Abstract
The recently discovered p53-related genes, p73 and p63, express multiple splice variants and N-terminally truncated forms initiated from an alternative promoter in intron 3. To date, no alternative promoter and multiple splice variants have been described for the p53 gene. In this study, we show that p53 has a gene structure similar to the p73 and p63 genes. The human p53 gene contains an alternative promoter and transcribes multiple splice variants. We show that p53 variants are expressed in normal human tissue in a tissue-dependent manner. We determine that the alternative promoter is conserved through evolution from Drosophila to man, suggesting that the p53 family gene structure plays an essential role in the multiple activities of the p53 family members. Consistent with this hypothesis, p53 variants are differentially expressed in human breast tumors compared with normal breast tissue. We establish that p53β can bind differentially to promoters and can enhance p53 target gene expression in a promoter-dependent manner, while Δ133p53 is dominant-negative toward full-length p53, inhibiting p53-mediated apoptosis. The differential expression of the p53 isoforms in human tumors may explain the difficulties in linking p53 status to the biological properties and drug sensitivity of human cancer.
Keywords: Splice, promoter, Drosophila, cancer, p73, p63
p53 was discovered 25 years ago as a protein interacting with the oncogenic T antigen from SV40 virus (De Leo et al. 1979; Kress et al. 1979; Lane and Crawford 1979; Linzer and Levine 1979; Melero et al. 1979). Over the last decade, it was appreciated that the p53 protein is the product of a pivotal tumor-suppressor gene, whose inactivation is the most frequent single gene event in human cancer, and that germline mutations in the human p53 gene are a cause of enhanced risk of developing cancer. Since then, p53 has become the focus of intensive basic and clinical research (for review, see Oren 2003).
Recently two p53-related genes (p63 and p73) were discovered. The p73 and p63 proteins, which they encode, share sequence homology with p53, enabling them to bind to p53 DNA-binding sites, to transactivate p53-responsive genes, and to induce cell cycle arrest or apoptosis.
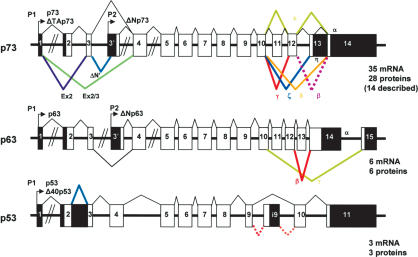
The p63 gene expresses at least three alternatively spliced C-terminal isoforms. The p73 gene expresses at least seven alternatively spliced C-terminal isoforms (α, β, γ, δ, ε, ζ, η) and at least four alternatively spliced N-terminal isoforms, which contain different parts of the transactivation domain. Moreover, both p73 and p63 genes can be transcribed from an alternative promoter located in the intron 3 (Fig. 1). This leads to the expression of N-terminally truncated isoforms (ΔNp73 and ΔNp63). Such isoforms can exert dominant-negative effects over p53, p73, and p63 activities (Benard et al. 2003; for review, see Melino et al. 2003) but were also shown to directly activate specific gene targets (Dohn et al. 2001; Wu et al. 2003; Liu et al. 2004). Altogether, the p63 gene expresses six mRNA variants that encode six different p63 protein isoforms, and the p73 gene expresses at least 35 mRNA variants that can encode theoretically 28 different p73 protein isoforms. So far, 14 different p73 protein isoforms have been described.
Figure 1.
Structure of the human p53, p63, and p73 genes. p63 and p73 express multiple splice variants and contain an internal promoter, while no internal promoter or multiple splice variants have been described for p53. (Black) Noncoding sequence; (white) coding sequence.
To date, the p53 gene structure is much simpler. The mouse p53 gene contains only one promoter and transcribes only two mRNA variants with an alternative splicing of the intron 10 (Wolf et al. 1985). The human p53 gene contains only one promoter, and transcribes three mRNA splice variants encoding, respectively, full-length p53, p53i9 (Flaman et al. 1996), and Δ40p53 (Courtois et al. 2002; Yin et al. 2002; Ghosh et al. 2004). p53i9 encoded by alternative splicing of the intron 9 was reported as a p53 protein isoform truncated of the last 60 amino acids of p53, defective in transcriptional activity and devoid of DNA-binding activity. No evidence was presented that the endogenous p53i9 isoform was expressed at the protein level. Δ40p53 (also named p47 or ΔNp53), is an N-terminally truncated p53 isoform deleted of the first 40 amino acids. Δ40p53 protein can be generated either by an alternative splicing of intron 2 or by alternative initiation of translation. The Δ40p53 protein still contains a part of the p53 transactivation domain, and it can activate gene expression after transfection through a second transactivation domain located between amino acids 43 and 63 (Zhu et al. 1998, 2000; Harms and Chen 2005). Δ40p53 can also act, after transfection, in a dominant-negative manner toward wild-type p53, inhibiting both p53 transcriptional activity and p53-mediated apoptosis (Courtois et al. 2002; Ghosh et al. 2004). Δ40p53 can modify p53 cell localization and inhibits p53 degradation by MDM2 (Ghosh et al. 2004).
The current knowledge of the p53 gene structure and of the p63/p73 gene structures is not consistent with our current understanding of the evolution of the p53 gene family. Mammalian genomes contain three members of the p53 family, while only one member has been identified in invertebrates, suggesting that the mammalian p53 family members are derived from the triplication of one ancestral gene (Yang et al. 2002). Given the complexity of p63 and p73 genes and that the p53 gene transcription pattern has not been studied since its discovery (before the PCR revolution), we questioned whether the p53 gene structure was in reality more complex than has been previously thought. We revisited p53 transcription using modern technology.
Here, we report that the p53 gene, like the p73 and p63 genes, contains an internal promoter and can transcribe six different mRNAs in normal human tissue, which can encode at least six p53 protein isoforms. p53 isoforms are expressed in several normal human tissues in a tissue-dependent manner. p53 isoforms are differentially expressed in breast tumors compared with normal breast tissue. p53 gene structure is conserved in Drosophila, suggesting that the p53 family gene structure plays an essential role in the activities of the p53 family members.
The endogenous p53β protein isoform binds differentially to p53-inducible promoters and enhances specifically p53 transcriptional activity on the BAX promoter but not on the p21 promoter. The differential expression of p53 isoforms in human tumors may provide some explanation of the disparate published findings about the prognostic value of p53 status in cancer.
Results
Human p53 gene contains an internal promoter in intron 4
To assess the transcription initiation site of the human p53 gene, we performed GeneRacer PCR on RNA extracted from normal human colon. GeneRacer PCR ensures that only capped transcripts are amplified and thereby allows the determination of the transcription initiation site. Given that the p63 and p73 genes contain an internal promoter in intron 3, we designed primers derived from exon 4 and exon 5 of p53 to try to detect the presence of such a promoter (Fig. 2A). By using primers from the beginning (RE4a) or from the end (RE4b) of exon 4, we amplified two distinct bands (a and b), which were cloned and sequenced (Fig. 2A). They corresponded to human p53 mRNA starting from two distinct transcription initiation sites separated by 54 bp in exon 1.
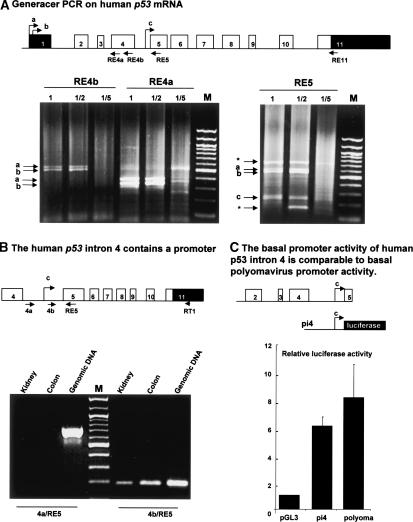
Figure 2.
The human p53 gene contains an internal promoter. (A) GeneRacer PCR on human p53 gene. Schema of the human p53 gene: (black) noncoding sequence; (white) coding sequence. Total RNA extract from normal human colon was treated for GeneRacer PCR according to the manufacturer's protocol to allow specific amplification of capped mRNA. The reverse transcription was performed by using primer RE11. A first PCR was performed using the GeneRacer primer-1 and one of the p53-specific primers (RE4a or RE4b or RE5). One microliter of the first PCR diluted (1/2, 1/5) or neat (1) was used to perform a semi-nested PCR using GeneRacer primer-2 and the appropriate p53-specific primer (RE4b or RE4a or RE5). The product of the second PCR was analyzed on 1% agarose gel, and the DNA was stained with ethidium bromide. After cloning and sequencing, bands a and b were found to contain sequences from exon 1 of human p53 while band c contained sequences from intron 4 of the human p53 gene. (★) Nonspecific bands. (B) The human p53 intron 4 contains a promoter. The RT-PCR was performed from total RNA extracted from kidney and colon human tissues using primer RT1. The PCR was performed using the primer RE5 with the primers 4a or 4b. PCR from human genomic DNA was performed using the same primers to control experimental conditions. (C) Luciferase reporter assay. The basal promoter activity of intron 4 of the human p53 gene is comparable to basal polyomavirus promoter activity. The human p53 intron 4 (pi4) was cloned upstream of the luciferase gene in the promoterless plasmid pGL3 basic and transfected with the Renilla luciferase expression vector into H1299 cells. The Renilla vector is used as an internal control of transfection efficiency. Cells were harvested 24 h after transfection, and the luciferase and Renilla activities were analyzed simultaneously. Luciferase activity was normalized to Renilla activity. Results shown are the average of four independent experiments performed in triplicate. SDs are indicated as error bars.
