Abstract
The goal of this study is to use the model system described earlier to make direct measurements of the enthalpy of helix formation at different temperatures. For this we studied model alanine peptides in which helix formation can be triggered by metal (La3+) binding. The heat of La3+ interaction with the peptides at different temperatures is measured by isothermal titration calorimetry. Circular dichroism spectroscopy is used to follow helix formation. Peptides of increasing length (12-, 16-, and 19-aa residues) that contain a La3+-binding loop followed by helices of increasing length, are used to separate the heat of metal binding from the enthalpy of helix formation. We demonstrate that (i) the enthalpy of helix formation is −0.9 ± 0.1 kcal/mol; (ii) the enthalpy of helix formation is independent of the peptide length; (iii) the enthalpy of helix formation does not depend significantly on temperature in the range from 5 to 45°C, suggesting that the heat capacity change on helix formation is very small. Thus, the use of metal binding to induce helix formation has an enormous potential for measuring various thermodynamic properties of α-helices.
The year 2001 marked the 50th anniversary of the α-helix, the first proposed protein secondary structure (1). However, despite numerous efforts (see refs. 2–5 and references therein), the detailed thermodynamic basis for helix formation and for the helix propensities of the amino acids is not yet well understood. Accurate values for such basic thermodynamic parameters as the changes in enthalpy and heat capacity on helix formation are still under debate. Recently Bierzynski and coworkers (6, 7) developed a peptide system for inducing helix formation by adding a metal ion. They took a 12-residue sequence, analogous to a Ca2+-binding loop from calmodulin (peptide P1), which forms a short and very stable C-terminal helix, containing three to four residues, when the peptide binds La3+. The C-terminal segment of La3+-bound P1 provides a stable helical nucleus for helix propagation when additional residues are added at the C terminus. Bierzynski and coworkers also made a longer peptide (P2) with three additional alanine residues plus one C-terminal glutamine residue, and determined the NMR structure of the La3+-ligated form. Their model system should allow accurate determination of the enthalpy of helix formation by using ITC (isothermal titration calorimetry) to monitor stepwise, La3+-induced helix formation, and we exploit this potential here.
Helix–coil transitions of short peptides in water cannot usually be described by a two-state model because the helix propagation constant is not large enough. For this reason, and also because the enthalpy change on helix formation is not large, temperature-induced helix unfolding spans a very broad temperature range, which makes it difficult to determine the baseline. Analysis of thermal helix–coil transitions can be based on the Zimm–Bragg or Lifson–Roig theories (e.g., see ref. 2), but the enthalpy values determined in this way must be tested by direct calorimetric measurements. The calorimetric measurements, on the other hand, are hampered by the broad transition curves and uncertain baselines: fully helical structures in typical peptide systems are never attained in aqueous solutions, even at 0°C. Moreover, the change in heat capacity on helix formation, ΔCp, is a critical parameter in determining the enthalpy change and ΔCp cannot be measured at present by scanning calorimetry because of the broadness of the thermal transition curves.
If a small change in solvent composition can induce helix formation, the enthalpy of helix formation can be measured using ITC, and the calmodulin-derived peptide system (6, 7) offers this possibility. The basic motif (P1-peptide) consists of 12-aa residues analogous to the calcium-binding loop III of rat calmodulin; the residues that coordinate Ca2+ in calmodulin are 1, 3, 5, 9 and 12 (8). Although P1 does not bind Ca2+ with high affinity, it does bind La3+ reasonably well (K = ≈105 M−1). In the NMR structure of the 16-residue La3+-ligated peptide P2 (1NKF, ref. 7), which contains four additional helix-forming residues at the C-terminal end, seven to eight C-terminal residues are helical (Fig. 1). CD data corroborate the structural results and, furthermore, CD data indicate that P2 does not have a high helical content in the absence of La3+, and therefore La3+ binding triggers helix formation. Consequently, helix formation induced by lanthanum binding can be a useful model system to study the thermodynamics of helix formation at constant temperature. In addition to determining more accurate values of the enthalpy changes on helix formation for various peptides, it should be possible to determine ΔCp by measuring ΔH as a function of temperature.
