Abstract
Hfq-binding antisense small RNAs of Escherichia coli, SgrS and RyhB, mediate the destabilization of target mRNAs in an RNase E-dependent manner. SgrS, whose expression is induced in response to phosphosugar stress, act on the ptsG mRNA encoding a major glucose transporter, while RyhB, whose expression is induced in response to Fe depletion, acts on several mRNAs encoding Fe-binding proteins. In this report, we addressed the question of how SgrS and RyhB RNAs cooperate with RNase E to destabilize the target mRNAs. We demonstrate that Hfq along with SgrS and RyhB copurified with RNase E but not with truncated RNase E. In addition, we show that RNase E but not other degradosome components copurified with Hfq. Taken together, we conclude that RNase E forms variable ribonucleoprotein complexes with Hfq/small RNAs through its C-terminal scaffold region. These complexes, distinct from the RNA degradosome, may act as specialized RNA decay machines that initiate the degradation of mRNAs targeted by each small RNA. The present finding has uncovered the mechanical basis of mRNA destabilization mediated by bacterial small RNAs. The formation of ribonucleoprotein complexes containing RNases could be a general way by which small RNAs destabilize target mRNAs in both prokaryotes and eukaryotes.
Keywords: Small RNA, Hfq, RNase E, ribonucleoprotein complex, regulated mRNA degradation
RNase E of Escherichia coli is a major endoribonuclease responsible for the degradation and/or processing of mRNAs and stable RNAs. It forms a multiprotein complex called the RNA degradosome with a 3′-exoribonuclease (polynucleotide phosphorylase, PNPase), a DEAD-box RNA helicase (RNA helicase B, RhlB), a glycolytic enzyme (enolase), and several other proteins (Carpousis et al. 1994; Miczak et al. 1996; Py et al. 1996). The RNase E polypeptide is composed of three domains, an N-terminal catalytic region, a central RNA-binding domain, and a C-terminal scaffold region responsible for binding of the associated proteins (McDowall and Cohen 1996; Vanzo et al. 1998; Carpousis 2002). It is believed that the RNA degradosome acts as a general RNA decay machine in which the components of the degradosome cooperate during the decay of many RNAs. In fact, it is reported that the major components of the degradosome can functionally interact with each other in the degradation of several RNAs either in vivo or in vitro (Py et al. 1996; Coburn et al. 1999; Khemici and Carpousis 2004; Prud'homme-Genereux et al. 2004).
We found previously that the ptsG mRNA encoding the membrane component of the major glucose transporter in E. coli is markedly destabilized in an RNase E-dependent fashion when the glycolytic pathway is blocked either by mutations at its early stages or by treatment with a nonmetabolizable glucose analog (Kimata et al. 2001). Accumulation of glucose-6-phosphate (G6P), fructose 6-phosphate, or α-methylglucoside 6-phosphate (αMG6P) triggers the RNase E-mediated destabilization of ptsG mRNA (Morita et al. 2003). More recently, we have discovered that the C-terminal scaffold region of RNase E as well as enolase is required for the rapid degradation of ptsG mRNA in response to phosphosugar stress (Morita et al. 2004). This destabilization of ptsG mRNA has been shown to be dependent on an RNA chaperone Hfq (Morita et al. 2004; Kawamoto et al. 2005). Hfq is known to stimulate base-pairing between various small regulatory RNAs and their target mRNAs to regulate mRNA translation and stability (Gottesman 2004; Storz et al. 2004; Valentin-Hansen et al. 2004). Therefore, it was suggested that an Hfq-binding small RNA may be involved in the destabilization of ptsG mRNA. Indeed, Vanderpool and Gottesman have discovered that a small RNA called SgrS (RyaA), initially identified by its binding to Hfq (Zhang et al. 2003), mediates the destabilization of ptsG mRNA (Vanderpool and Gottesman 2004). They have shown that SgrS is induced in response to phosphosugar accumulation, leading to the degradation of ptsG mRNA, presumably through SgrS-ptsG pairing. Furthermore, it has been shown that mRNA localization to the inner membrane coupled with the membrane insertion of nascent peptide is required for the Hfq/SgrS-dependent ptsG mRNA destabilization by reducing subsequent rounds of translation (Kawamoto et al. 2005; Vanderpool and Gottesman 2005).