The transcription initiation site (b) corresponds to the one previously identified (Bienz-Tadmor et al. 1985; Lamb and Crawford 1986). The transcription initiation site (a) has never been described. When we used a primer derived from exon 5 (RE5), we amplified three bands containing p53 sequences. Two p53 bands corresponded to the initiation sites described above (a and b), and one contained sequences from intron 4 (band c), suggesting the presence of an internal promoter in intron 4 of the human p53 gene (Fig. 2A).
To verify the existence of a transcription initiation site in intron 4, we performed RT-PCR from total RNA extracted from normal human kidney and colon tissues using primers (4a and 4b) derived, respectively, from the 5′ or from the 3′ end of intron 4. By using primer 4b, we amplified one band corresponding to human p53 mRNA initiated in intron 4 (Fig. 2B). However, when we performed a similar RT-PCR using primer 4a, we could not amplify any cDNA, although the PCR could be performed on genomic DNA. This indicates that the cDNA containing the 3′-end of intron 4 of the human p53 gene derives from an mRNA transcript and not from genomic DNA contamination.
To confirm the presence of a promoter in intron 4 of the human p53 gene, we performed luciferase reporter assays by transfecting H1299 cells, null for p53, with a reporter plasmid containing intron 4 of the human p53 gene cloned upstream of the luciferase gene in the pGL3 promoterless plasmid (Fig. 2C). The basal transcriptional activity of intron 4 is sixfold higher than the transcriptional activity produced by the promoterless plasmid pGL3 and is comparable to the basal polyomavirus promoter activity. This confirms that intron 4 of the human p53 gene has promoter activity. The mRNA transcribed from this internal promoter contains only one open reading frame that would encode for an N-truncated p53 protein initiated at codon 133 (Δ133p53) distinct from Δ40p53. The Δ133p53 isoform would not contain the transactivation domain or the proline domain, and would contain only part of the DNA-binding domain; while Δ40p53 contains as described above the second transactivation domain, the proline domain, and the DNA-binding domain.
Intron 9 of the human p53 gene can be alternatively spliced
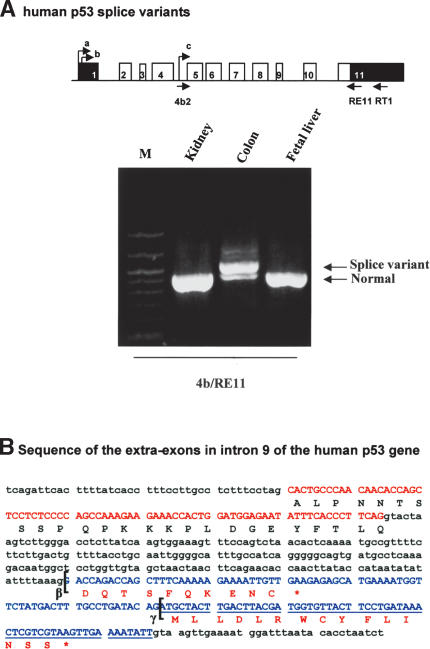
Using primers derived from intron 4 and exon 11 of the human p53 gene, we amplified by RT-PCR two bands in normal human colon (Fig. 3A). After cloning and sequencing, we established that the bands corresponded to three different splices of intron 9 of the human p53 mRNA initiated in intron 4. The lower band corresponds to the complete splice of intron 9 to exon 10. The upper band contains two alternative splice variants of an additional exon in intron 9 (exon 9b), one splice (β) leading to the expression of a truncated p53 protein terminating with 10 additional amino acids (DQTSFQKENC) and another splice (γ) leading to the expression of a truncated p53 protein terminating with 15 additional amino acids (MLLDLRWCYFLINSS) (Fig. 3B). Both splice variants would encode for p53 protein deleted of the oligomerization domain because of stop codons contained in exon 9b. Variant β correspond to p53i9 previously identified and reported as encoding a p53 protein isoform devoid of DNA-binding activity and defective in transcription. No further activities were described (Flaman et al. 1996). Endogenous variant β has never been shown to be expressed at the protein level.
Figure 3.
Intron 9 of the human p53 gene can be alternatively spliced. (A) RT-PCR. Human p53 splice variants. The reverse transcriptions of total RNA extracted from normal human tissue (colon, kidney, and fetal liver) were performed using primer RT1. Two successive PCRs were performed, the first using the primers 4b and RE11, followed by a second PCR using primers 4b2 and RE11. The arrows indicate the fragments corresponding to p53 variants initiated in intron 4. (B) Sequence of the extra-exons in intron 9 of the human p53 gene. Schema of the human p53 gene with the alternative splices (β and γ): (black) noncoding sequence; (white) coding sequence. Sequences of exon 9 and intron 9: (red) sequence of exon 9; (blue) sequence of the extra-exons in intron 9. The brackets indicate the splice sites giving p53β and p53γ protein isoforms.
Altogether, the human p53 gene can theoretically transcribe nine different p53 protein isoforms: p53, p53β, p53γ, Δ133p53, Δ133p53β, and Δ133p53γ due to alternative promoters in the intron 4 and alternative splicing of the intron 9, but also Δ40p53, Δ40p53β, and Δ40p53γ due to alternative splicing of intron 2 and alternative initiation of translation (Fig. 4).
Figure 4.
Schemas of the human p53 gene structure and p53 protein isoforms. (A) Schema of the human p53 gene structure. (B) Schema of the p53 protein isoforms theoretically encoded by the human p53 gene.
Tissue-specific expression of p53 isoforms
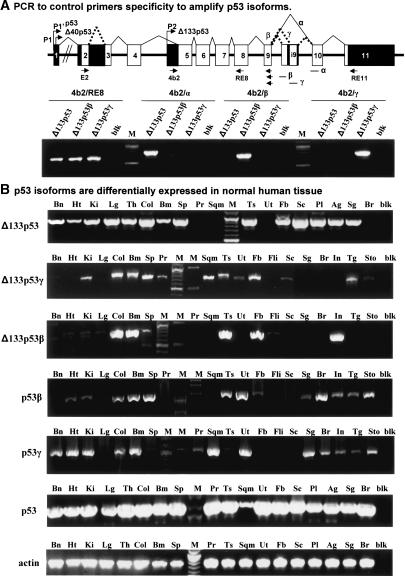
To determine whether the human p53 mRNA splice variants are expressed in normal human tissues, we designed primers specific to each p53 isoform to amplify p53, p53β, p53γ, Δ133p53, Δ133p53β, and Δ133p53γ splice variants by RT-PCR on total RNA (Fig. 5A,B). The Δ133p53 mRNA variant is expressed in most normal tissue but cannot be detected in prostate, uterus, skeletal muscle, and breast. The Δ133p53γ mRNA variant is expressed in most normal tissue but is not detected in brain, heart, lung, fetal liver, salivary gland, breast, and intestine, while the Δ133p53β mRNA variant is expressed in few normal tissues, being present only in colon, bone marrow, testis, fetal brain, and intestine.
Figure 5.
Expression of the p53 isoforms in normal human tissue. (A) PCR to control primers' specificity to amplify p53 isoforms. Primers were designed to be amplified specifically by normal splicing, splice β, and splice γ and were assessed by PCR on cloned cDNA of Δ133p53, Δ133p53β, and Δ133p53γ. (B) p53 isoforms are differentially expressed in normal human tissue. p53 mRNA variants were amplified specifically by PCR from total RNA extracted from normal human tissue (Clontech). (Bn) Brain; (Ht) heart; (Ki) kidney; (Lg) lung; (Th) thymus; (Col) colon; (Bm) bone marrow; (Sp) spleen; (Pr) prostate; (Ts) testis; (Sqm) skeletal muscle; (Ut) uterus; (Fb) fetal brain; (Sc) spinal cord; (Pl) placenta; (Ag) adrenal gland; (Sg) salivary gland; (Br) breast; (Fli) fetal liver; (In) intestine; (Tg) thyroid gland; (Sto) stomach; (blk) blank. Actin is shown as a control.
The p53β isoform is expressed in most normal tissue but cannot be detected in brain, lung, prostate, muscle, fetal brain, spinal cord, and fetal liver. The p53γ mRNA variant is expressed in most normal tissue but cannot be detected in lung, spleen, testis, fetal brain, spinal cord, and fetal liver.
