Abstract
Here we examine the function of ATM and ATR at telomeres in Arabidopsis. Although plants lacking ATM or ATR display wild-type telomere length homeostasis, chromosome end protection is compromised in atm atr mutants. Moreover, atm tert Arabidopsis experience an abrupt, early onset of genome instability, arguing that ATM is required for protection of short telomeres. ATR, by contrast, is required for maintenance of telomeric DNA as telomere shortening is dramatically accelerated in atr tert mutants relative to tert plants. Thus, ATM and ATR make essential and distinct contributions to chromosome end protection and telomere maintenance in higher eukaryotes.
Keywords: Telomere, DNA damage, telomerase, chromosome fusion, plant
Several factors involved in the repair of double-strand breaks in DNA are physically associated with telomeres and have essential roles in chromosome stability (d'Adda di Fagagna et al. 2004). Among them are the phosphati-dyl-inositol-3-kinase-like protein kinases (PIKKs) ATM/Tel1p and ATR/Mec1p/Rad3p, which coordinate the repair, cell-cycle checkpoint, and apoptotic responses to DNA damage (Kastan and Lim 2000; Shiloh 2003). ATM also protects against telomere fusions and participates in telomere length homeostasis as atm/tel1 mutants display shorter telomeres in both mammals and yeast (Lustig and Petes 1986; Smilenov et al. 1997). A telomeric role for ATR has not been explored in mammals because mutants are inviable (Brown and Baltimore 2000; de Klein et al. 2000). However, in yeast mec1-21 mutants, telomeres are shortened (Ritchie et al. 1999). In addition, recent data show that Mec1p facilitates loading of the single-strand telomere-binding protein Cdc13p (Takata et al. 2005). Yeast lacking both Tel1p and Mec1p fail to recruit telomerase and experience progressive telomere erosion (Naito et al. 1998; Ritchie et al. 1999; Chan et al. 2001).
Here we explore the role of ATM and ATR at telomeres in Arabidopsis thaliana, a higher eukaryote that tolerates disruption of both of these genes. We find that while neither ATM nor ATR is individually required for telomere length homeostasis, chromosome end protection is impaired in atm atr mutants. Our data further indicate that ATM is involved in protecting chromosome ends in cells with short telomeres, while ATR is required for maintenance of telomeric DNA.
Results and Discussion
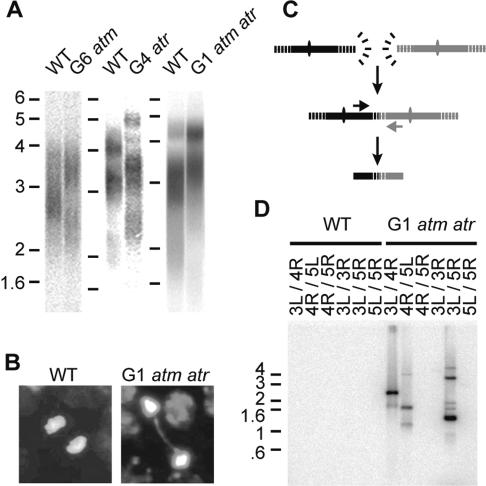
To examine the role of ATM and ATR in telomere length regulation, we performed telomere restriction fragment (TRF) analysis on genomic DNA extracted from atm and atr mutants that carry a homozygous T-DNA disruption in the corresponding genes (see Materials and Methods). In yeast and mammals, the absence of ATM/Tel1p leads to shortening of telomeres until they reach a new stable set point (Lustig and Petes 1986; Smilenov et al. 1997). In contrast, atm or atr Arabidopsis displayed wild-type telomere length, even after multiple generations (Fig. 1A). The same result was obtained with mutants doubly deficient in ATM and ATR (Fig. 1A). As previously noted (Culligan et al. 2004), atm atr mutants exhibited no growth defects but were completely sterile. Hence, neither ATM nor ATR is required for telomere length homeostasis in Arabidopsis.
Figure 1.