The degradation of mRNAs encoding Fe-binding or Fe-storage proteins in response to Fe depletion provides another example for the regulated mRNA degradation under a stress condition (Masse and Gottesman 2002; Masse et al. 2003). In this case, the degradation of target mRNAs is mediated by RyhB RNA, another Hfq-binding small regulatory RNA. The RyhB-mediated mRNA degradation also occurs in an RNase E-dependent manner and is coupled with RyhB turnover (Masse et al. 2003). In addition, the C-terminal scaffold region of RNase E apparently participates in the RyhB-mediated degradation of target mRNAs (Masse et al. 2003).
Despite the significant progress mentioned above, the mechanisms by which Hfq/small RNAs mediate the destabilization of target mRNAs have remained unclear. In particular, involvement of both RNase E and Hfq/small RNAs in the regulated mRNA degradation has raised the intriguing question of how RNase E cooperates with a small RNA and Hfq to destabilize selectively the target mRNAs. In the present study, we report experimental results that provide an answer to this question. We have found that Hfq is stably associated with the C-terminal scaffold region of RNase E. More importantly, we have found that SgrS and RyhB associate with RNase E through Hfq. These findings have led us to conclude that RNase E forms variable ribonucleoprotein complexes with various small RNAs along with Hfq to act as specialized RNA decay machines that degrade the target mRNAs guided by each small RNA. The formation of ribonucleoprotein complexes containing RNases could be a general way by which small RNAs destabilize target mRNAs in both prokaryotes and eukaryotes.
Results
The C-terminal scaffold region of RNase E is required for the destabilization of ptsG mRNA in response to αMG6P accumulation
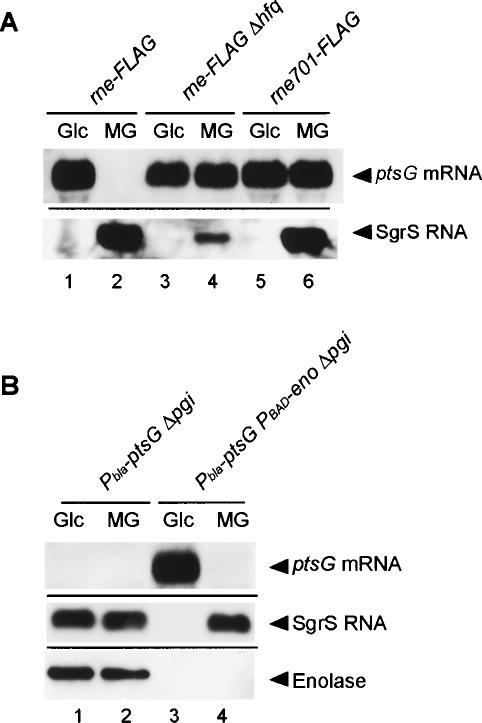
SgrS is expressed upon phosphosugar stress and acts with Hfq to destabilize the ptsG mRNA in an RNase E-dependent manner (Morita et al. 2004; Vanderpool and Gottesman 2004; Kawamoto et al. 2005). The C-terminal scaffold region of RNase E is required for the destabilization of ptsG mRNA mediated by SgrS in response to G6P accumulation (Morita et al. 2004). We first examined whether this is also true when cells are exposed to αMG. We used strains in which the chromosomal rne gene was replaced by the rne-Flag encoding the C-terminally Flag-tagged RNase E (RNase E-Flag) or the rne701-Flag encoding the Flag-tagged truncated RNase E lacking the C-terminal scaffold region (RNase E701-Flag). When the rne-Flag strain was exposed to αMG, SgrS RNA was induced, resulting in the destabilization of ptsG mRNA (Fig. 1A, lanes 1,2). When the rne gene was replaced by the rne701-Flag allele, the degradation of ptsG mRNA in response to the αMG addition was abolished without losing the induction of SgrS (Fig. 1A, lanes 5,6). Thus, the C-terminal scaffold region of RNase E is necessary for the rapid degradation of ptsG mRNA in response to αMG6P accumulation. The degradation of ptsG mRNA mediated by SgrS no longer occurred in a newly constructed strain totally lacking the hfq gene (Fig. 1A, lanes 3,4). The SgrS level in the hfq strain was significantly decreased presumably because of its reduced stability in the absence of Hfq as observed in other Hfq-binding small RNAs (Masse et al. 2003).