Therefore, the p53 splice variants and the alternative promoter-initiated products are expressed in a wide range of normal tissues but in a tissue-dependent manner, suggesting that the internal promoter and the splicing of p53 can be regulated.
p53 alternative promoter is conserved in Drosophila p53 gene
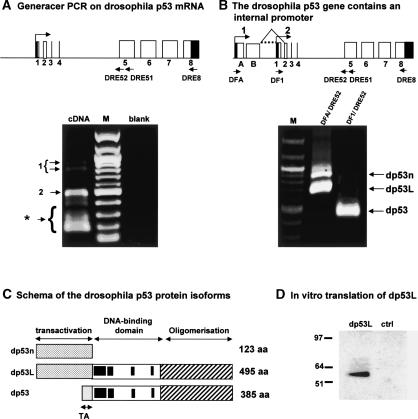
Our results demonstrate that the p53 gene has a gene structure similar to its related genes, p63 and p73, containing an internal promoter and expressing several splice variants. It is generally understood that the mammalian p53 gene family derives from the triplication of one ancestral p53 gene (Yang et al. 2002). This suggests that the ancestral p53 gene should also contain an internal promoter; however, only one transcript has been described for the Drosophila p53 gene (Brodsky et al. 2000; Jin et al. 2000; Ollmann et al. 2000). To assess this hypothesis, we performed GeneRacer PCR on total RNA extracted from Drosophila embryos. We amplified three variants of Drosophila p53 mRNA (Fig. 6A). All PCR fragments were cloned and sequenced. One form (dp53) is initiated at exon 1 and is identical to the one previously published. The second form (dp53L) contains two additional exons (exons A and B) and is initiated 580 bp upstream of exon 1. The third form (dp53n) is a splice variant of dp53L with no splicing of intron B (Fig. 6B). By RT-PCR using specific primers derived from exon 1 and exon A, we confirmed the GeneRacer PCR results (Fig. 6B). The Drosophila p53 gene would hence encode three Drosophila p53 protein isoforms: dp53L (495 amino acids), dp53 (385 amino acids), and dp53n (110 amino acids) (Fig. 6C). To determine whether the mRNA initiated at exon A (dp53L) could lead to protein expression, it was cloned and fused to a Flag-tag in the T7 expression vector to be expressed in vitro (Fig. 6D). It encodes a Drosophila p53 protein of 495 amino acids (110 amino acids longer than the dp53 form initially published) containing a longer N-terminal domain. Therefore, the previously published dp53 protein is an N-terminally truncated isoform produced from an internal promoter.
Figure 6.
The Drosophila p53 gene contains an internal promoter. (A) GeneRacer PCR on Drosophila p53 mRNA. Schema of the Drosophila p53 gene: (black) noncoding sequence; (white) coding sequence. Total RNA extract from Drosophila embryos was treated according to the manufacturer's protocol to allow amplification of capped mRNA. The reverse transcription was performed by using primer DRE8. The first PCR was performed using the GeneRacer primer-1 and the Drosophila p53-specific primer DRE51. One microliter of the first PCR was used to perform a nested PCR using the GeneRacer primer-2 and the p53-specific primer DRE52. The product of the second PCR was analyzed on a 1% agarose gel, and the DNA was stained with ethidium bromide. After cloning and sequencing, the bands (1) contained sequences from two additional exons (exon A and B) upstream of the previously described exon 1 of the Drosophila p53 gene. The upper band contained a splice variant. Band 2 contains sequences from the previously described exon 1 of Drosophila p53. (★) Nonspecific bands. (B) The Drosophila p53 gene contains an internal promoter. The reverse transcription was performed using primer DRE8 on total Drosophila embryo RNA extracts. A first PCR was performed using primers derived from exon A (DFA) or from exon 1 (DF1) and the reverse primer DRE51 followed by a semi-nested PCR using primer DFA or DF1 with primer DRE52. The arrows indicate the PCR fragments corresponding to Drosophila p53. Fragment dp53n corresponds to the Drosophila p53 mRNA not spliced at intron B and initiated at the first site (1) of initiation of transcription. Fragment dp53L contains the Drosophila p53 mRNA initiated at the first site (1) of initiation of transcription with splicing of intron B. Fragment dp53 contains the Drosophila p53 mRNA initiated at the second site of transcription initiation. (C) Schema of the Drosophila p53 protein isoforms. The Drosophila p53 gene can encode, theoretically, three different protein isoforms. The splice form dp53n may encode a truncated form of Drosophila p53 containing only the N-terminal end (123 amino acids). The form dp53L may encode a Drosophila p53 of 495 amino acids containing a large N-terminal domain. The form dp53 encodes the published Drosophila p53 protein. (D) In vitro translation of dp53L. The dp53L cDNA was fused to a Flag-tag and cloned in a T7-driven promoter expression vector. dp53L protein was produced by in vitro translation using a reticulocytes lysates kit and revealed by Western blot using anti-Flag antibody.
By aligning the protein sequences of full-length p53, p63, and p73 proteins from mammalian, bird, frog, mollusk, and fish, we observed that the only domain conserved through evolution within the N-terminal end, which is required to activate gene transcription, is the sequence FxxψW (Supplementary Figure S1). The dp53L isoform but not the dp53 isoform contains the domain FxxψW, indicating that dp53L is homologous to full-length p53, p63, and p73 and strongly suggests that the dp53L protein is able to transactivate genes.
We therefore identified two additional variants of Drosophila p53 mRNA and showed that the Drosophila p53 previously published was initiated from an internal promoter. This demonstrates that the internal promoter is conserved through evolution from Drosophila to man and strongly suggest that the internal promoter has an essential role in the control of p53 gene family activities.
p53 mRNA variants lead to expression of p53 protein isoforms
To determine whether the p53 mRNA variants can lead to protein expression, they were cloned in mammalian expression vector and transfected into H1299 cells that lack an endogenous p53 gene. The cDNA encoding for Δ40p53 was generated by mutagenesis of the first ATG of full-length p53 so that translation is initiated at the second methionine at position 40. To determine the translation initiation start of the mRNA initiated in intron 4 (Δ133p53), we mutated the first ATG of the open reading frame, corresponding to codon 133, to generate the expression vector Δ133p53mut. p53 protein expression was assessed 24 h after transfection by Western blot using sheep polyclonal (sp) anti-human p53 antibody (Midgley et al. 1992), which recognizes epitopes both in the first 100 amino acids and the last 60 amino acids of p53.
All p53 isoforms expression vectors lead to p53 protein isoform expression (Fig. 7A). Δ40p53, p53β, and p53γ proteins all have an apparent size of 45 kDa. p53β can also give an additional band at 40 kDa that can be detected on Western blots by the 1801 monoclonal anti-human p53 antibody but not by the DO-1 monoclonal anti-human p53 antibody (data not shown). DO-1 has its epitope between amino acids 20 and 25. Therefore, this band contains an N-terminally truncated p53 protein that could correspond to Δ40p53β protein generated by alternative initiation of translation.
Figure 7.
Endogenous p53 isoforms are expressed at the protein level. (A) Transfection of p53 isoforms in H1299. A p53 variant can lead to p53 protein isoform expression. H1299 cells, devoid of p53 expression, were transfected with p53 cDNA variants cloned in mammalian expression vectors as indicated. Proteins were extracted 24 h after transfection, and analyzed by Western blot using either sheep polyclonal (sp) anti-human p53 or mouse monoclonal (DO-12) anti-human p53 (Vojtesek et al. 1995). (B) Endogenous p53 protein isoforms can be detected in human cell lines. Proteins from human cell lines expressing wild-type p53 (293) or mutant p53 (T47D, SW620, HT-29), or devoid of p53 expression (H1299, Saos-2) were extracted as described in Materials and Methods. Ten micrograms of protein extract were analyzed by Western blot. p53 expression was revealed using DO-12 mouse monoclonal antibody. (C) Anti-p53β antibody (KJC8) is specific for p53β and Δ133p53β. H1299 cells, devoid of p53 expression, were transfected with p53, p53β, Δ133p53β, or empty expression vectors. Protein extracts were analyzed by Western blot using either CM1 rabbit polyclonal anti-human p53 or peptide affinity-purified KJC8 rabbit polyclonal anti-p53β antibody. (D) U2OS cells express endogenous p53β and Δ133p53β protein isoforms. U2OS cells were transfected with 400 nM final concentration of small interference RNA oligonucleotides specific for p53 (Smartpool p53siRNA; Dharmacon) or control (Smartpool controlsiRNA; Dharmacon). Cells were harvested 96 h after transfection, and proteins were analyzed by Western blot using CM1 rabbit polyclonal anti-human p53 or KJC8 rabbit polyclonal anti-p53β. The anti-actin antibody was used to control loading and transfer efficiency. (E) p53β and Δ133p53β are not accumulated in MCF7 tumor cells in response to Actinomycin-D treatment. MCF7 cells expressing wild-type p53 were untreated or treated with 60 ng/mL of the DNA-damaging drug Actinomycin-D (Act-D), a potent p53 inducer. Proteins were extracted 6 h after treatment and analyzed (5 μg) by Western blot using either DO-1 mouse monoclonal anti-human p53 or peptide affinity-purified KJC8 rabbit polyclonal anti-p53β antibody. The anti-actin antibody was used to control loading and transfer efficiency.