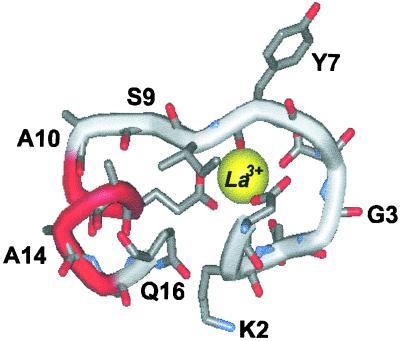
Figure 1.
Cartoon representation of the structure of P2-peptide in La3+-ligated form. Selected amino acid residues are labeled. La3+-ion shown in the CPK representation. One representative structure of 30 models in 1NFK entry (7) to Protein Data Bank was used for illustration.
To test the usefulness of this model system, we measured the enthalpies of La3+ binding to P1, P2 (P1 with additional four residues at the C terminus), and P3 (P1 with additional seven residues at the C terminus) using ITC. These experiments, when combined with CD-based estimates of the changes in fraction helix on La3+ binding, give estimates of the enthalpy of helix formation from 5–45°C.
Materials and Methods
Peptide Synthesis, Purification, and Sample Preparation.
Peptides P1 (Ac-DKDGDGYISAAE-NH2), P2 (Ac-DKDGDGYISAAEAAAQ-NH2), and P3 (Ac-DKDGDGYISAAEAAAAAAQ-NH2) were synthesized by the solid phase method, using fluorenylmethoxycarbonyl (Fmoc) chemistry as described (9). P6N5 peptide (Ac-AAKAAY-NH2) was a gift from J. M. Scholtz (Texas A&M University, College Station, TX). The peptides were purified by C18 reverse-phase column chromatography. The identity of peptides was confirmed by mass spectrometry and purity was shown to be better than 95% by analytical C18 reverse-phase column chromatography. For the La3+ titration experiments, the stock solutions of peptides were dialyzed extensively against at least two changes of corresponding buffer containing either 1 mM sodium cacodylate or Tris, and 100 mM NaCl (pH 6.9). Spectrapor CE dialysis membranes with a 1,000-Da molecular weight cutoff were used for dialyses. The peptide stock solution was diluted to a working concentration of ≈50 μM in the last change of the dialysis buffer. The peptide concentration was measured spectrophotometrically by using a molar extinction of 1,450 M−1⋅cm−1 at 275 nm determined as described (9). The LaCl3 (Alfa Aezar, Ward Hill, MA) solutions for ITC experiments in the last change of corresponding buffer were prepared by weight. The LaCl3 concentration was kept around 2 mM.
Circular Dichroism Spectroscopy.
The CD spectra of peptides in the presence or absence of LaCl3 were recorded using 1-mm pathlength cells in an Aviv 62 DS spectropolarimeter equipped with a Peltier temperature control unit (Aviv Associates, Lakewood, NJ) or in a Jasco J-715 spectropolarimeter (Jasco, Easton, MD) equipped with a Neslab RTE-140 temperature control unit (Neslab Instruments, Portsmouth, NH) as described (9). CD samples contained 100 μM of peptide in 1 mM sodium cacodylate/100 mM NaCl (pH 6.9) buffer, with or without 2.5 mM LaCl3. The thermal transition data were recorded every 1°C with an equilibration time of 2 min (Aviv 62 DS) or 6–8 min (Jasco J-715). The CD spectra of these short helices will be reported elsewhere (D.H.C., R. W. Woody, C. A. Rohl, and R.L.B., unpublished work), and compared with theoretical calculations by R. W. Woody. In comparing CD results with theory, ellipticity is expressed per mole of peptide groups rather than amino acid residues (10), and the same convention is used here.
Isothermal Titration Calorimetry.
