed by stepping the voltage to −60 mV and then −120 mV. (The gating is somewhat slower than average in this membrane but well within typical limits.) At the third arrow, 6 μg of CTE2 was added to the trans side, and the same series of voltage pulses was repeated, after the 1-min break. Note that the channels still closed. (D) Effect of trans avidin on ColA-ImmE2-biotin. Biotinylated hybrid (1 μg) was added, and ≈2 ns of conductance was turned on by a brief +60-mV pulse, at the first arrow. After closing these channels by stepping the membrane to −60 mV, avidin (25 μg) was added to the trans compartment (second arrow, during a 40-sec break in trace shown). Channels that were opened shortly after the break by a brief 60-mV pulse failed to close (note change in scale). (E) Effect of trans ImmE2 on ColA-CTE2. After adding 0.3 μg of the hybrid colicin, channels were opened with a brief +80-mV pulse and closed with a −90-mV pulse. At the first arrow, 7 μg of ImmE2 was added to the trans solution. Channels were then opened with the +80-mV pulse at the second arrow. After 10 sec the voltage was again stepped to −90 mV and then most of the channels were trapped in the open state. (F) Single channels of ColA-CTE2. Buffer is 1 M KCl/20 mM Hepes/5 mM CaCl2/1 mM EDTA, pH 7. Protein (0.1 μg) had been added earlier. (G) Single channels of ColA-CTE2 preincubated with 20× molar excess of ImmE2. Buffer is 1 M KCl/20 mM Hepes/5 mM CaCl2/1 mM EDTA, pH 7. Protein (0.03 μg colicin) had been added earlier. Translocation of the inserted CTE2 does not occur under these conditions. (H) Irreversibility of trans ImmE2 biding to ColA-CTE2. Before the part of the record shown, normal gating of 0.3 μg of colicin was confirmed and 2 μg of ImmE2 was added to the trans side. At the initial +60-mV pulse, ≈100 pA of current was turned on. When the voltage was stepped to −60 mV, the channels did not close (because they were held open by binding to the trans ImmE2). At the first arrow, 10 μg of CTE2 was added to the trans side to inactivate the ImmE2. The next pulse to −60 mV (≈7 min after the addition of trans CTE2) shows that the channels were still locked open. After another 30 min, they remained locked open. The next two short +60-mV pulses turned on new channels (note the scale changes), and then a −60-mV pulse turned off all of the new conductance, leaving the original, locked-on channels, still open.
The effect of CTE2 on colA-ImmE2 channels is consistent with the normal binding of these domains seen in the E2/ImmE2 complex. The effect is specific for the ImmE2 chimera: trans CTE2 has no effect on wt colicin A channels. Nor can it be attributed to nonspecific binding between the acidic ImmE2 insert and the basic CTE2 because the highly basic protein avidin had not effect (not shown). An excess of trans ImmE2 blocks the effect of trans CTE2 (Fig. 2B). If colA-ImmE2 is either incubated with CTE2 in solution before adding it to the bilayer chamber, or, if closed colA-ImmE2 channels are exposed to cis CTE2, qualitatively normal, voltage-gated channels are still formed, but they are insensitive to trans CTE2 (Fig. 2C and data not shown). Thus, binding of the insert in the closed state to CTE2 prevents the translocation of the insert from the cis to the trans side, but not the formation of voltage-gated channels.
As another measure of the translocation of the inserted ImmE2, we biotinylated it at its lone cysteine residue with HPDP-biotin (6), at position 23 of ImmE2 (the colicin A host protein has no cysteines) and observed the effect of avidin (which binds tightly to biotin) on colA-ImmE2-biotin channels. As expected, trans avidin had no effect on unbiotinylated ColA-ImmE2 channels or on closed ColA-ImmE2-biotin channels, but it prevented the closure of open ColA-ImmE2-biotin channels, presumably by binding to the biotin moiety that had been translocated, along with the inserted ImmE2, when the channel opened (Fig. 2D). This effect could be reversed with trans tris(2-carboxyethyl)phosphine (TCEP), a water-soluble reducing agent that can cut the disulfide bond linking Cys-23 of the inserted ImmE2 to the biotin derivative, thus showing that the avidin molecule must remain bound for blockage of closure to be maintained under our conditions. (The fact that the channel can close if avidin is removed shows that it, and presumably the E2 peptides, do not act by coaxing the channel into a natural, deep, open state.) Cis avidin had little effect on gating but rendered trans avidin ineffective. In fact, as long as the insert was held on the cis side, neither trans avidin nor CTE2 had any effect.