The Δ133p53 expression vector expresses a p53 protein of 35 kDa, while the Δ133p53mut expression vector expresses a p53 protein of 30 kDa, indicating that the translation initiation start of the Δ133p53 mRNA initiated in intron 4 is at ATG 133. The protein isoforms Δ133p53γ and Δ133p53β have an apparent size of 28 kDa and can only be detected by DO-12 monoclonal antibody, which recognizes an epitope located between amino acids 256 and 270 in the DNA-binding domain of p53 (Vojtesek et al. 1995).
p53 protein isoforms are expressed under standard cell culture conditions
To determine whether endogenous p53 protein isoforms could be expressed under standard cell culture conditions, we extracted proteins from different cell lines expressing mutant or stabilized p53 protein or no p53 proteins, and we analyzed p53 content by Western blot using DO-12 monoclonal antibody anti-human p53. DO-12 recognizes an epitope common to all p53 isoforms. DO-12 revealed several bands ranging from 35 to 53 kDa in cell lines expressing p53 gene but revealed no bands in cell lines devoid of p53 gene (Fig. 7B). This suggests that p53 protein isoforms may be expressed at the protein level under standard cell culture conditions.
None of the available p53 antibodies allows the specific identification of endogenous p53 protein isoforms, as they cannot distinguish isoforms from full-length p53 forms that have been subject to post-translational modification or protein degradation. We tried to raise antibodies specific for Δ133p53 or p53β or p53γ isoforms. However, we have failed so far to raise antibodies against several peptides derived from the peptide (γ). Moreover, it is difficult to raise an antibody specific to Δ133p53 as full-length p53 contains all of the predicted amino acid sequences of Δ133p53. The antibody will need to recognize the unique N terminus of Δ133p53 to be specific. Therefore, we could not study the expression of endogenous p53γ or Δ133p53 protein isoforms. We have, however, successfully raised a polyclonal antibody (KJC8) against the peptide DQTSFQKENC specific to p53β and Δ133p53β isoforms.
H1299 cells were transfected with p53, p53β, and Δ133p53β expression vectors to assess the KJC8 anti-body specificity by Western blot (Fig. 7C). The CM1 polyclonal antibody raised against full-length human p53 is specific to p53 and p53β but not Δ133p53β as this isoform does not contain the epitopes recognized by CM1. The affinity-purified rabbit polyclonal KJC8 anti-body recognized only p53β and Δ133p53β (28 kDa) but not p53, indicating that the KJC8 antibody is specific to the p53β protein isoforms.
We determined whether p53 protein isoforms are expressed under normal cell culture conditions using U2OS cells (Fig. 7D). CM1 detected p53 and a band of lower mobility (45 kDa) in U2OS cells. KJC8 revealed two bands (45 and 28 kDa) in U2OS cells. To confirm that the bands recognized by CM1 and KJC8 antibodies in U2OS protein extract were p53 isoforms, we transfected U2OS cells with a Smartpool of p53siRNA oligonucleotides that target sequences in all the isoforms of p53. As the unique sequences are too short, we could not design p53siRNAs that would specifically inhibit expression of p53 isoforms. The intensity of the bands revealed by CM1 and KJC8 antibodies was reduced in U2OS cells transfected with p53siRNA but not in cells transfected with a control siRNA, confirming that both bands (45 and 28 kDa) represent p53 isoforms containing the β epitope. Therefore, p53 protein isoforms p53β and Δ133p53β are expressed under standard cell culture conditions.
p53β isoforms are not accumulated in MCF7 tumor cells in response to the chemotherapeutic drug Actinomycin D
MCF7 cells expressing endogenous wild-type p53 were treated or not treated with Actinomycin D to determine whether p53β isoforms could be accumulated in response to stress. Proteins were extracted 6 h after treatment and analyzed by Western blot using DO-1 or KJC8 antibodies. DO-1 can detect p53, p53β, and p53γ but no other p53 isoforms. As expected, p53 protein is accumulated in response to Actinomycin D treatment, but no accumulation of p53β or Δ133p53β was detected in response to stress using KJC8 antibody in the MCF7 tumor cell protein extracts (Fig. 7E). Similar results were obtained in U2OS osteosarcoma cell lines treated with Actinomycin D (data not shown). However, further studies will determine whether p53 isoforms can be accumulated in response to stress in normal primary cells.
Subcellular localization
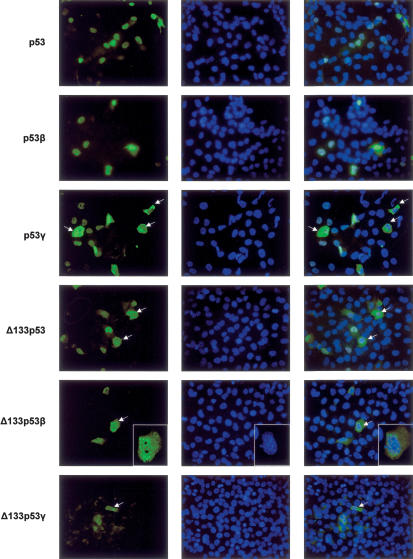
p53 isoforms were transfected into H1299 cells to determine their subcellular localization. p53γ-, p53β-, and Δ133p53-transfected cells were stained with CM1 anti-body. Δ133p53γ-transfected cells were stained with DO-12, while the Δ133p53β-transfected cells were stained with KJC8 antibody (Fig. 8). Full-length p53 is localized in the nucleus. Δ133p53 and p53β are localized mainly in the nucleus with a minor staining in the cytoplasm. p53γ is localized in most cells in the nucleus but can be localized in the cytoplasm in some cells, suggesting that p53γ could be shuttling between the nucleus and the cytoplasm.
Figure 8.
Subcellular localization of the p53 protein isoforms. H1299 cells transfected with p53, p53β, p53γ, or Δ133p53 expression vectors were stained by indirect immunofluorescence (FITC) using rabbit polyclonal CM1 anti-human p53 antibody. H1299 cells transfected with Δ133p53β expression vector were stained by indirect immunofluorescence (FITC) using rabbit polyclonal KJC8 anti-p53β antibody. H1299 cells transfected with Δ133p53γ expression vector were stained by indirect immunofluorescence (FITC) using mouse monoclonal DO-12 anti-p53 antibody. DNA was stained by DAPI. The merge is shown. The cells with a particular staining are indicated with an arrow.
In most cells, Δ133p53β protein is localized in the nucleus and the cytoplasm, indicating that the nuclear localization is dependent on the N terminus and the C terminus of p53. However, in some cells, Δ133p53β forms foci in the nucleus that are different from nucleoli.
Δ133p53β and Δ133p53γ differ only by the last 15 amino acids of the C-terminal end. Contrary to Δ133p53β, Δ133p53γ is localized only in the cytoplasm, indicating that the C-terminal amino acids encoded by the alternative exon 9b can modify the subcellular localization of these isoforms.
p53 and p53β bind differentially to p53-responsive promoters
p53 has many biochemical and biological activities. It is thus difficult to determine the role of each p53 isoform in each activity of p53. As we raised an antibody specific to the p53β isoforms, we decided to limit our study to the characterization of p53β DNA-binding activity and transcriptional activity.
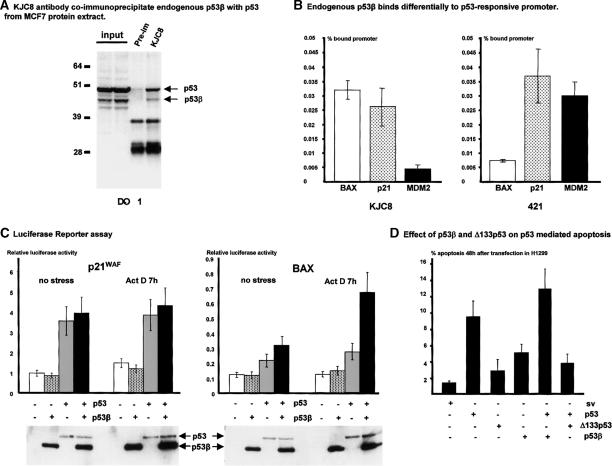
First, we determined that KJC8 antibody can immunoprecipitate endogenous p53β from a protein extract of MCF7 cells cultured under standard conditions (Fig. 9A). Endogenous full-length p53 is coimmunoprecipitated with p53β using KJC8 antibody, suggesting that p53 and p53β can form a protein complex.