Telomere length homeostasis and chromosome end protection in plants lacking ATM and ATR. (A) TRF analysis of genomic DNA from wild type (WT) and from atm, atr, and atm atr mutants. A representative sample for each line is shown. Molecular weight markers (in kilobases) are indicated. (B) Cytogenetic analysis of anaphase figures from wild type (WT) and atm atr mutants. DNA is stained with DAPI. (C) Schematic of fusion PCR strategy. Primers specific for subtelomeres directed toward chromosome ends are used to amplify chromosome fusion junctions. PCR products are detected by Southern blot using either a telomeric or a subtelomeric probe. (D) Detection of chromosome end fusions by telomere fusion PCR. The subtelomeric primers used for PCR are indicated (L and R designate left and right arm of the chromosome, respectively). For 3L, the primer used was 3L-F1, except for the 3L/5R reaction where 3L-1 was used. Telomeric DNA was visualized by hybridization using a (TTTAGGG)4 probe. Reactions were conducted on pooled DNA samples from three plants.
We gauged the status of chromosome end protection in atm and atr mutants by examining mitotic figures from dissected pistils. Bridged anaphase chromosomes are a hallmark of dysfunctional telomeres, reflecting the formation of dicentric chromosomes that arise from fusion of deprotected chromosome ends (Hande et al. 1999; Artandi et al. 2000). Among >400 anaphases examined from consecutive generations of atm and atr single mutants, no bridged chromosomes were detected (Table 1). In contrast, 42/826 (5.1%) of the anaphases from atm atr double mutants showed bridged chromosomes (Table 1; Fig. 1B). To determine whether the bridges involved telomeres, we assayed for fusion of chromosome ends using a PCR approach that exploits the unique sequences abutting Arabidopsis telomeres (Heacock et al. 2004). PCR products are amplified when telomeres form covalent associations with each other (Fig. 1C). As expected, no products were detected with DNA samples from wild type or atm or atr mutants (Table 1; Fig. 1D). However, some rare, but persistent, PCR products were obtained in DNA samples from atm atr double mutants (Table 1; Fig. 1D). The few PCR products we could clone and sequence displayed telomere-to-telomere fusions involving very short telomere tracts of ∼500 bp (data not shown). This architecture is reminiscent of chromosome fusion junctions amplified from late-generation telomerase mutants, where unprotected telomeres are subjected to exonucleolytic attack prior to fusion (Heacock et al. 2004). We conclude that while bulk telomere length is wild type, a fraction of chromosome ends in atm atr mutants become deprotected and recruited into end-joining reactions.
Table 1.
Genome instability measured by the incidence of anaphase bridges and telomere fusion PCR products in different genetic backgrounds
| Generation
|
Anaphases
|
Fusion PCR products
|
|||
|---|---|---|---|---|---|
| Line | Total | Bridges | Ratio% | ||
| WT | G1-G4 | >400 | 0 | 0 | − |
| atm | G1-G6 | >400 | 0 | 0 | − |
| atr | G1-G4 | >400 | 0 | 0 | − |
| atm atr | G1 | 826 | 42 | 5.1 | +/− |
| tert | G4 | >400 | 0 | 0 | − |
| G5 | 385 | 1 | 0.3 | +/− | |
| atm tert | G3 | 533 | 0 | 0 | − |
| G4 | 430 | 0 | 0 | +/− | |
| G5a | 446 (314) | 97 (0) | 21.8 (0) | +/− | |
| art tert | G2 | 403 | 0 | 0 | ++ |
| G3a | 181 (518) | 137 (75) | 43.1 (12.6) | ++ | |
(+/−) Rare but persistent PCR products; (++) abundant heterogeneous products.
Growth defects were relatively heterogeneous among siblings, as previously observed for tert plants (Riha et al. 2001), and hence cytogenetic analysis was performed in sic k plants and their wild-type-looking siblings from the same parent (in parentheses). Values are for pools of three plants. For telomere fusion PCR data, six primer pair combinations were used.