Figure 1.
(A) Effects of the C-terminal truncation of RNase E and hfq deletion on the expression of SgrS RNA and ptsG mRNA. TM338 (rne-Flag-cat), TM618 (rne-Flag-cat Δhfq), and TM528 (rne701-Flag-cat) cells were grown in LB medium. At A600 = 0.6, the culture was split, 1% glucose (Glc) or α-methyl-glucoside (αMG) was added to each culture, and incubation was continued for 20 min. Total RNAs were prepared and 15 or 5 μg of each RNA sample was subjected to Northern blot analysis using a ptsG or sgrS probe, respectively. (B) Effect of enolase depletion on the expression of SgrS RNA and ptsG mRNA. TM461 (Δpgi Pbla-ptsG) and TM509 (Δpgi Pbla-ptsG PBAD-eno) cells were grown in M9 medium containing 0.2% succinate and 0.02% glycerol. At A600 = 0.6, crude extracts and total RNAs were prepared. The crude extracts corresponding to 0.01 A600 unit were subjected to Western blot analysis using anti-enolase antibody. Each RNA sample (15 or 5 μg) was subjected to Northern blot analysis using a ptsG or sgrS probe, respectively.
Enolase is required for the destabilization of ptsG mRNA in response to G6P but not αMG6P accumulation
In our previous study, we found that depleting enolase but not PNPase or RhlB blocked the rapid degradation of ptsG mRNA in response to G6P accumulation (Morita et al. 2004). To examine the effect of enolase depletion on the rapid degradation of ptsG mRNA in cells exposed to αMG, strains lacking the pgi gene encoding phosphoglucose isomerase, TM461 (Δpgi Pbla-ptsG) and TM509 (Δpgi Pbla-ptsG PBAD-eno), were grown in the M9 minimum medium containing succinate plus glycerol without arabinose. Both the Pbla-ptsG and PBAD-eno are on the chromosome in these strains. We used the PBAD-eno strain because we failed to construct the eno disruption mutant. Glucose or αMG was added to the culture, and the incubation was continued for 30 min to allow the accumulation of G6P or αMG6P. The total proteins and RNAs prepared were subjected to Western and Northern blot analyses. As expected, the ptsG mRNA was destabilized in the presence of enolase when cells were exposed to either glucose or αMG (Fig. 1B, lanes 1,2). In addition, we confirmed that the rapid degradation of ptsG mRNA was eliminated under enolase depletion in the pgi cells exposed to glucose (Fig. 1B, lane 3). To our surprise, however, the destabilization of ptsG mRNA was observed upon the addition of αMG even when enolase was depleted (Fig. 1B, lane 4). Interestingly, SgrS was not detectable in pgi cells under the enolase depletion upon the addition of glucose, while it was highly expressed upon the addition of αMG (Fig. 1B, lanes 3,4). When enolase was present, SgrS was efficiently expressed in cells exposed to either αMG or glucose (Fig. 1B, lanes 1,2). The Hfq level was essentially the same in four different conditions (data not shown). These results imply that the role of enolase in the rapid degradation of ptsG mRNA in pgi cells exposed to glucose is to allow the efficient expression of SgrS. We also observed that SgrS accumulates in the pgi rne701-Flag cells upon the glucose addition (data not shown). Thus, the enolase requirement is apparently independent of the degradosome. Although it is intriguing how enolase acts to stimulate the SgrS expression in response to G6P accumulation, we did not pursue this issue further in the present study.