Figure 9.
p53β binds the p53-responsive promoter and enhances p53 transcriptional activity on the BAX promoter but not on the p21 promoter. (A) KJC8 antibody coimmunoprecipitates endogenous p53β with p53 from MCF7 protein extract. Protein from MCF7 cells cultured under standard cell culture conditions were extracted and treated for immunoprecipitation as described in Materials and Methods. Proteins (100 μg) were immunoprecipitated with KJC8 antibody or with preimmune serum, and p53 content was revealed after Western blot using mouse monoclonal antibody (DO-1) anti-human p53. Ten percent of the input was loaded as a control. (B) Endogenous p53β binds differentially to the p53-responsive promoter. Protein from MCF7 cells cultured under standard cell culture conditions were extracted and treated for ChIP assay as described in Materials and Methods. Extracts were immunoprecipitated with KJC8 antibodyb mouse monoclonal 421 anti-p53 antibody, or nonrelated rabbit polyclonal antibody as a negative control. The amounts of BAX and p21 promoter contained in the input or immunoprecipitated with each of the antibodies were quantified by real-time PCR as described (Kaeser and Iggo 2002). The amount of promoter nonspecifically immunoprecipitated with the nonrelated rabbit polyclonal antibody was subtracted from the amount of promoter immunoprecipitated with KJC8 or 421 antibody. The results are expressed as the percentage of promoter specifically immunoprecipitated by KJC8 or 421 antibody compared with the total amount of promoter contained in the input. The results shown are the average of four independent experiments. (C) Luciferase reporter assay. p53β enhances p53 transcriptional activity on the BAX promoter but not the p21 promoter. H1299 cells (20,000 cells/well) were seeded on a 24-well plate. Cells were cotransfected in duplicate with the plasmids bax-luc or p21-luc containing the natural promoters driving the luciferase reporter gene, with a trace amount of pSVrenilla as an internal control, and 25 ng/well of p53 and/or 50 ng/well of p53β as indicated. Cells were treated 18 h after transfection with 60 ng/mL of Actinomycin D (ActD) for 6 h. The amount of SV-40 promoter was maintained constant in each transfection by adding empty SV-40 expression vector. Cells were harvested 24 h after transfection. Luciferase activity was normalized to Renilla activity. The results shown are the average of four independent experiments performed in duplicate. SDs are indicated as error bars. To take account of the transfection efficiency, luciferase assay protein extracts were analyzed by Western blot after normalization for Renilla activity. p53 and p53β expressions were revealed using DO-1 monoclonal antibodies. (D) Effect of p53β and Δ133p53 on p53-mediated apoptosis. H1299 cells were transfected with 5 μg/mL of the indicated expression vector or empty SV-40 expression vector (sv). In the experiments of coexpression of p53 with p53 isoforms, H1299 cells were cotransfected with 5 μg/mL of p53 expression vector and 5 μg/mL of the indicated p53 isoform expression vectors. The amount of SV-40 promoter was maintained constant at 10 μg/mL in each transfection by adding empty SV-40 expression vector. The DNA content of each transfected population was determined by flow cytometry analysis. The percentage of sub-G1 DNA content represents the percentage of apoptotic cells 48 h after transfection. The histogram represents the average of three independent transfections. SDs are reported as error bars.
Then, we asked whether endogenous p53β could bind to endogenous promoters in MCF7 cells using the chromatin immunoprecipitation assay (ChIP assay). We performed ChIP assay using quantitative PCR (Kaeser and Iggo 2002) on BAX, p21, and MDM2 endogenous promoters after immunoprecipitation with KJC8 antibody or 421 monoclonal anti-p53 antibody or a nonrelated polyclonal antibody as negative control. The 421 antibody has its epitope between amino acids 372 and 382 encoded by exon 11 and thus cannot recognize p53β or p53γ isoforms.
As Figure 9B shows, KJC8 antibody immunoprecipitates six times more BAX and p21 promoters than MDM2 promoters, indicating that endogenous p53β protein binds preferentially to the BAX or p21 promoters rather than the MDM2 promoter. The 421 antibody immunoprecipitated five times more p21 and MDM2 promoters than BAX promoters, indicating that full-length p53 binds poorly to the BAX promoter compared with the p21 and MDM2 promoters.
As p53β binds differentially to endogenous p53-inducible promoters and p53 binds preferentially to p21 and MDM2 promoters rather than the BAX promoter, it suggests that p53β could affect p53 transcriptional activity on the BAX promoter.
Transcriptional activity
We could not design a p53β siRNA or antisense plasmid able to knock down specifically p53β expression. Therefore, to determine the specific effect of p53β on transcription and to avoid the effects of the other p53 isoforms expressed in the MCF7 cell, we cotransfected H1299 cells with p53β or/and p53 expression vectors and the luciferase reporter plasmids driven by the p21 promoter or the BAX promoter (Fig. 9C). Transfection of p53β, in the absence of p53, has no effect on p21 or BAX promoter transactivation, while p53 induces transactivation of BAX and p21 promoters. Cell treatment with Actinomycin D, an activator of p53, does not increase p53 transcriptional activity. This is probably because transfection already induced a cellular stress sufficient to activate p53 transcriptional activity (Renzing and Lane 1995). p53β cotransfected with p53 enhances BAX promoter activity. This effect is increased by Actinomycin D treatment. However, cotransfection of p53β with p53 has no effect on p21 promoter activity even after treatment with Actinomycin D.
To determine whether the effect of p53β on the BAX promoter is due to an increase in p53 transcriptional activity or an increase in p53 protein level, we performed a Western blot on luciferase assay protein extract after normalization by Renilla luciferase activity to take account of the transfection efficiency. The p53 protein level is not increased in the presence of p53β, indicating that p53β specifically enhances p53 transcriptional activity on the BAX but not on the p21 promoter.
Apoptosis
To assess proapoptotic activity of p53β and Δ133p53, H1299 cells were transfected with each of the p53 isoforms, and the percentage of cells in the sub-G1 population was determined by FACS analysis as described (Bourdon et al. 2002). Δ133p53 is defective in promoting apoptosis, while p53β has reduced proapoptotic activity compared with FL-p53 (Fig. 9D).
Cotransfection of p53β with p53 does not significantly enhance p53-mediated apoptosis in H1299 cells. However, p53-mediated apoptosis is impaired in cells cotransfected with wild-type p53 and Δ133p53, indicating that Δ133p53 is dominant-negative toward wild-type p53.
These results suggest that the biochemical and biological functions of p53 can be mediated by the interplay between p53 isoforms and full-length p53 protein and thus that regulation of p53 function in normal and tumor tissues in man is likely to be more complex than has been hitherto appreciated.
Differential expression of p53 isoforms in breast tumors
As p53β and Δ133p53 have different biological activities, we asked whether p53 isoforms are differentially expressed in neoplastic human tissues compared with normal tissue.
We detected p53, p53β, and p53γ but not Δ133p53, Δ133p53β, and Δ133p53γ mRNA in a pool of total RNA extracted from normal breast tissue from eight donors (Fig. 5B).
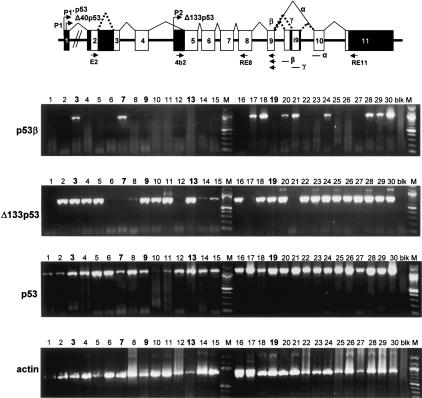
We performed RT-PCR from total RNA extracted from 30 randomly selected human breast tumors (Fig. 10). p53 mRNAs were amplified in all tumor samples and sequenced between exon 4 and exon 9. Only five tumors out of 30 express mutant p53. The tumors 7 and 19 express p53 mutated at codon 248 (CGG → CAG, Arg → Gln). Tumor 13 expresses p53 mutated at codon 216 (GTG → ATG, Val → Met). Tumors 3 and 9 express p53 mutated at codon 132 (tumor 3: AAG → AGG, Lys → Arg; tumor 9: AAG → AAT, Lys → Asn).
Figure 10.
p53 mRNA variants are differentially expressed in tumors from patient to patient. Total RNA from 30 breast tumors were provided by Tayside Tumour Bank. Reverse transcription was performed as described in Materials and Methods using primer RT1 and 400 ng of total RNA. The p53 cDNAs were amplified by PCR using appropriate primers as described in Material and Methods and on schema of the p53 gene structure. p53γ, Δ133p53β, and Δ133p53γ mRNAs were not detected (data not shown). Actin was used a control. Tumors expressing mutant p53 are marked in bold: tumor 3 (mutant p53 K132 → R), tumor 7 (mutant p53 R248 → Q), tumor 9 (mutant p53 K132 → N), tumor 13 (mutant p53 V216 → M), and tumor 19 (mutant p53 R248 → Q). All the other tumors express wild-type p53.
None of the 30 breast tumors expressed the same panel of p53 isoforms as normal breast tissue. Contrary to normal breast tissue, p53γ was not detected in any of the 30 human breast tumors analyzed. p53β was detected in only 10 out of 30 tumors, while Δ133p53 mRNA was detected in 24 out of 30 breast tumors. As in normal breast tissue, Δ133p53γ and Δ133p53β mRNA was not detected in human breast tumors.
Therefore, p53, p53β, p53γ, and Δ133p53 are differentially expressed in normal and tumor breast tissue. Moreover, p53 isoform expression is different from tumor to tumor. Some tumors (#3 and #9) can express a mutant p53 but a wild-type Δ133p53.
As only five out of 30 tumors express mutant p53, it suggests that failure of appropriate regulation of expression of p53 isoforms may have a role in tumor formation. In particular, these results suggest that the expression or loss of expression of certain p53 isoforms could impair p53 function in cells that do not harbor inactivating mutations of the parental p53 gene. This could explain the difficulties in human breast tumors to link p53 status to prognosis and drug sensitivity.