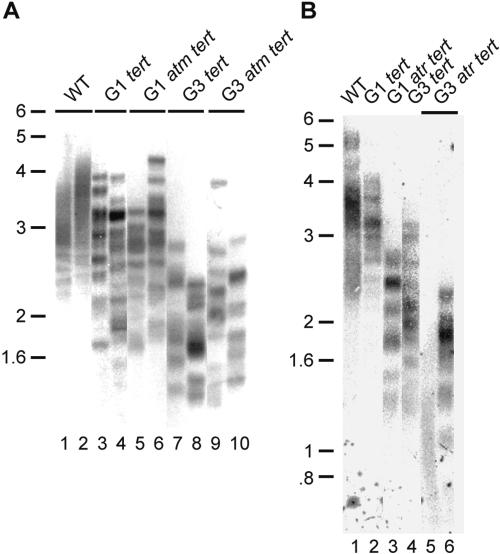
ATM plays a dual role at telomeres, activating the DNA damage response program at dysfunctional telomeres and yet preventing this activation at functional telomeres (Karlseder et al. 1999; Takai et al. 2003). To examine the role of ATM at short telomeres in Arabidopsis, mutants deficient in both ATM and TERT, the catalytic subunit of telomerase, were monitored during five generations of growth. The rate of telomere shortening was indistinguishable between atm tert and tert mutants, with telomeres declining by ∼500 bp per plant generation in both settings (Fig. 2A; data not shown).
Figure 2.
Telomere maintenance in tert, atm tert, and atr tert mutants. TRF analysis of atm tert (A) and atr tert (B) at different generations. Double and single mutants in G1 were derived from the same double-heterozygous parent and were self-pollinated to follow progeny through successive generations.
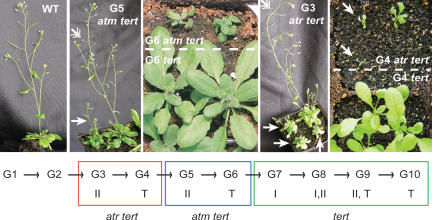
In Arabidopsis, telomerase inactivation ultimately leads to cell proliferation defects that manifest as growth and development impairment in the sixth (G6) or seventh (G7) generation (type I defects) and then worsen in subsequent generations (type II defects) until mutants reach, in G9-G10, a terminal phenotype as miniature, grossly deformed, and sterile plants (Riha et al. 2001). As expected, tert mutants displayed no outward phenotype for the first six generations of growth (data not shown). In contrast, most atm tert mutants exhibited a type II or terminal phenotype in G5 and were characterized by misshapen leaves, a lack of apical dominance, late flowering, and significantly reduced fertility (Fig. 3).
Figure 3.
Early onset of growth and developmental defects in atm tert and atr tert mutants. Representative G5 atm tert and G3 atr tert mutants are shown. Arrows indicate plants with gross developmental defects. Double arrowheads denote abnormal or undeveloped siliques (seed pods). The diagram at the bottom summarizes the phenotypes of tert, atm tert, and atr tert mutants. See text for description of type I, II, and terminal (T) phenotypes. The mutants shown are representative of each line and each generation.
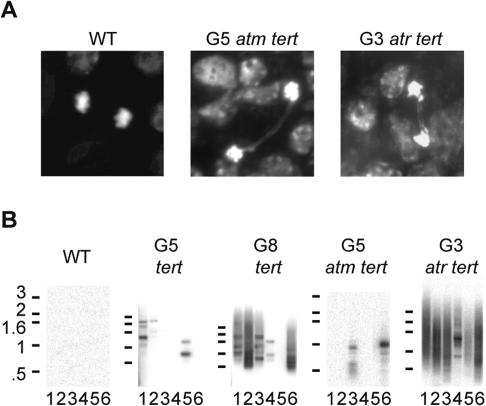
To investigate the molecular basis for the accelerated onset of cell proliferation defects, we performed cytogenetic analysis. As expected, there was no evidence for genome instability in the first five generations of tert, or in the first four generations of atm tert mutants (Table 1; data not shown). However, ∼20% of the anaphase nuclei from the most severely affected G5 atm tert plants harbored bridged chromosomes compared with <1% in G5 tert (Table 1; Fig. 4A). This high frequency of anaphase bridges in G5 atm tert is comparable to G7-G8 tert Arabidopsis (Riha et al. 2001). Thus, unlike the gradual onset of genome instability that occurs over multiple generations (G4-G8) in tert (Riha et al. 2001), atm tert mutants abruptly destabilized their genomes in a single generation.
Figure 4.
Chromosome instability and telomere fusions in atm tert and atr tert. (A) Cytogenetic abnormalities in anaphases from G5 atm tert and G3 atr tert mutants. Multiple anaphase bridges were observed in G3 atr tert. An example of a double bridge is shown for this mutant. Slides were stained with DAPI. (B) Detection of telomere fusions by PCR with the following primer combinations: 3L/4R (lane 1); 4R/5L (lane 2); 4R/5R (lane 3); 3L/3R (lane 4); 3L/5R (lane 5); 5L/5R (lane 6). Conditions were the same as for Figure 1.