The overall procedure for ITC experiments was similar to that described (11). The VP-ITC from Microcal (Northampton, MA) was used in all experiments. Briefly, 3 μl of the LaCl3 solution at concentrations ≈2 mM were injected into the cell containing peptide solution at concentration ≈50 μM. The blank injections of solution into corresponding buffer were used to account for the heats of mixing and dilution. In each experiment 40 to 45 injections are made, so that the final molar ratio of LaCl3 to peptide is 3.5 to 4. The heat of the reaction, Q, is obtained by integrating the peak after each injection of peptide ligand using ORIGIN software provided by the manufacturer (Microcal, Northampton, MA). The heat of the reaction at each injection is related to the calorimetric enthalpy of binding, ΔHcal, and the other thermodynamic parameters as (12):
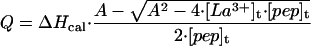 |
1 |
where A = [pep]t + [La3+]t + 1/Ka, n is the stoichiometry of the La3+/peptide complex, Ka is the association constant, [pep]t is the concentration of a peptide, and [La3+]t is the total concentration of LaCl3. Alternatively, ΔHcal was calculated by summing individual heat increments corrected for dilution and dividing it by the total number of moles of peptide in the cell.
Results
Isothermal Titration Calorimetry.
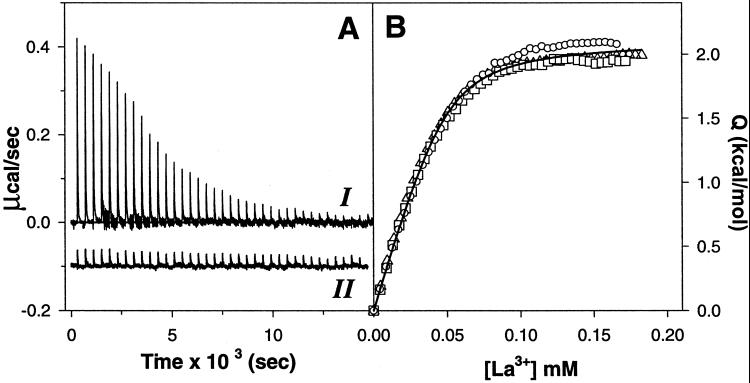
Fig. 2A shows representative ITC titration of the solution of P1-peptide with LaCl3 at 25°C. The interaction is endothermic and the titration of La3+ goes to completion within 30 injections. Fig. 2B plots the sum of the measured heat effects after each injection, normalized by the concentration of P1-peptide in the cell, for two independent experiments performed in sodium cacodylate buffer and for one experiment performed in Tris⋅HCl buffer. The agreement between titration profiles performed in the two different buffers (sodium cacodylate and Tris⋅HCl) is remarkably good. These two buffers have significantly different ionization enthalpies: −0.5 kcal/mol for cacodylate and 11.3 kcal/mol for Tris (13). Agreement of the titration profiles performed in buffers with different ionization enthalpies provides experimental evidence for the absence of linked protonation effects when La3+ binds to P1-peptide (11, 14). Similar results have been obtained for peptides P2 and P3 (data not shown). Consequently, the measured heat effects do not include any contribution from buffer ionization linked to protonation of P1-peptide.
Figure 2.
Isothermal titration calorimetry. (A) Representative ITC experiment. Heat effects recorded as a function of time during 40 successive 3-μl injections of 2.01 mM solution of LaCl3 into the cell containing either 50 μM solution of P1-peptide (profile I) or buffer solution (profile II) at 25°C. Buffer conditions: 1 mM sodium cacodylate, 100 mM NaCl, pH 6.9. (B) Comparison of three independent LaCl3 titration profiles for P1-peptide at 25°C. The buffer conditions are sodium cacodylate (□ and ○) and Tris (▵). The solid line represents the nonlinear regression of all three experiments to Eq. 1 with the parameters n = 0.99 ± 0.03; Kd = (5.3 ± 0.6) × 10−6 M; ΔH = 2.1 ± 0.1 kcal/mol.