Interestingly, the biotinylated hybrid is still susceptible to trans CTE2, showing that the presence of the cysteine bearing the HPDP-biotin adduct does not block binding by CTE2. ImmE9, a well characterized E colicin immunity protein largely homologous to ImmE2, can tolerate the attachment of large reagents, such as 5,5′-dithiobis(2-nitrobenzoic acid (DTNB), to its Cys-23, which is at the edge of its binding region, without losing its activity, but modification with N-iodoacetyl-N′-(8-sulfo-1-naphthyl)ethylenediamine (IASN) prevents its binding to its natural substrate, colicin E9 (25). HPDP-biotin is larger than IASN, but the structures of the corresponding cysteine derivatives are rather similar: a disulfide bond followed by a spacer arm ending in a double ring. The longer spacer arm of HPDP-biotin may account for its inability to prevent binding to CTE2.
Translocation of CTE2.
After establishing that the 86-residue ImmE2 was translocated, we reversed the proteins: We inserted the 134-residue CTE2 into the helix 3/4 (H3/4) loop, and we used ImmE2 as the probe (Fig. 1). We used a CTE2 containing the double point mutation R544A/H575A, which lacks DNase activity but retains its ability to bind ImmE2 (26) (Fig. 1 B and D), thus allowing its synthesis in the absence of immunity protein. In planar bilayers, this pair of proteins behaved much like the inverse pair. Trans ImmE2 blocked the closing of open ColA-CTE2 channels (Fig. 2E), an effect blocked by the previous addition of an excess of trans CTE2. Cis ImmE2 prevented the trans ImmE2 effect (not shown), evidently by preventing the translocation of the insert. Such trapping of the CTE2 insert on the cis side did not result in any qualitative alteration in the properties of single ColA-CTE2 channels, shown without (Fig. 2F) and with (Fig. 2G) cis ImmE2. (In 1 M KCl at pH 7, the conductances were, respectively, 13.0 ± 1.6 pS and 12.7 ± 5.5 pS). Once locked open with trans ImmE2, an excess of trans CTE2 does not reverse the blockage for over 30 min, but channels that open subsequently close normally, showing that no free trans ImmE2 remains to bind to the inserted CTE2 (Fig. 2H).
pH Dependence.
Translocation of the inserted proteins was observed at pH 5 and pH 6 but not at pH 7, even though voltage-gated channels were formed at all these pHs. Thus channel formation can be uncoupled from translocation with a simple change in pH: It is not necessary to artificially restrict the H3/4 region to the cis side by attaching it to a large protein. The failure to transport the inserts at pH 7 reflects a pH dependence of the efficiency of translocation, not of channel formation (although channel formation is itself pH-dependent). This effect of pH is likely to be an effect on the colicin A, rather than on the inserted protein, because the two inserts bear opposite net charges. In vivo, both hybrids are active toxins against sensitive cells but less so than wt colicin. In contrast to the bilayer results, ColA-CTE2 was totally inactive in vivo at pH 7, whereas the activity of ColA-ImmE2 was reduced by a factor of only 2 compared with wt colicin A. At pH 5 (a pH that permits high activity and efficient translocation in vitro), however, both hybrids are almost as active as wt. The different behaviors observed in vitro and in vivo are probably caused by the interactions with cellular components that precede channel formation in vivo.
Discussion
The inferred tight binding of the exogenous insert to its partner on the trans side argues that the insert is presented intact to the trans solution. The structure of the bound complex of the heterodimer colicin E9/ImmE9, which is 80%/60% homologous to colicin E2/ImmE2, shows that the immunity protein makes contact with the colicin DNase domain through residues of two of its four helices, making use of a dual recognition mechanism (17, 23, 27–30). Nineteen residues from both proteins are involved in 12 direct hydrogen bonds between them, in addition to 5 hydrogen bonds mediated by water molecules involving seven additional residues. A mismatch of the binding site on only one of the two helices, as occurs when colicin E9 DNase binds to the noncognate ImmE2, causes the binding constant to fall by six orders of magnitude (23). Assuming that the colicin E2/ImmE2 contact is similarly constructed, the effectively irreversible binding we observed in the presence of excess complementary protein strongly suggests that both proteins are essentially correctly folded on the trans side and form a high-affinity complex. Only a few incorrect contacts would be expected to reduce the affinity to levels where the blocking effect on channel closure would be easily reversed in the presence of an excess of the appropriate competitor. The fact that it is not implies that, in both cases, the entire protein was translocated and was present on the trans side in its native form.