While telomere fusion PCR products were generated from DNA samples derived from G4 and G5 atm tert mutants, their abundance was remarkably low and similar to the signals obtained with G5 tert mutants, which display no genome instability (Table 1; Fig. 4B; data not shown). In contrast, DNA from G8 tert plants with a comparable number of anaphase bridges to G5 atm tert mutants gave rise to a large number of telomere fusion PCR products (Fig. 4B). The low abundance of fusion PCR products from atm tert mutants could reflect fusion events distinct from those observed in tert. For example, chromosome fusions involving long telomeres or sister chromatids would form extensive palindromes that would not be amplified in our PCR assay. Likewise, we would miss telomeres subjected to catastrophic shortening prior to fusion if the binding site for the subtelomeric primer were eliminated. Our initial attempts to detect fusion products using primers located further upstream in the subtelomeric regions were unsuccessful. Therefore, extensive nucleolytic processing of the ends remains a possibility. Finally, we would also fail to detect fusions involving internal double-strand breaks. Such fusions could result from a checkpoint defect that would allow additional rounds of breakage-fusion-breakage cycles, and as a consequence, increase the probability of secondary fusion events involving internal double-strand breaks.
Mice lacking the RNA component of telomerase and ATM display a phenotype similar to atm tert Arabidopsis, with an early onset of telomere dysfunction without accelerated telomere shortening (Qi et al. 2003). Similarly, the telomeres of tel1 tlc1 yeast mutants shorten at the same rate as tlc1 mutants, but double mutants show dramatic increases in the frequency of telomere fusions to artificial double-strand breaks induced by HO endonuclease (Chan and Blackburn 2003) and in the frequency of chromosomal rearrangements (Myung et al. 2001). Thus, a role for ATM in stabilizing the genomes of cells with shortened telomeres is conserved.
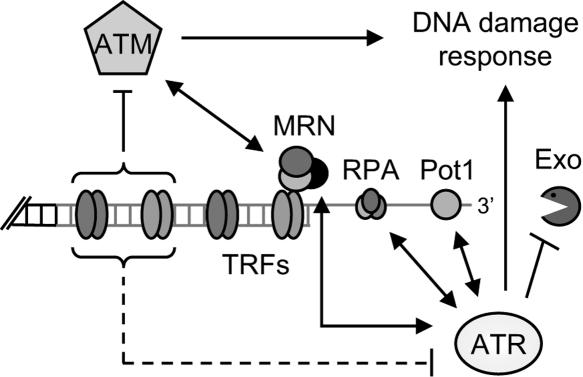
The telomere defects associated with ATM deficiency may simply reflect an impaired DNA damage checkpoint; however, evidence that ATM plays a more direct role at telomeres is growing (Fig. 5). Tel1p localizes to telomeres in yeast (Takata et al. 2004), and since ATM is recruited to double-strand breaks by Mre11, a member of the Mre11/Rad50/Nbs1 complex, this could also occur at telomeres (Lee and Paull 2005). In mammals, ATM associates with the double-strand telomere proteins TRF1 and TRF2 (Kishi et al. 2001; Karlseder et al. 2004), and it is hypothesized that ATM association with telomeric chromatin blocks induction of a DNA damage response (d'Adda di Fagagna et al. 2004). In the absence of telomerase, progressive shortening of telomeres would ultimately reduce the occupancy of TRF1/TRF2 proteins, promoting activation of ATM and the ensuing cell cycle arrest. Thus, it is possible that the response to short telomeres is inadequate in plants lacking ATM, culminating in an early onset of genome instability.
Figure 5.
Model illustrating possible interactions between ATM and ATR and telomere components in Arabidopsis. Interactions denoted by solid arrows have been described in yeast or mammals. Dashed lines show hypothetical interactions. We and others (d'Adda di Fagagna et al. 2004; this study) speculate that the interaction of ATM and ATR with telomere proteins inhibits activation of the DNA damage response. In plants lacking ATM and telomerase, progressive loss of telomeric DNA leads to an inadequate DNA damage response and an abrupt accumulation of cells with profound genome instability. ATR plays a different role at telomeres. The lack of ATR in plants deficient in telomerase results in an increased rate of telomere shortening. Accelerated telomere erosion may reflect greater vulnerability to nucleolytic attack due to diminished binding of ATR-associated telomere proteins (see text for details).