Discussion
The recently discovered p53-related genes, p63 and p73, transcribe multiple splice variants and can initiate transcription from an alternative promoter located in intron 3. The human p73 gene is transcribed for 35 different mRNA, which would encode, theoretically, 28 different p73 protein isoforms. To date, 14 p73 proteins isoforms have been described. The human p63 gene is transcribed for six different mRNA splice variants encoding six different p63 protein isoforms. The activities of each p73 or p63 isoform are still under investigation, but it appears from most recent studies that each p73 or p63 isoform has specific and distinct activities (Wu et al. 2003; Liu et al. 2004; Goldschneider et al. 2005).
Up until now, no alternative promoter has been reported for the p53 gene. In mice, only one alternative splicing of intron 10—affecting the p53 C terminus—has previously been described, which leads to a p53 protein (p53AS) containing the oligomerization domain but devoid of the last 30 amino acids (Wolf et al. 1985). The p53AS isoform is constitutively active for sequence-specific DNA binding, can transactivate p53 target genes, but inhibits full-length p53-mediated apoptosis (Wu et al. 1994; Almog et al. 2000). For the human p53 gene, two mRNA splice variants have been previously reported: (1) p53i9, alternative splicing of intron 9; and (2) Δ40p53 (also named p47 or ΔNp53), an alternative splicing of intron 2. p53i9 encodes a C-truncated p53 protein isoform of 341 amino acids for which no biochemical activities have been reported. The p53i9 protein lacks the p53 tetramerization domain, fails to bind DNA in vitro, and has a transcriptional defect in vivo in both yeast and mammalian cells (Flaman et al. 1996). It has not been reported that endogenous p53i9 is expressed at the protein level. Δ40p53 encodes an N-terminally truncated p53 isoform deleted of the first 40 amino acids (Ghosh et al. 2004). Δ40p53 protein can also be generated by alternative initiation of translation (Courtois et al. 2002; Yin et al. 2002). Δ40p53 contains a large part of the transactivation domain and can still activate gene expression after transfection through a second transactivation domain located between amino acids 43 and 63 (Zhu et al. 1998, 2000; Harms and Chen 2005). Δ40p53 can also act, after transfection, in a dominant-negative manner toward wild-type p53, inhibiting both p53 transcriptional activity and p53-mediated apoptosis (Courtois et al. 2002; Ghosh et al. 2004). Δ40p53 can modify p53 cell localization and inhibits p53 degradation by MDM2 (Ghosh et al. 2004).
In this study, we have shown that the p53 gene has a complex transcriptional expression pattern encoding different p53 mRNA variants, through both the use of alternative splicing and the existence of an internal promoter in intron 4. The C terminus can be alternatively spliced to produce three isoforms—p53, p53β, and p53γ—where the p53β and p53γ isoforms lack the oligomerization domain. The alternative promoter leads to the expression of an N-terminally truncated p53 protein (Δ133p53), which is initiated at codon 133. Therefore, the human p53 gene can encode at least nine different p53 protein isoforms—p53, p53β, p53γ, Δ133p53, Δ133p53β, and Δ133p53γ due to alternative splicing of intron 9 and usage of the alternative promoter in intron 4, but also Δ40p53 due to alternative splicing of intron 2 and alternative initiation of translation (Courtois et al. 2002; Yin et al. 2002; Ghosh et al. 2004).
All p53 isoforms are expressed in normal tissue at the mRNA level. Moreover, the pattern of expression is tissue specific, indicating that their expression can be selectively regulated.
It is generally understood that the vertebrate p53 gene family derives from the triplication of one ancestral p53 gene (Yang et al. 2002). Human p63, p73, and p53 genes contain an internal promoter and express several splice variants, while only one transcript has been described for the Drosophila p53 gene. To determine whether the Drosophila p53 gene contains an internal promoter, we performed GeneRacer PCR on Drosophila p53 mRNA. We have identified two additional Drosophila p53 mRNAs. Both mRNAs contain two additional exons (440 bp) and are initiated 580 bp upstream of the previously defined transcription initiation site of the Drosophila p53 gene. We are currently studying the transcriptional activity and the biological activities of the Drosophila p53 protein isoforms. The previously published Drosophila p53 mRNA is therefore a variant initiated from an internal promoter and encodes an N-terminally truncated isoform deleted of the first 110 amino acids (dp53). This indicates that the internal promoter is conserved among the p53 gene family and through evolution from Drosophila to man. It strongly suggests that the internal promoter plays an essential role in the multiple activities of the p53 family members.
We cannot specifically assess the endogenous expression of p53 protein isoforms by any existing p53 antibodies as they cannot distinguish isoforms from full-length p53 forms that have been subject to post-translational modification or protein degradation. We are currently trying to raise antibodies specific for the Δ133p53 or p53γ isoforms. However, raising antibodies against several peptides derived from the peptide (γ) has so far been unsuccessful, probably because it is not immunogenic enough. It is difficult to raise an antibody specific to Δ133p53 as full-length p53 contains all of the linear amino acid sequence present in Δ133p53. The antibody will need to recognize the unique N terminus of Δ133p53 to be specific.
We raised an antibody (KJC8) specific to the p53β protein isoform. Using this affinity-purified polyclonal antibody and p53 siRNA, we show that the endogenous p53β and Δ133p53β isoforms are expressed at the protein level, confirming that the alternative promoter is active and can lead to protein expression. Therefore, we would anticipate that all the other p53 isoforms are expressed as proteins in human tissue.
p53β and Δ133p53β protein levels are not increased in response to Actinomycin D treatment in MCF7 or in U2OS cells. However, we noticed that transfected p53β is accumulated like full-length p53 in H1299 cells after Actinomycin D treatment (Fig. 9C), suggesting that p53β protein expression may be abnormally regulated in MCF7 and in U2OS cells. Further studies will determine whether the p53β protein level can be regulated in response to cellular stress in normal primary cells.
p53 isoforms transfected in H1299 cells have different subcellular localization. Moreover, p53γ, Δ133p53, Δ133p53β, and Δ133p53γ have different subcellular localizations in a minor but significant subpopulation of transfected cells, suggesting that p53 isoform biological activities can be modulated in relation to the cell cycle by regulating their subcellular localization.
p53 protein isoforms are less abundant than full-length p53 protein. However, p53 protein is subject to multiple post-translational modifications including phosphorylation, acetylation, methylation, ubiquitination, sumoylation, and neddylation (for review, see Oren 2003; Xirodimas et al. 2004). It is thus unclear what subfractions of total p53 are active in the different biochemical and biological functions of p53 (gene expression, cell cycle arrest, senescence, apoptosis, differentiation, replication, control of chromosome segregation, mitosis, meiosis, DNA repair, etc.). Moreover, p53 abundance is not necessarily associated with p53 transcriptional activity. Hence, we have isolated p53 mutants that are expressed at a low level but have a far higher specific transcriptional activity than the wild-type protein (Liu et al. 2001). In a mouse model of aging, very low level expression of fragments of p53 has caused profound phenotypes dependent on their effect in trans on wild-type p53 (Tyner et al. 2002; Maier et al. 2004). Therefore, p53 protein isoforms may be abundant and active enough to regulate the activities of a specific pool of post-translationally modified p53. This hypothesis is supported by our ChIP experiments. We showed that endogenous p53β and p53 proteins bind differentially to promoters. p53β binds preferentially to BAX and p21 promoters rather than the MDM2 promoter, while p53 binds preferentially (five times more) to p21 and MDM2 promoters than to the BAX promoter. This suggested that p53β could interfere with p53 transcriptional activity on the BAX but not the p21 promoter. To assess this hypothesis, we performed gene reporter assay in cells devoid of p53 expression to avoid any effects of the other endogenous p53 isoforms expressed in MCF7 cells. We determined thus that coexpression of p53 with p53β enhances BAX promoter activity but not p21 promoter activity in response to stress. However, p53β has no transcriptional activity on the BAX or p21 promoter in the absence of p53. We cannot rule out that p53β can directly transactivate specific promoters independently of p53, and further experiments will determine whether p53β can modulate p73 or p63 transcriptional activity.
By Western blot with the luciferase assay protein extract, we have shown that the p53 protein level is not increased in the presence of p53β, indicating that p53β enhances BAX promoter activity by increasing the intrinsic p53 transcriptional activity. The coimmunoprecipitation of p53 with p53β suggests that p53β interferes with p53 transcriptional activity by forming a protein complex with p53. The p53/p53β complex may bind preferentially to specific p53-responsive elements, and therefore only a subset of p53-inducible promoters will be responsive to p53β. We are currently identifying the promoters preferentially induced by p53β and studying how the p53/p53β complex can be formed, as p53β has lost the oligomerization domain.
p53β corresponds to p53i9 previously identified and reported as encoding a p53 protein isoform devoid of DNA-binding activity and defective in transcription (Flaman et al. 1996). The discrepancy between our results is probably due to the fact that we performed different and additional experiments using different promoters.