Hfq associates with RNase E through its C-terminal scaffold region
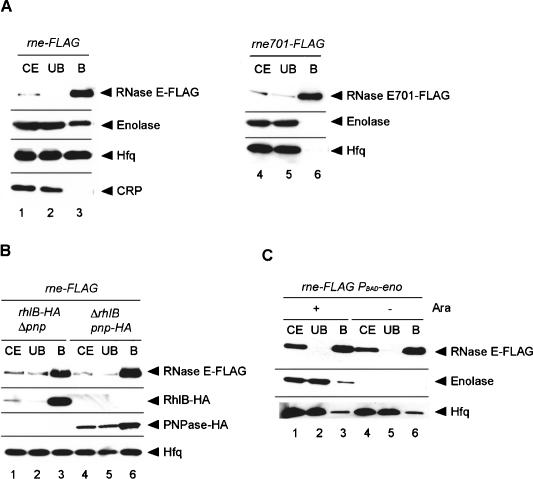
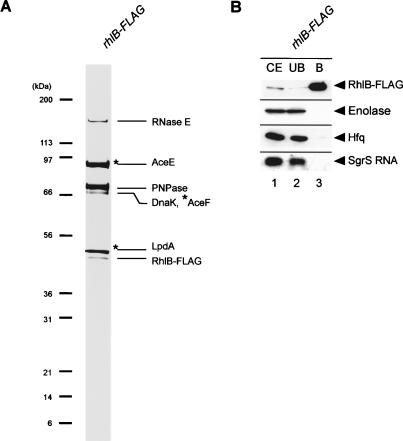
The truncation of the C-terminal scaffold region of RNase E eliminates the rapid degradation of ptsG mRNA without affecting the induction of SgrS under phosphosugar stress. An important question is why the ptsG mRNA becomes sensitive to the full-length RNase E but not to the truncated RNase E. This can be achieved if the full-length RNase E but not the truncated RNase E is recruited to the ptsG mRNA depending on Hfq and/or SgrS RNA. To test whether this is the case, a pull-down assay was carried out by using a cell extract prepared from the rne-Flag strain grown in LB medium. The cell extract was incubated with anti-Flag M2-agarose beads, and proteins were eluted from the beads. Proteins in the bound fraction along with those in the crude extract and in the unbound fraction were analyzed by Western blotting using several different antibodies (Fig. 2A, lanes 1-3). The anti-Flag-probed Western blot revealed that RNase E-Flag was efficiently recovered in the bound fraction (Fig. 2A, lane 3). Enolase, one of the major components of the RNA degradosome, but not a control CRP protein copurified with RNase E-Flag (Fig. 2A, lane 3) as shown previously (Morita et al. 2004). Importantly, we newly found that Hfq copurified with RNase E-Flag (Fig. 2A, lane 3). It was estimated that at least 10%-20% of total Hfq is associated with RNase E-Flag. The pull-down assay was also performed with a cell extract prepared from the rne701-Flag strain (Fig. 2A, lanes 3,4). Although RNase E701-Flag was efficiently recovered in the bound fraction, neither Hfq nor enolase copurified with RNase E701-Flag (Fig. 2A, lane 6). These results imply that Hfq is associated with RNase E through its C-terminal scaffold region.
Figure 2.
(A) Physical interaction between RNase E and Hfq. TM338 (lanes 1-3) and TM528 (lanes 4-6) were grown in 200 mL of LB medium to A600 = 0.6. Crude extracts were prepared and subjected to the pull-down assay using anti-Flag agarose. The crude extract (CE), unbound fraction (UB), and bound fraction (B) were analyzed by Western blotting. Anti-Flag, anti-CRP, anti-enolase, and anti-Hfq antibodies were used to detect RNase E-Flag (RNase E701-Flag), CRP, enolase, and Hfq, respectively. (B) Effect of deletion of pnp or rhlB gene on the Hfq-RNase E interaction. TM581 (rhlB-HA Δpnp rne-Flag) and TM583 (ΔrhlB pnp-HA rne-Flag) cells were grown in LB medium to A600 = 0.6. Crude extracts were prepared and subjected to the pull-down assay using anti-Flag agarose. The crude extract, unbound fraction, and bound fraction were analyzed by Western blotting. Anti-Flag, anti-HA, and anti-Hfq antibodies were used to detect RNase E, PNPase-HA/RhlB-HA, and Hfq, respectively. (C) Effect of enolase depletion on the Hfq-RNase E interaction. TM452 (rne-Flag cat-PBAD-eno) cells were grown in M9 medium containing 0.2% succinate and 0.02% glycerol. At A600 = 0.4, the culture was split and 1% arabinose was added to one culture, and incubation was continued for 1 h. Crude extracts were prepared and subjected to the pull-down assay using anti-Flag agarose. The crude extract, unbound fraction, and bound fraction were analyzed by Western blotting. Anti-Flag, anti-enolase, and anti-Hfq antibodies were used to detect RNase E, enolase, and Hfq, respectively.