Apparently, translocation proceeds from the C terminus of the translocated segment toward the N-terminus, and its arrest at H3/4 is without import because the channel is formed before helices 1–4 are translocated. In a model of gating developed for colicin Ia, it was proposed that helices 2–5 cross the membrane when the channel opens, but that it is still possible to form channels, albeit with altered conductance and gating, even if a part of the translocated domain is compelled to remain on the cis side of the membrane (by attaching it to a large, soluble protein) (6). In the case of colicin Ia, the presence of H1 as a transmembrane element is an important determinant of channel properties (6, 24), but in the present case it appears that channels prevented from translocating helices 1–3 are not critically altered. This result is, however, consistent with the observation that helices 1–4 are not important for the colicin A channel (13), and thus may reflect a difference between colicins A and Ia.
The ability of colicin A to tolerate these foreign proteins in its own translocated domain and to translate them along with it has implications for the gating mechanism of these channels. The transfer of helices 2–5 of colicin Ia across the bilayer was originally established by identifying a series of residues in this region that cross the membrane in association with gating (6). There is, however, another interpretation of such data that could not be definitively ruled out. The H2–5 region might be highly mobile within the bilayer, such that a brief fluctuation could cause any particular residue to momentarily appear on the trans side, where it could be bound by an experimental probe, detected, and counted as translocated, even though other parts of the domain might be in the membrane, or even on the cis side, at that moment. That is, the experiment itself would be responsible for holding the translocated domain on the trans side. The present results argue against this because they establish that colicin is able to present a large sequence of protein stably to the trans compartment.
The mechanism of translocation used by colicin is far from clear, as is any obvious rationale for such a function in a toxin that kills by forming a low-conductance (<10-pS) channel. All evidence suggests that the channel is a monomer and that the pore itself is formed by, at most, four transmembrane segments. Because colicin does not form large, multimeric pores, as do many protein-translocating toxins, we surmise that it works differently (7) and that the translocated cargo is not simply passing through the ion-conducting channel. A few other toxins have been identified that may work similarly, the best understood of which is diphtheria toxin (31, 32), although in these other cases a translocated domain of the toxin is the lethal agent. Perhaps colicin evolved from such a toxin and has lost a primordial intracellular function, leaving only the channel, which is sufficient to kill its small target cell.
Our results show that the gating machinery of colicin A is capable of moving exogenous proteins across lipid bilayers and presenting them in functional form to the trans solution (Fig. 3). Either ImmE2 or CTE2, inserted into colicin A, was moved across the membrane, where it was recognized by its partner, much as it would be recognized in solution. Although either insert is readily translocated, the bound pair is not. This may be because of the larger size of the complex, but is more likely because of the stable structure formed by the tightly bound pair. Perhaps the inserts must fully or partially unfold to cross the membrane, but, if so, they evidently refold correctly on the trans side. Other than the lack of disulfide bonds, which would prevent complete unfolding, the two proteins used as inserts here do not have any properties that should obviously predispose them to cross bilayers and are in fact quite dissimilar to each other. (In vivo, the C-terminal domain, at least, of colicin E2 is transported into the cytoplasm, presumably by target cell systems.) ImmE2 is largely α-helical and is highly negatively charged, whereas CTE2 has both α and β structure and is highly positively charged. Given that the colicin channel can translocate such dissimilar proteins, it may well be able to transport any number of diverse cargoes.