Fig. 2B shows also the results of the simultaneous fit of all three experiments to Eq. 1. The stoichiometry of binding is 0.99 ± 0.03, indicating that one molecule of La3+ binds to P1-peptide, as expected. The dissociation constant is 5 μM, similar to earlier estimates for the synthetic peptide analogues of calcium-binding loops (7, 8).
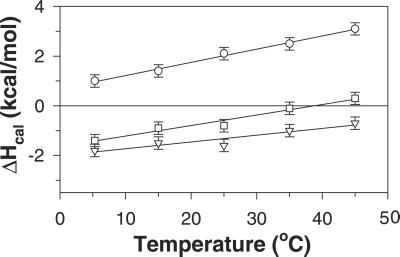
Fig. 3 shows the temperature dependence of the experimental enthalpy, ΔHcal, that accompanies La3+ binding to P1, P2, and P3 peptides. The enthalpy values differ not only in magnitude but also in the sign of the observed heat effect, which is endothermic for the P1 peptide but exothermic for P2 and P3 peptides.
Figure 3.
Temperature dependence of the calorimetric enthalpy of interaction of peptides P1 (○), P2 (□), and P3 (▿) with La3+-ions. Error bars represent the standard deviation of multiple experiments. The lines through the points are linear fits of the data and are shown to guide the eye.
The enthalpy measured experimentally for each peptide on adding La3+ [ΔHcal(P1), ΔHcal(P2), ΔHcal(P3)] consists of several terms:
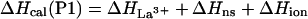 |
2 |
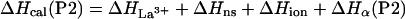 |
3 |
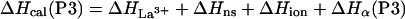 |
4 |
where ΔHLa3+ represents the enthalpy of binding La3+ to the calcium binding loop; ΔHion is the enthalpy linked to protonation effects, caused by changes in pKa values on complex formation; ΔHns is the enthalpy of reorganization of the calcium binding loop (i.e., first 12 residues); and ΔHα is the enthalpy of helix formation in each of the peptides. Of these four terms, the enthalpy of ionization appears to be very small—i.e., ΔHion ≈ 0. The enthalpy of rearrangement of the first 12 residues involves conformational change in the calcium-binding loop on La3+ binding and also helix formation by three to four C-terminal residues; it is represented by the sum of ΔHLa3+ and ΔHns. This term is assumed to be the same for all three peptides and therefore equals the experimentally measured enthalpy of P1-peptide, ΔHcal(P1). The enthalpies of helix formation in the P2 and P3 peptides are written as follows:
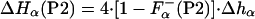 |
5 |
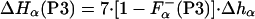 |
6 |
where F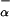 represents the fraction of each peptide that is already in helical conformation before adding La3+ and Δhα represents the enthalpy of helix formation per residue, which becomes helical upon La3+ binding. Note that peptides P2 and P3 contain four and seven additional residues, respectively, that are not included into the ΔHns term.
represents the fraction of each peptide that is already in helical conformation before adding La3+ and Δhα represents the enthalpy of helix formation per residue, which becomes helical upon La3+ binding. Note that peptides P2 and P3 contain four and seven additional residues, respectively, that are not included into the ΔHns term.
From Eqs. 2–6 it follows that the enthalpy of helix formation, on a per residue basis, Δhα, can be calculated as:
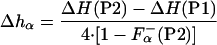 |
7 |
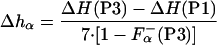 |
8 |
provided that the fraction of the peptide forming helical structure upon La3+ binding is known. This fraction can be measured using circular dichroism spectroscopy.
Circular Dichroism Spectroscopy.