Neither PNPase, RhlB, nor enolase is involved in the Hfq-RNase E interaction
PNPase, RhlB, and enolase are associated with the C-terminal scaffold region of RNase E as major components of the degradosome (Vanzo et al. 1998; Carpousis 2002). To examine whether PNPase or RhlB is required for the Hfq-RNase E interaction, the pull-down assay was carried out by using cell extracts lacking either of these two proteins. In these experiments, rne-Flag rhlB-HA Δpnp and rne-Flag ΔrhlB pnp-HA strains were used. The pnp-HA and rhlB-HA alleles encode HA-tagged PNPase and RhlB, respectively (Morita et al. 2004). The presence or absence of each of the degradosome components was confirmed by Western blotting using anti-Flag and anti-HA antibodies (Fig. 2B). The anti-Hfq-probed Western blot revealed that Hfq copurified with RNase E-Flag even in the absence of PNPase or RhlB, indicating that neither PNPase nor RhlB is required for the RNase E-Hfq interaction (Fig. 2B, lanes 3,6). To examine whether enolase is involved in the Hfq-RNase E interaction, the rne-Flag PBAD-eno strain in which the eno gene is under the control of an arabinose-inducible promoter PBAD (Morita et al. 2004) was grown in the presence or absence of arabinose. As expected, enolase was produced only in the presence of arabinose (Fig. 2C). The pull-down assay was performed with anti-Flag M2-agarose beads (Fig. 2C). Hfq copurified with RNase E-Flag in the presence and absence of enolase, indicating that enolase is not required for the RNase E-Hfq interaction (Fig. 2C, lanes 3,6). These results imply that neither PNPase, RhlB, nor enolase is involved in the Hfq-RNase E interaction.
SgrS RNA associates with RNase E through Hfq
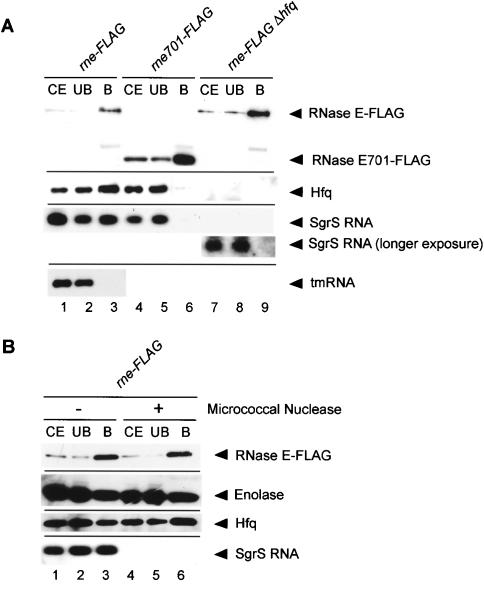
The finding that Hfq is tightly associated with RNase E has raised the attractive possibility that SgrS RNA also interacts with RNase E through Hfq when it is expressed. To test this, the pull-down assay was performed with a cell extract prepared from the rne-Flag cells grown in the presence of α-MG. Hfq again copurified with RNase E-Flag, indicating that the phosphosugar stress does not affect the RNase E-Hfq interaction (Fig. 3A, lanes 1-3). Protein samples in three fractions were treated with phenol and subjected to Northern blotting. A significant amount of SgrS RNA was detected in the affinity-purified RNase E-Flag sample (Fig. 3A, lane 3). Semiquantitative analyses of SgrS RNA in three fractions indicate that ∼10%-20% of total SgrS RNA is associated with RNase E-Flag. However, tmRNA, a representative of non-Hfq-binding RNAs, was not detected in the affinity-purified RNase E-Flag sample (Fig. 3A, lane 3). This suggests that the association of SgrS with RNase E-Flag through Hfq is not due to the nonspecific RNA binding ability of RNase E and/or Hfq. As expected, no SgrS RNA was detected in the RNase E701-Flag sample prepared from the rne701-Flag cells grown in the presence of α-MG (Fig. 3A, lane 6) or in the RNase E-Flag sample prepared from the Δhfq cells grown in the presence of α-MG even when the gel was overexposed (Fig. 3A, lane 9). These results strongly suggest that SgrS associates with RNase E through Hfq.