As p53 isoforms can bind differentially to promoters, can modulate p53 target gene expression and p53-mediated apoptosis, the differential expression of the p53 isoforms in human breast tumors could explain the difficulties in many clinical studies to link p53 status to the biological properties and drug sensitivity of human cancer. Furthermore, it suggests that failure of appropriate regulation of expression of p53 isoforms may have a role in tumor formation since attenuation of the wild-type p53 response would render the cells more susceptible to further genetic damage and therefore neoplastic transformation and tumor progression. It will be essential to extend our study to a larger series of tumors.
In conclusion, the structure of the p53 gene is similar to the p63 and p73 genes, revealing an unforeseen complex regulation. The interplay between p53 isoforms and p53 on specific targets may play a major role in controlling the activity of p53-related proteins.
Materials and methods
GeneRacer PCR
GeneRacer PCRs were performed according to the manufacturer's protocol (InVitrogen). Total RNA extracts from normal human tissue were purchased from Clonetech. Drosophila total RNA extracts were provided by Dr. John Abrams's laboratory. RT-PCR experiments were performed using AMV reverse transcriptase at 42°C using the primers indicated in the figure legends. The primers were RE4a, 5′-ACCTGGGTCTTCAGTG AACCATTG-3′; RE4b, 5′-ATGCAAGAAGCCCAGACGGAA ACC-3′; RE5, 5′-TTGGCAAAACATCTTGTTGAGGGC-3′; RE11, 5′-AATGTCAGTCTGAGTCAGGCCCTTCTG-3′; RT1, GACGCACACCTATTGCAAGCAAGGGTTC-3′; 4a, 5′-GATT CCTCTTCACCCTTAGGCTTC-3′; 4b, 5′-TAGACGCCAACT CTCTCTAG-3′; and 4b2, 5′-TGGGTTGCAGGAGGTGCTTA CAC-3′. The human p53 alternative promoter was amplified from human genomic DNA using the forward primer 5′-ACAT TGGAAGAGAGAATGTGAAGC-3′ and RE5. p53 splice variants were amplified using E2, 5′-ATGGAGGAGCCGCAGT CAGAT-3′ and reverse primers Ri91, 5′-TTTCAACTTACGAC GAGT-3′ followed by a second PCR using primers E2 and β, 5′-TTGAAAGCTGGTCTGGTCCTGA-3′ to amplify specifically p53β; or using primers E2 and γ, 5′-TCGTAAGTCAAGT AGCATCTGAAGG-3′ to amplify specifically p53γ. Δ133p53 was amplified using primers 4b and RT1 followed by a second PCR using primers 4b2 and α, 5′-CTCACGCCCACGGA TCTGA-3′. Δ133p53β/γ were amplified using primers 4b and Ri91 followed by a second PCR using primers 4b2 and β for Δ133p53β or primers 4b2 and γ for Δ133p53γ.
Luciferase reporter assay and transfection
On a 24-well plate, 6 × 104 H1299 cells were seeded per well and transfected in duplicate by Fugene (Boehringer) containing 250 ng/well of luciferase reporter plasmids, 10 ng/well of SV-Renilla luciferase, 25 ng/well of p53 expression vector driven by the SV40 promoter, and/or 50 ng/well of p53β expression vector driven by the SV40 promoter. For each transfection mix, the concentration of SV40 promoter was balanced with SV40 empty expression vector. Cells were incubated for 24 h at 37°C, washed, and lysed by adding 100 μL/well of passive lysis buffer (Promega). Cells were treated for 7 h with 60 ng/mL Actinomycin D (final concentration). Twenty microliters of protein extract was analyzed in a Microlumat LB 96V luminometer (Berthold EG&G Instrument) using the Dual Luciferase Reporter Assay kit (Promega).
FACScan analysis
H1299 cells (8 × 105) were seeded in a 10-cm Petri dish and transfected using Fugene. After 48 h of incubation at 37°C, cells were fixed in 70% ethanol and immunostained for CM1 or KJC8 anti-p53 antibody. DNA was stained by propidium iodide. Cells were analyzed by flow cytometry (FACScan; Becton Dickinson) using a three-parameter analysis (Deguin-Chambon et al. 2000).
siRNA
The small interference RNAs were purchased from DHARMACON SMARTpool p53siRNA and siCONTROL nontargeting siRNA. U2OS cells were transfected with 400 nM siRNA (final concentration) with the jetSI siRNA transfection reagent (poly-plus-transfection). Cells were harvested 96 h after transfection.
Human tissue samples
Samples of normal colonic mucosa and colorectal tumor were obtained through the MRC/CR-UK Tayside tissue/Tumour Bank with appropriate informed consent and ethical approval. The samples of normal and tumor tissue were snap-frozen in liquid nitrogen immediately upon receipt in the Pathology Department and thereafter stored at -80°C until required for analysis.
Western blot and immunoprecipitation
After treatment, cells were scraped and washed in PBS. Proteins were extracted in RIPA buffer (0.1% SDS, 1% NP-40, 0.5% DCA, 1 mM EDTA, 0.2 mg/mL RNase, 0.2 mg/mL Dnase, 1 mg/mL protease inhibitor; Roche complete Pefabloc). Following incubation on ice for 15 min, the crude lysate was briefly sonicated. After centrifugation at 10,000g at 4°C, the supernatant was decanted into a chilled Eppendorf tube and either used immediately or snap-frozen in liquid nitrogen and stored at -80°C.
Protein extracts were quantified by Bradford assay and analyzed on SDS-PAGE as previously described (Bourdon et al. 2002).
For immunoprecipitation, MCF7 cells were scraped and washed in PBS. Proteins were extracted in lysis buffer (50 mM Tris at pH 7.5, 10% glycerol, 100 mM NaCl, 0.2 mM EDTA, 0.1% NP-40, 0.2 mg/mL RNase, 0.2 mg/mL Dnase, 1 mg/mL protease inhibitor; Roche complete Pefabloc) and treated as above. One-hundred micrograms of total protein extract was incubated overnight with 5 μg of KJC8 antibody at 4°C; protein-A Sepharose beads (60 μL of slurry) were blocked overnight in BSA buffer (10% BSA; PBS). The next day, blocked protein-A Sepharose beads were incubated with antibody and protein extract for 1 h at 4°C. Beads were precipitated at 3000 rpm for 5min. After supernatant was removed, beads were washed three times. Protein content was then analyzed by Western blot as previously described (Bourdon et al. 2002).
Immunofluorescence staining
H1299 cells (3 × 105) seeded on two-well glass chamber slides (Lab Tek chamber slide) were transfected as described in the figure legends, fixed by 1% formaldehyde, and permeabilized with 0.1% Triton X-100 in PBS. Cells were incubated for 1 h at room temperature with primary antibody diluted in 10% FCS-DMEM. After washing with PBS, cells were incubated with Fluorescein (FITC)-conjugated anti-IgG (Jackson Immunochemicals) diluted in 10% FCS-DMEM. DNA was stained with DAPI.
Production and affinity purification of the KJC8 polyclonal antibody
The peptide TLQDQTSFQKENC corresponding to the carboxyl-end of human p53β protein was conjugated to the carrier protein KLH and inoculated into a rabbit as described in Harlow and Lane (1988). The anti-p53β antibody (KJC8) was affinity-purified using a peptide column and was adsorbed on H1299 protein extract.
Antibodies
Rabbit polyclonal anti-p53 antibody (CM1) and sheep polyclonal anti-p53 antibody (SP) were described previously (Midgley et al. 1992); mouse monoclonal anti-p53 (DO-1) and (421) antibodies were described (Stephen et al. 1995). Mouse monoclonal anti-p53 antibody (DO-12) was described previously (Vojtesek et al. 1995). Actin (Ab1) antibody was purchased from Calbiochem.
ChIP assay
MCF7 cells (2 × 106) were seeded per 15-cm plate and cultured under standard conditions. Cells were fixed with 1% formaldehyde for 24 h after seeding and then scraped and washed in PBS according to the protocol previously described (Bates et al. 2005). Primers and probes for quantitative PCR were described (Kaeser and Iggo 2002). Immunoprecipitation was carried out as described above using KJC8 antibody or 421 antibody or nonrelated rabbit polyclonal antibody as a negative control.
Acknowledgments
We thank Dr. Borek Vojtesek for the production of the KJC8 antibody. We thank Dr. Ron Hay and Dr. Fabienne Aillet for their help. We thank Dr. John Abrams for providing Drosophila total RNA extract. The Tayside Tissue/Tumour Bank is supported jointly by the Medical Research Council and Cancer Research-UK. We thank Mrs. Helen McDowell, Mrs. Susan Bray, and Mrs. Debbie Wilton for their technical assistance with human tissue samples. We thank Dr. Emma Warbrick and Dr. Alison Sparks for critical readings. This work was supported by Cancer Research-UK. D.P.L. is a Gibb Fellow of Cancer Research-UK.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1339905.
Corresponding authors.