The fraction of helical residues in peptides P2 and P3 in the absence of La3+ can be calculated from the experimentally measured ellipticity of these peptides, [Θ−(P2)] and [Θ−(P3)], as
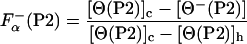 |
9 |
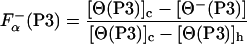 |
10 |
where F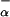 (P2) and F
(P2) and F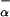 (P3) are fraction helix in the absence of La3+, [Θ(P2)]c and [Θ(P3)]c are the ellipticities of peptides P2 and P3 in the unfolded (“coil”) state, respectively, and [Θ(P2)]h and [Θ(P3)]h are the ellipticities of peptides P2 and P3 in the fully helical state, respectively. Note that F
(P3) are fraction helix in the absence of La3+, [Θ(P2)]c and [Θ(P3)]c are the ellipticities of peptides P2 and P3 in the unfolded (“coil”) state, respectively, and [Θ(P2)]h and [Θ(P3)]h are the ellipticities of peptides P2 and P3 in the fully helical state, respectively. Note that F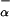 (P2) and F
(P2) and F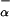 (P3) refer only to those additional residues that are forming helical structure besides the first 12 residues of peptide P1.
(P3) refer only to those additional residues that are forming helical structure besides the first 12 residues of peptide P1.
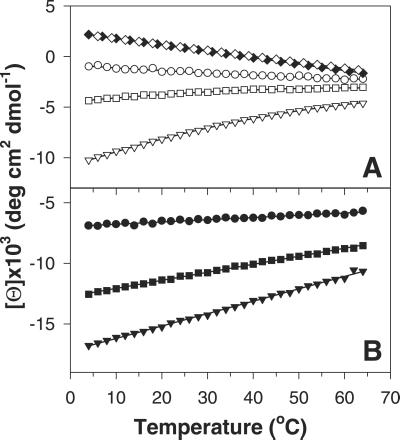
Fig. 4 A and B shows the dependence on temperature of the ellipticity of peptides P1, P2, and P2 in the presence and absence of La3+. Note that the ellipticity of each peptide is larger in the presence of La3+ (Fig. 4B) than in its absence (Fig. 4A) and that the plots with La3+ present are straight lines, whereas the plots with La3+ absent curve downward for peptides P2 and P3. The temperature dependence of the ellipticity of a short model peptide P6N5 (see sequence in Materials and Methods) is also given in Fig. 4A. Analysis of these results (D.H.C., R. W. Woody, C. A. Rohl, and R.L.B., unpublished work) indicates that, in the presence of La3+, the helix contents of peptides P1, P2, and P3 are independent of temperature from 4–65°C, and are presumed to be >95% helical in this range. The NMR structure of P2 at 2°C indicates that residues Ser-9 to Ala-15 are helical and that Gln-16 is partly helical, whereas chemical shift data indicate that residues Ala-10 to Gln-16 are helical (7). In analyzing our ITC data for both peptides P2 and P3 in the presence of La3+, residues past the calcium-binding loop are treated as being fully helical. Therefore, it is assumed that [Θ(P2)]h = [Θ⊕(P2)] and [Θ(P3)]h = [Θ⊕(P3)], where [Θ⊕(P2)] and [Θ⊕(P3)] represent the ellipticity of P2 or P3 peptides in the presence of La3+. The curved plots for peptides P2 and P3 in the absence of La3+ (Fig. 4A) suggest that some helical structure is present. The ellipticities for the unfolded states of P2 and P3 peptides can be obtained as follows:
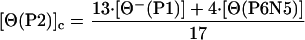 |
11 |
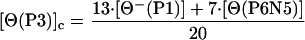 |
12 |
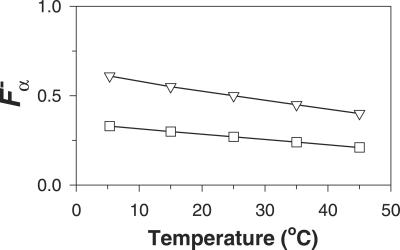
where [Θ−(P1)] is the ellipticity of P1 in the absence of La3+, and [Θ(P6N5)] is the ellipticity value for the model peptide. The reason for using the CD spectra of two peptides (P1 and P6N5) to represent the unfolded CD spectra of peptides P2 and P3 is that “unfolded” peptides do in fact have CD spectra, and therefore it is desirable to model the unfolded CD spectrum of a peptide by using matching sequences. The P1 peptide matches the sequence of the first 12 residues (or 13 peptide groups) of peptides P2 and P3, whereas the alanine-based peptide P6N5 matches approximately the remaining alanine-based sequence. It is possible that the CD spectrum of unfolded P1 contains a small contribution from partly helical residues. We assume that any partly helical residues are contained in residues 1–8, which do not become helical on adding La3+. The fraction helix for peptides P2 and P3 in the absence of La3+ is shown in Fig. 5. As expected, both peptides P2 and P3 have some residual helix content in the absence of La3+ at low temperature. This residual helix content gradually melts with increasing temperature.