Figure 3.
(A) Physical interaction between RNase E and SgrS RNA. TM338 (rne-Flag-cat), TM618 (rne-Flag-cat Δhfq), and TM528 (rne701-Flag-cat) cells were grown in LB medium. At A600 = 0.6, 1% αMG was added to each culture, and incubation was continued for 20 min. For analysis of proteins associated with RNase E, crude extracts were prepared and subjected to the pull-down assay using anti-Flag agarose. The crude extracts, unbound fractions, and bound fractions were analyzed by Western blotting using anti-Flag and anti-Hfq antibodies. For analysis of RNAs associated with RNase E analysis, 5 μL of deproteinized crude extracts, unbound fractions, and bound fractions (see Materials and Methods) were subjected to Northern blotting using the sgrS and ssrA probes. (B) Effect of nuclease treatment on the Hfq-RNase E interaction. (Lanes 4-6) A crude extract was prepared from TM338 (rne-Flag-cat), treated with micrococcal nuclease, and subjected to the pull-down assay using anti-Flag agarose. (Lanes 1-3) As a control, the extract without micrococcal nuclease was also subjected to the pull-down assay.
Hfq directly associates with RNase E
Hfq is an RNA-binding protein (Valentin-Hansen et al. 2004). In addition, it is known that RNase E-based degradosome contains a significant amount of heterogeneous RNAs primarily consisting of ribosomal RNAs and their fragments (Bessarab et al. 1998; Lin-Chao et al. 1999). This raises the question of whether the interaction between RNase E and Hfq is a direct protein-protein interaction or the two proteins associate via RNAs. To address this question, a cell extract of the rne-Flag strain grown in the presence of α-MG was incubated with micrococcal nuclease in the presence of Ca2+ and then subjected to the pull-down assay using anti-Flag M2-agarose beads. Proteins were analyzed by Western blotting using anti-Flag, anti-Hfq, and anti-enolase antibodies. The treatment with micrococcal nuclease did not affect the RNase E-Hfq interaction (Fig. 3B, lanes 4-6). SgrS RNA was no longer detected in all three fractions, indicating that RNAs were efficiently degraded by the nuclease treatment (Fig. 3B, lanes 4-6). The incubation without micrococcal nuclease did not affect the association of SgrS with RNase E/Hfq (Fig. 3B, lanes 1-3). These results suggest that Hfq directly associates with RNase E.
Analysis of proteins associated with affinity-purified RNase E
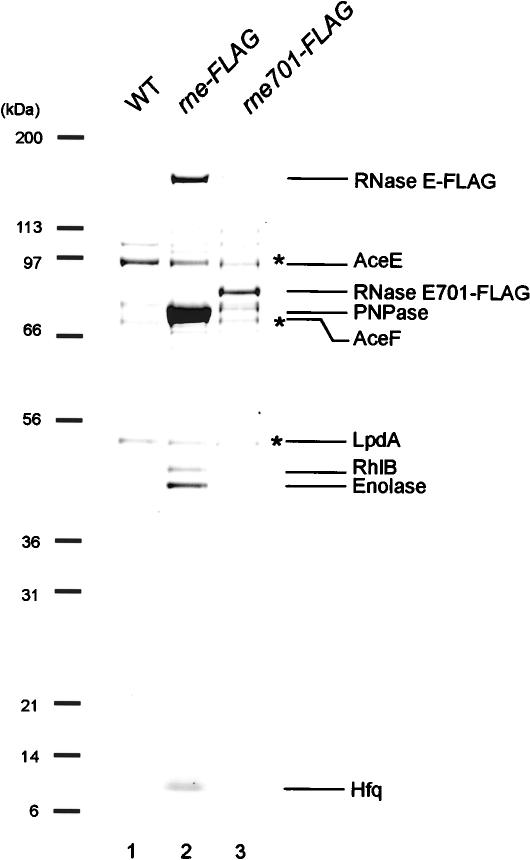
In order to characterize further the interaction between RNase E and Hfq/SgrS, a cell extract was prepared in the presence of a detergent (0.1% Tween 20) and subjected to affinity-purification. Most RNase E was recovered in the soluble fraction in our experimental conditions. The protein components of the affinity-purified RNase E-Flag and RNase E701-Flag prepared from cells grown in the presence of α-MG were analyzed by SDS-PAGE and Coomassie staining. As a control, proteins bound to the anti-Flag M2-agarose beads from a cell extract lacking the Flag fusion protein were also analyzed. A polypeptide corresponding to Hfq was clearly detected along with proteins corresponding to RNase E-Flag, PNPase, RhlB, and enolase in the affinity-purified RNase E-Flag (Fig. 4A, lane 2). The identity of these proteins was verified by Western blotting and/or by mass spectrometry analysis. As expected, these proteins were no longer detected in the affinity-purified