Due to the lethality of ATR deletion in mammals, the contribution of this gene to telomere biology has not been explored in higher eukaryotes. Therefore, we asked whether ATM and ATR play a similar role at short telomeres in Arabidopsis. Unexpectedly, we discovered that ATR contributes to telomere integrity in a fundamentally different manner than ATM. Telomeres in atr tert Arabidopsis suffer a dramatically accelerated rate of loss relative to tert and atm tert mutants (Fig. 2A,B). Telomere tracts in G1 atr tert were approximately the same length as those in G3 tert mutants (Fig. 2B, lanes 3,4); in the most severely affected G3 atr tert individuals, the shortest telomeres were <600 bp (Fig. 2B, lane 5). Consistent with the increased rate of telomere shortening, atr tert mutants reached the terminal phenotype much earlier than their tert or atm tert counterparts. Although no outward defects were apparent in G1 and G2, by G3 atr tert plants were almost completely sterile and extremely small with misshapen dark-green leaves (Fig. 3). Also consistent with rapid telomere attrition, atr tert mutants displayed profound genome instability in G3. Up to 43% of the anaphases harbored bridged chromosomes, with several involving multiple chromosomes (Table 1; Fig. 4A). In addition, abundant extremely heterogeneous telomere fusion PCR products were generated in DNA samples from G2 and G3 atr tert mutants (Table 1; Fig. 4B; data not shown). Cloning of 10 of these products revealed a profile of chromosome junctions similar to late generation tert mutants. The proportion of telomere-to-telomere fusions (80%) and the extent of microhomology at the fusion junction are more similar to fusion events in tert mutants than in ku tert or ku tert mre11 (Supplementary Tables 1, 2; Supplementary Fig. 1; Heacock et al. 2004). Taken together, our data argue that the genome instability associated with atr tert mutants reflects an increased deficiency in telomeric DNA maintenance, instead of chromosome end protection. Thus, ATR plays a different role at Arabidopsis telomeres than ATM and acts in concert with telomerase to maintain telomeric DNA tracts.
ATR has been shown to function in resolving stalled replication forks (Cha and Kleckner 2002). Consequently, it is conceivable that the replication of highly repetitive telomeric DNA is most efficient in the presence of ATR. However, in atr Arabidopsis, telomere length is unperturbed, implying that replication of the duplex region of the telomere tract is not compromised by ATR deficiency. An alternative, more speculative model is that ATR is physically associated with the chromosome terminus and acts in telomere maintenance by prohibiting illegitimate exonuclease attack or by controlling processing of the telomeric C-strand (Fig. 5). As for ATM, telomere association of ATR could proceed through interactions with Mre11 (Takata et al. 2005). Alternatively, since RPA recruits ATR to stalled replication forks (Zou and Elledge 2003), RPA may facilitate ATR localization to telomeres. In support of this idea, RPA mutation leads to progressive telomere shortening (Smith et al. 2000; Schramke et al. 2004). In yeast, Mec1p is known to promote loading of the single-strand telomere-binding protein Cdc13p (Takata et al. 2005), whose functions include protection of the G-rich telomeric DNA strand. By analogy, at higher eukaryotic telomeres, ATR may be involved in the recruitment of Pot1, the presumed ortholog of Cdc13p (Smogorzewska and de Lange 2004). We speculate that the telomere maintenance defect we observe in plants lacking both TERT and ATR reflects a combined failure of telomerase to maintain the telomeric G-rich strand and ATR to protect against inappropriate nuclease digestion.
In summary, these data provide the first insight into the role of ATR at higher eukaryotic telomeres. Further analysis of ATR and ATM interactions in the genetically tractable Arabidopsis model should help to clarify the contribution of these proteins to chromosome end protection and telomere maintenance.