Figure 4.
Temperature dependence of the ellipticity of peptides in the presence and in the absence of LaCl3: P1 at 215 nm (○, no La3+; ●, with La3+); P2 at 217 nm (□, no La3+; ■, with La3+); P3 at 219 nm (▿, no La3+; ▾, with La3+). The opened and closed diamonds (⋄, ♦) in A show the temperature dependence of the ellipticity of P6N5 peptide (Ac-AAKAAY-NH2) monitored at 219 and 217 nm, respectively.
Figure 5.
Temperature dependence of the fraction helix of peptides P2 (□) and P3 (▿) in the absence of La3+-ions. The lines through the points are shown only to guide the eye.
Enthalpy of Helix Formation.
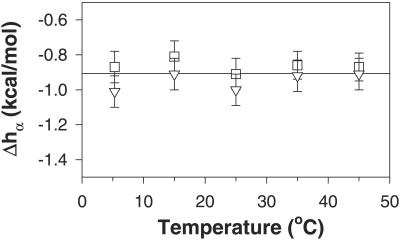
The enthalpy of helix formation per residue, Δhα, can now be calculated by combining Eqs. 9 and 10 and 7 and 8. The results of these calculations at different temperatures are presented in Fig. 6. There are three important results. First, the values of Δhα calculated from peptide P2 are not very different from the values calculated for peptide P3. This fact indicates that the assumptions made in calculating Δhα values are probably correct. Second, the value of Δhα is −0.9 ± 0.1 kcal/mol of amino acid residues, which is close to previous estimates of the enthalpy of helix formation (see Discussion). Third, the Δhα value does not change significantly with temperature, according to these data. This result means that ΔCp is too small to be detected by enthalpy measurements as a function of temperature in this model system.
Figure 6.
Temperature dependence of the enthalpy of helix formation Δhα calculated per mole residue found for peptide P2 (□) and P3 (▿). The solid horizontal line shows the average of all data points.
Discussion
The most recent calorimetric measurements of the enthalpy of helix formation are those of Richardson et al. (15) for a 31-residue peptide with the sequence Ac-Y(MEARA)6-NH2. They give ΔH equal to −0.84 ± 0.1 kcal per mole of amino acid residues. CD measurements of the thermal helix–coil transition curves for a set of six alanine-based peptides, varying in chain length from 14–50 residues, gave an estimate of −1.0 kcal/mol (2) when analyzed by either the Zimm–Bragg or Lifson–Roig theories (see also ref. 9). The thermal unfolding curve of the 50-residue peptide was also measured calorimetrically and the measured enthalpy agreed within error with the value found from the CD-monitored unfolding curves (16). Although the calorimetric enthalpy measurement is direct, it is less precise than estimates of ΔH obtained by using helix–coil theory to analyze thermal unfolding curves, because the unfolding curve even of a 50-residue peptide is very broad and fitting the baseline is problematic. Early estimates of the enthalpy of helix formation using homopolymers of poly-L-lysine and poly-L-glutamate gave values between −1.1 and −0.88 kcal/mol residues (17, 18). All these values are similar to each other and somewhat higher than a recent estimate of −0.65 kcal/mol per residue by Taylor et al. (19). These authors studied a 29-residue helical peptide, which contains i to i + 4 chemical crosslinks (lactam bridges) at both the N and C termini to stabilize the helix covalently. Their results appear to fit the two-state model and they interpreted them by using this model, and by assuming that the cross-linked end residues induce more cooperative unfolding behavior.