RNase E701-Flag (Fig. 4A, lane 3). The affinity-purified RNase E-Flag and RNase E701-Flag contained three additional prominent polypeptides (marked with asterisks) that were also detected in the control (Fig. 4A, lane 12), suggesting that these proteins bind to the anti-Flag antibodies. Mass spectrometry analysis revealed that these polypeptides are subunits of pyruvate dehydrogenase. (Prud'homme-Genereux et al. 2004). To examine whether the subunits of pyruvate dehydrogenase affect the association of Hfq/small RNAs with RNase E and the small RNAs-mediated destabilization of target mRNAs, we constructed a strain lacking both the aceE and aceF genes encoding E1 and E2 subunits of pyruvate dehydrogenase. The analysis of affinity-purified RNase E-Flag revealed that the deletion of the aceE-aceF did not affect the RNase E-Hfq/SgrS interaction (data not shown).
Figure 4.
Analysis of protein composition of affinity-purified RNase E-Flag. IT1568 (wild-type), TM522 (rne-Flag), and TM611 (rne701-Flag) were grown in 1 L of LB medium to A600 = 0.6. Crude extracts were prepared, and RNase E-Flag and RNase E701-Flag were affinity-purified using anti-Flag agarose as described in Materials and Methods. The affinity-purified samples were analyzed by a 4%-12% polyacrylamide gradient-0.1% SDS gel electrophoresis and CBB staining. Each of the protein bands was excised from the stained gel and identified by mass spectrometry. The positions of size markers are shown on the left.
Affinity-purified Hfq contains RNase E but not RhlB helicase and enolase
The copurification experiment using RNase E-Flag has established that Hfq and SgrS RNA are associated with the C-terminal scaffold region of RNase E. However, it is not clear yet whether Hfq binds to RNase E together with other degradosome components. To address this question, we constructed a strain carrying the hfq-Flag rne-HA allele. The addition of α-MG to this strain caused the rapid degradation of ptsG mRNA along with the induction of SgrS RNA, indicating that Hfq-Flag is functional (data not shown). A cell extract prepared from the hfq-Flag rne-HA strain grown in LB medium was incubated with anti-Flag M2-agarose beads, and proteins bound to the beads were analyzed by SDS-PAGE and Coomassie staining (Fig. 5A, lane 1). RNase E clearly copurified with Hfq, confirming the specific interaction between Hfq and RNase E. Interestingly, RhlB helicase and enolase did not copurify with Hfq. This implies that the RNase E-Hfq complex no longer binds to enolase and RhlB helicase. On the other hand, a significant amount of PNPase copurified with Hfq. This may be due to the specific interaction between PNPase and Hfq (Mohanty et al. 2004). In fact, PNPase copurified with Hfq even when Hfq-Flag was affinity-purified from hfq-Flag rne701-HA cells expressing a truncated RNase E701-HA that lacks the C-terminal scaffold region (Fig. 5A, lane 2). Taken together, we conclude that the Hfq-RNase E complex no longer binds to other degradosome components. In addition, ribosomal S1 protein and several low-molecular-weight polypeptides of various sizes were also copurified with Hfq. Mass spectrometry revealed that most of these bands correspond to various ribosomal proteins, suggesting that ribosomal proteins or ribosome itself associate with Hfq. It should be noted that the association of S1 with Hfq is previously reported (Sukhodolets and Garges 2003).