Materials and methods
Mutant lines
The SALK T-DNA lines (Alonso et al. 2003) SALK_006953 and SALK_032841, that respectively carry an insertion in intron 64 of ATM (At3g48190) and in exon 10 of ATR (At5g40820), have been described previously as atm-2 (Garcia et al. 2003) and atr-2 (Culligan et al. 2004). The tert mutant and its phenotypes have also been described previously (Riha et al. 2001). All crosses were made between plants heterozygous for the desired mutations. Double-heterozygous F1 plants were identified by genotyping and then self-propagated to F2 to obtain double- and single-homozygous mutants and wild-type siblings. These plants (G1) were self-propagated for several generations. Two independent lines, coming from two individual F2 plants, were obtained for each genotype. For genotyping, genomic DNA was extracted from flower buds using a high-throughput method (Xin et al. 2003). Plants were grown at 23°C in an environmental chamber under a 16 h light/8 h dark photoperiod.
TRF analysis
Four to seven fully grown rosette leaves from 6-wk-old individual plants were ground in liquid nitrogen, resuspended in 3 mL of DNA extraction buffer (2% hexadecyltrimethyl-ammonium bromide, 1.4 mM NaCl, 20 mM EDTA, 100 mM Tris-Cl at pH 8.0, 10 mg/L RNAse A), and incubated for 1 h at 65°C. After phenol/chloroform extraction, genomic DNA was precipitated in isopropanol and resuspended in TE buffer. DNA (3 μg) was digested overnight at 65°C with Tru1I (Fermentas). Products were resolved on a 1% agarose gel. Telomeric signals were detected by Southern-blot using a 5′-32P-labeled (TTTAGGG)4 single-strand probe.
Cytogenetics experiments and telomere fusion PCR
DAPI (4′,6′-diamidino-2-phenylindole)-stained chromosome spreads were prepared from pistils as described (Martínez-Zapater and Salinas 1998). Telomere fusion PCR experiments including PCR conditions, cloning, sequencing, and program analysis were performed as described (Heacock et al. 2004). Primers were 3L-F1 (AGACGAGGAGACTAG GAACG), 3L-1 (CATAATTCTCACAGCAGCACCGTAGA), 3R-R1 (GT ATGGATGCCGGGAAAGTTGCAGACAA), 4R-F1 (CTTATCAACAT CAGCTGGTC), 5L-1 (AGGTAGAGTGAACCTAACACTTGGA), and 5R-1 (CAGGACGTGTGAAACAGAAACTACA).
Acknowledgments
We are grateful to Dustin Cox and Brice Wilson for technical assistance, and to Carolyn Price, Tom McKnight, and Jeff Kapler for critically reading the manuscript. We also thank members of the Shippen laboratory for many helpful discussions. This work was supported by National Institutes of Health Grant GM65383 to D.E.S.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1333805.
References
- Alonso J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653-657. [DOI] [PubMed] [Google Scholar]
- Artandi S.E., Chang, S., Lee, S.L., Alson, S., Gottlieb, G.J., Chin, L., and DePinho, R.A. 2000. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406: 641-645. [DOI] [PubMed] [Google Scholar]
- Brown E.J. and Baltimore, D. 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & Dev. 14: 397-402. [PMC free article] [PubMed] [Google Scholar]
- Cha R.S. and Kleckner, N. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602-606. [DOI] [PubMed] [Google Scholar]
- Chan S.W. and Blackburn, E.H. 2003. Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol. Cell 11: 1379-1387. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Chang, J., Prescott, J., and Blackburn, E.H. 2001. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11: 1240-1250. [DOI] [PubMed] [Google Scholar]
- Culligan K., Tissier, A., and Britt, A. 2004. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16: 1091-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F., Teo, S.H., and Jackson, S.P. 2004. Functional links between telomeres and proteins of the DNA-damage response. Genes & Dev. 18: 1781-1799. [DOI] [PubMed] [Google Scholar]
- de Klein A., Muijtjens, M., van Os, R., Verhoeven, Y., Smit, B., Carr, A.M., Lehmann, A.R., and Hoeijmakers, J.H. 2000. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr. Biol. 10: 479-482. [DOI] [PubMed] [Google Scholar]