References
- Almog N., Goldfinger, N., and Rotter, V. 2000. p53-dependent apoptosis is regulated by a C-terminally alternatively spliced form of murine p53. Oncogene 19: 3395-3403. [DOI] [PubMed] [Google Scholar]
- Bates G.J., Nicol, S.M., Wilson, B.J., Jacobs, A.M., Bourdon, J.C., Wardrop, J., Gregory, D.J., Lane, D.P., Perkins, N.D., and Fuller-Pace, F.V. 2005. The DEAD box protein p68: A novel transcriptional coactivator of the p53 tumour suppressor. EMBO J. 24: 543-553. [Epub 2005 January 20.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard J., Douc-Rasy, S., and Ahomadegbe, J.C. 2003. TP53 family members and human cancers. Hum. Mutat. 21: 182-191. [DOI] [PubMed] [Google Scholar]
- Bienz-Tadmor B., Zakut-Houri, R., Libresco, S., Givol, D., and Oren, M. 1985. The 5′ region of the p53 gene: Evolutionary conservation and evidence for a negative regulatory element. EMBO J. 4: 3209-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon J.-C., Renzing, J., Robertson, P.L., Fernandes, K.N., and Lane, D.P. 2002. Scotin, a novel p53-inducible proapoptotic protein located in the ER and the nuclear membrane. J. Cell. Biol. 158: 235-246. [Epub 2002 July 22.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky M.H., Nordstrom, W., Tsang, G., Kwan, E., Rubin, G.M., and Abrams, J.M. 2000. Drosophila p53 binds a damage response element at the reaper locus. Cell 101: 103-113. [DOI] [PubMed] [Google Scholar]
- Courtois S., Verhaegh, G., North, S., Luciani, M.G., Lassus, P., Hibner, U., Oren, M., and Hainaut, P. 2002. ΔN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene 21: 6722-6728. [DOI] [PubMed] [Google Scholar]
- Deguin-Chambon V., Vacher, M., Jullien, M., May, E., and Bourdon, J.C. 2000. Direct transactivation of c-Ha-Ras gene by p53: Evidence for its involvement in p53 transactivation activity and p53-mediated apoptosis. Oncogene 19: 5831-5841. [DOI] [PubMed] [Google Scholar]
- De Leo A.B., Jay, G., Appella, E., Dubois, G.C., Law, L.W., and Old, L.J. 1979. Detection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouse. Proc. Natl. Acad. Sci. 76: 2420-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohn M., Zhang, S., and Chen, X. 2001. p63α and ΔNp63α can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 20: 3193-3205. [DOI] [PubMed] [Google Scholar]
- Flaman J.-M., Waridel, F., Estreicher, A., Vannier, A., Limacher, J.-M., Gilbert, D., Iggo, R., and Frebourg, T. 1996. The human tumor suppressor gene p53 is alternatively spliced in normal cells. Oncogene 12: 813-818. [PubMed] [Google Scholar]
- Ghosh A., Stewart, D., and Matlashewski, G. 2004. Regulation of human p53 activity and cell localization by alternative splicing. Mol. Cell. Biol. 24: 7987-7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschneider D., Million, K., Meiller, A., Haddada, H., Puisieux, A., Benard, J., May, E., and Douc-Rasy, S. 2005. The neurogene BTG2TIS21/PC3 is transactivated by ΔNp73α via p53 specifically in neuroblastoma cells. J. Cell Sci. 118: 1245-1253. [Epub 2005 March 1.] [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane, D.P. 1988. Antibodies: A laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Harms K.L. and Chen, X. 2005. The C terminus of p53 family proteins is a cell fate determinant. Mol. Cell. Biol. 25: 2014-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Martinek, S., Joo, W.S., Wortman, J.R., Mirkovic, N., Sali, A., Yandell, M.D., Pavletich, N.P., Young, M.W., and Levine, A.J. 2000. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc. Natl. Acad. Sci. 97: 7301-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser M.D. and Iggo, R.D. 2002. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl. Acad. Sci. 99: 95-100. [Epub 2001 December 26.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress M., May, E., Cassingena, R., and May, P. 1979. Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. J. Virol. 31: 472-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb P. and Crawford, L. 1986. Characterization of the human p53 gene. Mol. Cell. Biol. 6: 1379-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D.P. and Crawford, L.V. 1979. T antigen is bound to a host protein in SV40-transformed cells. Nature 278: 261-263. [DOI] [PubMed] [Google Scholar]
- Linzer D.I.H. and Levine, A.J. 1979. Characterization of a 54 K dalton cellular SV40 tumor antigen present in SV40-transformed cells and in infected embryonal carcinoma cells. Cell 1: 43-52. [DOI] [PubMed] [Google Scholar]
- Liu W.L., Midgley, C., Stephen, C., Saville, M., and Lane, D.P. 2001. Biological significance of a small highly conserved region in the N terminus of the p53 tumour suppressor protein. J. Mol. Biol. 313: 711-731. [DOI] [PubMed] [Google Scholar]
- Liu G., Nozell, S., Xiao, H., and Chen, X. 2004. ΔNp73β is active in transactivation and growth suppression. Mol. Cell. Biol. 24: 487-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B., Gluba, W., Bernier, B., Turner, T., Mohammad, K., Guise, T., Sutherland, A., Thorner, M., and Scrable, H. 2004. Modulation of mammalian life span by the short isoform of p53. Genes & Dev. 18: 306-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melero J.A., Stitt, D.T., Mangel, W.F., and Carroll, R.B. 1979. Identification of new polypeptide species (48-55K) immunoprecipitable by antiserum to purified large T antigen and present in simian virus 40-infected and transformed cells. J. Virol. 93: 466-480. [DOI] [PubMed] [Google Scholar]
- Melino G., Lu, X., Gasco, M., Crook, T., and Knight, R.A. 2003. Functional regulation of p73 and p63: Development and cancer. Trends Biochem. Sci. 28: 663-670. [DOI] [PubMed] [Google Scholar]
- Midgley C.A., Fisher, C.J., Bartek, J., Vojtesek, B., Lane, D., and Barnes, D.M. 1992. Analysis of p53 expression in human tumours: An antibody raised against human p53 expressed in Escherichia coli. J. Cell Sci. 101: 183-189. [DOI] [PubMed] [Google Scholar]
- Ollmann M., Young, L.M., Di Como, C.J., Karim, F., Belvin, M., Robertson, S., Whittaker, K., Demsky, M., Fisher, W.W., Buchman, A., et al. 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101: 91-101. [DOI] [PubMed] [Google Scholar]
- Oren M. 2003. Decision making by p53: Life, death and cancer. Cell Death Differ. 1: 431-442. [DOI] [PubMed] [Google Scholar]
- Renzing J. and Lane, D.P. 1995. p53-dependent growth arrest following calcium phosphate-mediated transfection of murine fibroblasts. Oncogene 10: 1865-1868. [PubMed] [Google Scholar]
- Stephen C.W., Helminen, P., and Lane, D.P. 1995. Characterisation of epitopes on human p53 using phage-displayed peptide libraries: Insights into antibody-peptide interactions. J. Mol. Biol. 248: 58-78. [DOI] [PubMed] [Google Scholar]
- Tyner S.D., Venkatachalam, S., Choi, J., Jones, S., Ghebranious, N., Igelmann, H., Lu, X., Soron, G., Cooper, B., Brayton, C., et al. 2002. p53 mutant mice that display early ageing-associated phenotypes. Nature 415: 45-53. [DOI] [PubMed] [Google Scholar]
- Vojtesek B., Dolezalova, H., Lauerova, L., Svitakova, M., Havlis, P., Kovarik, J., Midgley, C.A., and Lane, D.P. 1995. Conformational changes in p53 analysed using new antibodies to the core DNA binding domain of the protein. Oncogene 10: 389-393. [PubMed] [Google Scholar]
- Wolf D., Harris, N., Goldfinger, N., and Rotter, V. 1985. Isolation of a full-length mouse cDNA clone coding for an immunologically distinct p53 molecule. Mol. Cell. Biol. 5: 127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Liu, Y., Lee, L., Miner, Z., and Kulesz-Martin, M. 1994. Wild-type alternatively spliced p53: Binding to DNA and interaction with the major p53 protein in vitro and in cells. EMBO J. 13: 4823-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Nomoto, S., Hoque, M.O., Dracheva, T., Osada, M., Lee, C.C., Dong, S.M., Guo, Z., Benoit, N., Cohen, Y., et al. 2003. ΔNp63α and TAp63α regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 63: 2351-2357. [PubMed] [Google Scholar]
- Xirodimas D.P., Saville, M.K., Bourdon, J.-C., Hay, R.T., and Lane, D.P. 2004. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118: 83-97. [DOI] [PubMed] [Google Scholar]
- Yang A., Kaghad, M., Caput, D., and McKeon, F. 2002. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 18: 90-95. [DOI] [PubMed] [Google Scholar]
- Yin Y., Stephen, C.W., Luciani, M.G., and Fahraeus, R. 2002. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat. Cell Biol. 4: 462-467. [DOI] [PubMed] [Google Scholar]
- Zhu J., Zhou, W., Jiang, J., and Chen, X. 1998. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J. Biol. Chem. 273: 13030-13036. [DOI] [PubMed] [Google Scholar]
- Zhu J., Zhang, S., Jiang, J., and Chen, X. 2000. Definition of the p53 functional domains necessary for inducing apoptosis. J. Biol. Chem. 275: 39927-39934. [DOI] [PubMed] [Google Scholar]