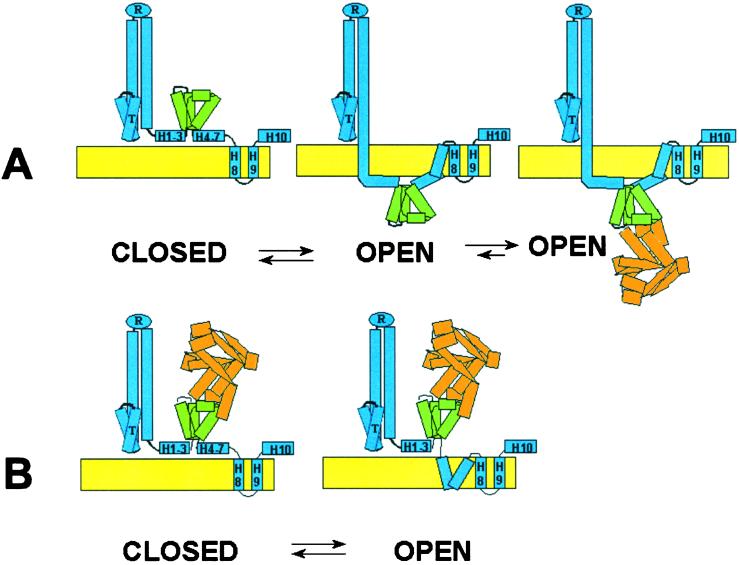
Figure 3.
Closed and open configurations of ColA-ImmE2. Colicin A, blue; ImmE2 insert, green; CTE2, orange. The crystal structure of CTE9/ImmE9 (28) was used as a guide to draw ImmE2 and CTE2. Colicin A is drawn schematically to conform to the structure of colicin Ia (10), except for the C-terminal domain. The transmembrane segments between ImmE2 and H8 in the open conformation are unlabeled to indicate the dearth of information concerning their structure. (A) Trans CTE2. (B) Cis CTE2.
Acknowledgments
We thank Colin Kleanthous (University of East Anglia, U.K.) for the generous gift of purified ImmE2 and CTE2. We thank Lazaro Molina and Daniele Cavard for the gift of ImmE2 Ab and colicin E2 Ab, respectively. We also thank Alan Finkelstein and Paul Kienker for helpful discussions, and James Sturgis for careful reading of the manuscript. This work was supported by National Institutes of Health Grant GM 29210.
Abbreviations
- ImmE2
colicin E2 immunity protein
- CTE2
the C-terminal domain of colicin E2
- ColA-ImmE2
colicin A bearing the ImmE2 insertion
- ColA-CTE2
colicin A bearing the CTE2 insertion
- Hn
helix n of the pore-forming domain
- cis
the side (of the membrane) to which colicin is added
- trans
the side opposite to cis
- wt
wild type
- HPDP-biotin
N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pfanner N, Geissler A. Nat Rev Mol Cell Biol. 2001;2:339–349. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- 2.Johnson A E, van Waes M A. Annu Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 3.Shannon J L, Fernandez R C. J Bacteriol. 1999;181:5838–5842. doi: 10.1128/jb.181.18.5838-5842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaustein R O, Koehler T M, Collier R J, Finkelstein A. Proc Natl Acad Sci USA. 1989;86:2209–2213. doi: 10.1073/pnas.86.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slatin S L, Qiu X-Q, Jakes K S, Finkelstein A. Nature (London) 1994;371:158–161. doi: 10.1038/371158a0. [DOI] [PubMed] [Google Scholar]
- 6.Qiu X-Q, Jakes K S, Kienker P K, Finkelstein A, Slatin S L. J Gen Physiol. 1996;107:313–328. doi: 10.1085/jgp.107.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakey J H, Slatin S L. In: Current Topics in Microbiology and Immunology. van der Goot F G, editor. Vol. 257. Berlin: Springer; 2001. pp. 131–161. [DOI] [PubMed] [Google Scholar]
- 8.Jakes K S, Kienker P K, Slatin S L, Finkelstein A. Proc Natl Acad Sci USA. 1998;95:4321–4326. doi: 10.1073/pnas.95.8.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker M W, Pattus F, Tucker A D, Tsernoglou D. Nature (London) 1989;337:93–96. doi: 10.1038/337093a0. [DOI] [PubMed] [Google Scholar]
- 10.Wiener M, Freymann D, Ghosh P, Stroud R M. Nature (London) 1997;385:461–464. doi: 10.1038/385461a0. [DOI] [PubMed] [Google Scholar]
- 11.Elkins P A, Song H Y, Cramer W A, Stauffacher C V. Proteins. 1994;19:150–157. doi: 10.1002/prot.340190208. [DOI] [PubMed] [Google Scholar]
- 12.Vetter I R, Parker M W, Tucker A D, Lakey J H, Pattus F, Tsernoglou D. Structure (London) 1998;6:863–874. doi: 10.1016/s0969-2126(98)00088-4. [DOI] [PubMed] [Google Scholar]