Most of these results are consistent with the assumption that the enthalpy of helix formation depends only on the peptide backbone and is the same for different amino acids, except perhaps for glycine and proline. However, the relative helix propensities of the nonpolar amino acids change with temperature (20), and the effect appears to be enthalpic, because it has the opposite sign to that expected for the ΔCp arising from burial of nonpolar surface area. The changes of the relative helix propensities with temperature have the sign expected if side chains reduce the access of water to the peptide backbone and interfere with backbone solvation (20, 21). According to this proposal, other nonpolar amino acids should have smaller (negative) enthalpies of helix formation than alanine and the free-energy differences between the helix propensities provide an upper limit to the enthalpy differences between alanine and other nonpolar amino acids.
Because of the broad thermal unfolding curves shown by peptide helices, it has been particularly difficult to obtain the temperature dependence of the enthalpy change and the partial heat capacity of the fully helical peptide. It is also difficult to obtain accurately the temperature dependence of the partial heat capacity of the fully unfolded peptide. The latter obstacle has been overcome recently, however, by calculating the heat capacity of the unfolded state from an additivity approach (22). Direct measurements of the heat capacities of fully unfolded peptides support the validity of the additivity approach (J. M. Richardson and G.I.M., unpublished work). The importance of knowing the value of ΔCp is illustrated by the work of Richardson et al. (15) who, by using the calculated heat capacity for the unfolded state, were able to measure the enthalpy of helix formation of the 31-residue peptide sequence Y(MEARA)6-NH2. Their value is based on the assumption that the heat capacity change on helix formation, ΔCp, is zero. Not only the value but even the sign of ΔCp for helix formation has been controversial. A structure-based thermodynamic calculation predicts that ΔCp for alanine-based helix folding should be small and positive (23). The opposite conclusion was reached from a calorimetric study of a short peptide with covalently crosslinked terminal residues (19), and also from a theoretical calculation (A. Garcia, personal communication). Our data demonstrate that the ΔCp of helix formation cannot be large. Using the maximal and minimal values of Δhα shown in Fig. 6 at the highest and lowest experimental temperatures (5 and 45°C) places limits on the allowed value of ΔCp, which then must be between −8 and 8 cal/K per mol residue. Fitting their DSC data, Taylor et al. (19) gave a value for ΔCp of −3.8 ± 0.7 cal/K per mole residue. A computational estimate gave a negative value of −6 ± 3 cal/K per mol residue (A. Garcia, personal communication). An early structural thermodynamics prediction by Ooi and Oobatake (24) gave a smaller value of −1.9 cal/K per mol residue.
Concluding Remarks.
The feasibility of obtaining thermodynamic information about helix formation by using metal binding to induce helix formation in peptide sequences based on calmodulin was demonstrated using isothermal titration calorimetry. The enthalpy of alanine helix formation is found to be −0.9 ± 0.1 kcal per mol residue and does not depend on temperature in the range 5–45°C. This model system has a great potential for measuring more detailed thermodynamic properties, such as the enthalpy of helix formation for various amino acid residues, the enthalpies of interactions between amino acid side chains, and their temperature dependencies.
Acknowledgments
We would like to express our deep appreciation to Dr. Andrzej Bierzynski for advice and discussion, and to Dr. Marty Scholtz both for his discussion and for providing P6N5 peptide, and Angel Garcia for sharing unpublished data. This work was supported in part by National Institutes of Health Grant GM54537 (to G.I.M.), an Individual Grant from National Science Council, The Executive Yuan, Taiwan, Republic of China Grant NSC 89-2113-M-018-011 (to D.-H.C.), and Laboratory Grant NHRI-EX90-8807BL from National Health Research Institutes, The Executive Yuan, Taiwan, Republic of China (to D.-H.C.).