- Garcia V., Bruchet, H., Camescasse, D., Granier, F., Bouchez, D., and Tissier, A. 2003. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hande M.P., Samper, E., Lansdorp, P., and Blasco, M.A. 1999. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J. Cell Biol. 144: 589-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock M., Spangler, E., Riha, K., Puizina, J., and Shippen, D.E. 2004. Molecular analysis of telomere fusions in Arabidopsis: Multiple pathways for chromosome end-joining. EMBO J. 23: 2304-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J., Broccoli, D., Dai, Y., Hardy, S., and de Lange, T. 1999. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283: 1321-1325. [DOI] [PubMed] [Google Scholar]
- Karlseder J., Hoke, K., Mirzoeva, O.K., Bakkenist, C., Kastan, M.B., Petrini, J.H., and de Lange, T. 2004. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2: E240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M.B. and Lim, D.S. 2000. The many substrates and functions of ATM. Nat. Rev. Mol. Cell. Biol. 1: 179-186. [DOI] [PubMed] [Google Scholar]
- Kishi S., Zhou, X.Z., Ziv, Y., Khoo, C., Hill, D.E., Shiloh, Y., and Lu, K.P. 2001. Telomeric protein Pin2/TRF1 as in important ATM target in response to double strand DNA breaks. J. Biol. Chem. 276: 29282-29291. [DOI] [PubMed] [Google Scholar]
- Lee J.H. and Paull, T.T. 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308: 551-554. [DOI] [PubMed] [Google Scholar]
- Lustig A.J. and Petes, T.D. 1986. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. 83: 1398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Zapater J.M. and Salinas, J. 1998. Arabidopsis protocols. Humana Press, Totowa, NJ.
- Myung K., Chen, C., and Kolodner, R.D. 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073-1076. [DOI] [PubMed] [Google Scholar]
- Naito T., Matsuura, A., and Ishikawa, F. 1998. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat. Genet. 20: 203-206. [DOI] [PubMed] [Google Scholar]
- Qi L., Strong, M.A., Karim, B.O., Armanios, M., Huso, D.L., and Greider, C.W. 2003. Short telomeres and ataxia-telangiectasia mutated deficiency cooperatively increase telomere dysfunction and suppress tumorigenesis. Cancer Res. 63: 8188-8196. [PubMed] [Google Scholar]
- Riha K., McKnight, T.D., Griffing, L.R., and Shippen, D.E. 2001. Living with genome instability: Plant responses to telomere dysfunction. Science 291: 1797-1800. [DOI] [PubMed] [Google Scholar]
- Ritchie K.B., Mallory, J.C., and Petes, T.D. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 6065-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke V., Luciano, P., Brevet, V., Guillot, S., Corda, Y., Longhese, M.P., Gilson, E., and Geli, V. 2004. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat. Genet. 36: 46-54. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. 2003. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer 3: 155-168. [DOI] [PubMed] [Google Scholar]
- Smilenov L.B., Morgan, S.E., Mellado, W., Sawant, S.G., Kastan, M.B., and Pandita, T.K. 1997. Influence of ATM function on telomere metabolism. Oncogene 15: 2659-2665. [DOI] [PubMed] [Google Scholar]
- Smith J., Zou, H., and Rothstein, R. 2000. Characterization of genetic interactions with RFA1: The role of RPA in DNA replication and telomere maintenance. Biochimie 82: 71-78. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A. and de Lange, T. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73: 177-208. [DOI] [PubMed] [Google Scholar]
- Takai H., Smogorzewska, A., and de Lange, T. 2003. DNA damage foci at dysfunctional telomeres. Curr. Biol. 13: 1549-1556. [DOI] [PubMed] [Google Scholar]
- Takata H., Kanoh, Y., Gunge, N., Shirahige, K., and Matsuura, A. 2004. Reciprocal association of the budding yeast ATM-related proteins Tel1 and Mec1 with telomeres in vivo. Mol. Cell 14: 515-522. [DOI] [PubMed] [Google Scholar]
- Takata H., Tanaka, Y., and Matsuura, A. 2005. Late S phase-specific recruitment of Mre11 complex triggers hierarchical assembly of telomere replication proteins in Saccharomyces cerevisiae. Mol. Cell 17: 573-583. [DOI] [PubMed] [Google Scholar]
- Xin Z., Velten, J.P., Oliver, M.J., and Burke, J.J. 2003. High-throughput DNA extraction method suitable for PCR. Biotechniques 34: 820-824, 826. [DOI] [PubMed] [Google Scholar]
- Zou L. and Elledge, S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542-1548. [DOI] [PubMed] [Google Scholar]