- 13.Nardi A, Slatin S L, Baty D, Duché D. J Mol Biol. 2001;307:1293–1303. doi: 10.1006/jmbi.2001.4524. [DOI] [PubMed] [Google Scholar]
- 14.Qiu X Q, Jakes K S, Finkelstein A, Slatin S L. J Biol Chem. 1994;269:7483–7488. [PubMed] [Google Scholar]
- 15.Jakes K S. In: Molecular Action of Toxins and Viruses. Cohen P, van Heyningen S, editors. Amsterdam: Elsevier; 1982. pp. 131–167. [Google Scholar]
- 16.Wallis R, Leung K Y, Pommer A J, Videler H, Moore G R, James R, Kleanthous C. Biochemistry. 1995;34:13751–13759. doi: 10.1021/bi00042a005. [DOI] [PubMed] [Google Scholar]
- 17.Wallis R, Leung K Y, Osborne M J, James R, Moore G R, Kleanthous C. Biochemistry. 1998;37:476–485. doi: 10.1021/bi971884a. [DOI] [PubMed] [Google Scholar]
- 18.Wallis R, Moore G R, James R, Kleanthous C. Biochemistry. 1995;34:13743–13750. doi: 10.1021/bi00042a004. [DOI] [PubMed] [Google Scholar]
- 19.Benedetti H, Frenette M, Baty D, Knibiehler M, Pattus F, Lazdunski C. J Mol Biol. 1991;217:429–439. doi: 10.1016/0022-2836(91)90747-t. [DOI] [PubMed] [Google Scholar]
- 20.Duché D, Parker M W, Gonzalez-Mañas J-M, Pattus F, Baty D. J Biol Chem. 1994;269:6332–6339. [PubMed] [Google Scholar]
- 21.Montal M. Methods Enzymol. 1974;32:545–554. doi: 10.1016/0076-6879(74)32053-8. [DOI] [PubMed] [Google Scholar]
- 22.Jakes K S, Abrams C K, Finkelstein A, Slatin S L. J Biol Chem. 1990;265:6984–6991. [PubMed] [Google Scholar]
- 23.Li W, Hamill S J, Hemmings A M, Moore G R, James R, Kleanthous C. Biochemistry. 1998;37:11771–11779. doi: 10.1021/bi9808621. [DOI] [PubMed] [Google Scholar]
- 24.Kienker P K, Jakes K S, Finkelstein A. J Gen Physiol. 2000;116:587–597. doi: 10.1085/jgp.116.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallis R, Moore G R, Kleanthous C, James R. Eur J Biochem. 1992;210:923–930. doi: 10.1111/j.1432-1033.1992.tb17496.x. [DOI] [PubMed] [Google Scholar]
- 26.Garinot-Schneider C, Pommer A J, Moore G R, Kleanthous C, James R. J Mol Biol. 1996;260:731–742. doi: 10.1006/jmbi.1996.0433. [DOI] [PubMed] [Google Scholar]
- 27.Kleanthous C, Kuhlmann U C, Pommer A J, Ferguson N, Radford S E, Moore G R, James R, Hemmings A M. Nat Struct Biol. 1999;6:243–252. doi: 10.1038/6683. [DOI] [PubMed] [Google Scholar]
- 28.Kuhlmann U C, Pommer A J, Moore G R, James R, Kleanthous C. J Mol Biol. 2000;301:1163–1178. doi: 10.1006/jmbi.2000.3945. [DOI] [PubMed] [Google Scholar]
- 29.Li W, Dennis C A, Moore G R, James R, Kleanthous C. J Biol Chem. 1997;272:22253–22258. doi: 10.1074/jbc.272.35.22253. [DOI] [PubMed] [Google Scholar]
- 30.Osborne M J, Wallis R, Leung K Y, Williams G, Lian L Y, James R, Kleanthous C, Moore G R. Biochem J. 1997;323:823–831. doi: 10.1042/bj3230823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senzel L, Gordon M, Blaustein R O, Oh K J, Collier R J, Finkelstein A. J Gen Physiol. 2000;115:421–434. doi: 10.1085/jgp.115.4.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finkelstein A, Oh K J, Senzel L, Gordon M, Blaustein R O, Collier R J. Int J Med Microbiol. 2000;290:435–440. doi: 10.1016/S1438-4221(00)80059-4. [DOI] [PubMed] [Google Scholar]