Abbreviation
- ITC
isothermal titration calorimetry
References
- 1.Pauling L, Corey R B, Branson H R. Proc Natl Acad Sci USA. 1951;37:205–210. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scholtz J M, Qian H, York E J, Stewart J M, Baldwin R L. Biopolymers. 1991;31:1463–1470. doi: 10.1002/bip.360311304. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabartty A, Baldwin R L. Adv Protein Chem. 1995;46:141–176. [PubMed] [Google Scholar]
- 4.Rohl C A, Baldwin R L. Methods Enzymol. 1998;295:1–26. doi: 10.1016/s0076-6879(98)95032-7. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin R L, Rose G D. Trends Biochem Sci. 1999;24:26–33. doi: 10.1016/s0968-0004(98)01346-2. [DOI] [PubMed] [Google Scholar]
- 6.Wojcik J, Goral J, Pawlowski K, Bierzynski A. Biochemistry. 1997;36:680–687. doi: 10.1021/bi961821c. [DOI] [PubMed] [Google Scholar]
- 7.Siedlecka M, Goch G, Ejchart A, Sticht H, Bierzyski A. Proc Natl Acad Sci USA. 1999;96:903–908. doi: 10.1073/pnas.96.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsden B J, Hodges R S, Sykes B D. Biochemistry. 1989;28:8839–8847. doi: 10.1021/bi00448a024. [DOI] [PubMed] [Google Scholar]
- 9.Luo P, Baldwin R L. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- 10.Woody R W. Methods Enzymol. 1995;246:34–71. doi: 10.1016/0076-6879(95)46006-3. [DOI] [PubMed] [Google Scholar]
- 11.Brokx R D, Lopez M M, Vogel H J, Makhatadze G I. J Biol Chem. 2001;276:14083–14091. doi: 10.1074/jbc.M011026200. [DOI] [PubMed] [Google Scholar]
- 12.Lopez M M, Yutani K, Makhatadze G I. J Biol Chem. 1999;274:33601–33608. doi: 10.1074/jbc.274.47.33601. [DOI] [PubMed] [Google Scholar]
- 13.Fukada H, Takahashi K. Proteins. 1998;33:159–166. [PubMed] [Google Scholar]
- 14.Baker B M, Murphy K P. Biophys J. 1996;71:2049–2055. doi: 10.1016/S0006-3495(96)79403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson J M, McMahon K W, MacDonald C C, Makhatadze G I. Biochemistry. 1999;38:12869–12875. doi: 10.1021/bi990724r. [DOI] [PubMed] [Google Scholar]
- 16.Scholtz J M, Marqusee S, Baldwin R L, York E J, Stewart J M, Santoro M, Bolen D W. Proc Natl Acad Sci USA. 1991;88:2854–2858. doi: 10.1073/pnas.88.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rialdi G, Hermans J., Jr J Am Chem Soc. 1966;88:5719–5720. doi: 10.1021/ja00976a007. [DOI] [PubMed] [Google Scholar]
- 18.Chou P Y, Scheraga H A. Biopolymers. 1971;10:657–680. doi: 10.1002/bip.360100406. [DOI] [PubMed] [Google Scholar]
- 19.Taylor J W, Greenfield N J, Wu B, Privalov P L. J Mol Biol. 1999;291:965–976. doi: 10.1006/jmbi.1999.3025. [DOI] [PubMed] [Google Scholar]
- 20.Luo P, Baldwin R L. Proc Natl Acad Sci USA. 1999;96:4930–4935. doi: 10.1073/pnas.96.9.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avbelj F, Luo P, Baldwin R L. Proc Natl Acad Sci USA. 2000;97:10786–10791. doi: 10.1073/pnas.200343197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makhatadze G I, Privalov P L. J Mol Biol. 1990;213:375–384. doi: 10.1016/S0022-2836(05)80197-4. [DOI] [PubMed] [Google Scholar]
- 23.Lee K H, Xie D, Freire E, Amzel L M. Proteins. 1994;20:68–84. doi: 10.1002/prot.340200108. [DOI] [PubMed] [Google Scholar]
- 24.Ooi T, Oobatake M. Proc Natl Acad Sci USA. 1991;88:2859–2863. doi: 10.1073/pnas.88.7.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]