Figure 5.
(A) Analysis of protein composition of affinity-purified Hfq-Flag. TM651 (hfq-Flag rne-HA) and TM652 (hfq-Flag rne701-HA) were grown in 1 L of LB medium to A600 = 0.6. Hfq-Flag was affinity-purified and analyzed by a 4%-12% polyacrylamide gradient-0.1% SDS gel electrophoresis and CBB staining. Each of the protein bands was excised from the stained gel and identified by mass spectrometry. The positions of size markers are shown on the left.(B) Western blot analysis of affinity-purified Hfq-Flag. TM651 (lanes 1-3) and TM652 (lanes 3-6) were grown in 200 mL of LB medium to A600 = 0.6. Crude extracts were prepared and subjected to the pull-down assay using anti-Flag agarose. The crude extract (CE), unbound fraction (UB), and bound fraction (B) were analyzed by Western blotting. Anti-Flag, anti-enolase, and anti-HA antibodies were used to detect Hfq-Flag, enolase, and RNase E-HA (RNase E701-HA), respectively.
The specific interaction between RNase E and Hfq was further investigated by the pull-down assay using the hfq-Flag rne-HA and hfq-Flag rne701-HA strains (Fig. 5B). Proteins bound to the anti-Flag M2-agarose beads were analyzed by Western blotting. As expected, RNase E-HA but not RNase E701-HA copurified with Hfq-Flag, confirming again that Hfq specifically associates with the C-terminal scaffold region of RNase E.
Affinity-purified RhlB contains RNase E but not Hfq
The experiments mentioned above strongly suggest that Hfq and other degradosome components do not associate with RNase E simultaneously. If this is the case, it is expected that RNase E but not Hfq copurifies with RhlB, for example. To test this prediction, we constructed the rhlB-Flag rne-HA strain. A cell extract was incubated with anti-Flag M2-agarose beads, and proteins bound to the beads were analyzed by SDS-PAGE and Coomassie staining (Fig. 6A). Proteins in the crude extract, in the unbound and bound fractions, were also analyzed by Western blotting (Fig. 6B). As expected, RNase E copurified with RhlB, confirming the specific interaction between RhlB and RNase E. On the other hand, no Hfq copurified with RhlB. In addition, Northern blot analysis revealed that SgrS did not copurify with RhlB (Fig. 6B). These results confirm that Hfq/SgrS does not bind to the RNA degradosome containing RhlB. Another unexpected observation was that the affinity-purified RhlB does not contain enolase (Fig. 6A,B), suggesting that the bindings of RhlB and enolase to RNase E are also mutually exclusive. This conclusion is consistent with a recent report that RhlB helicase did not bind to the enolase-bound C-terminal domain of RNase E (Callaghan et al. 2004).
Figure 6.
(A) Protein composition of affinity-purified RhlB-Flag. TM647 (rhlB-Flag) was grown in 1 L of LB medium to A600 = 0.6. RhlB-Flag was affinity-purified and analyzed by a 4%-12% polyacrylamide gradient-0.1% SDS gel electrophoresis and CBB staining. Each of the protein bands was excised from the stained gel and identified by mass spectrometry. The positions of size markers are shown on the left.(B) Western and Northern analyses of affinity-purified Hfq-Flag. TM647 (rhlB-Flag) was grown in 200 mL of LB medium. At A600 = 0.6, 1% αMG was added and incubation was continued for 20 min. A crude extract was prepared and subjected to the pull-down assay using anti-Flag agarose. The crude extract (CE), unbound fraction (UB), and bound fraction (B) were analyzed by Western blotting. Anti-Flag, anti-enolase, and anti-HA antibodies were used to detect Hfq-Flag, enolase, and RNase E-HA (RNase E701-HA), respectively. For analysis of SgrS RNA, deproteinized crude extracts, unbound fractions, and bound fractions were subjected to Northern blotting using the sgrS probe.
RyhB also associate with RNase E through Hfq
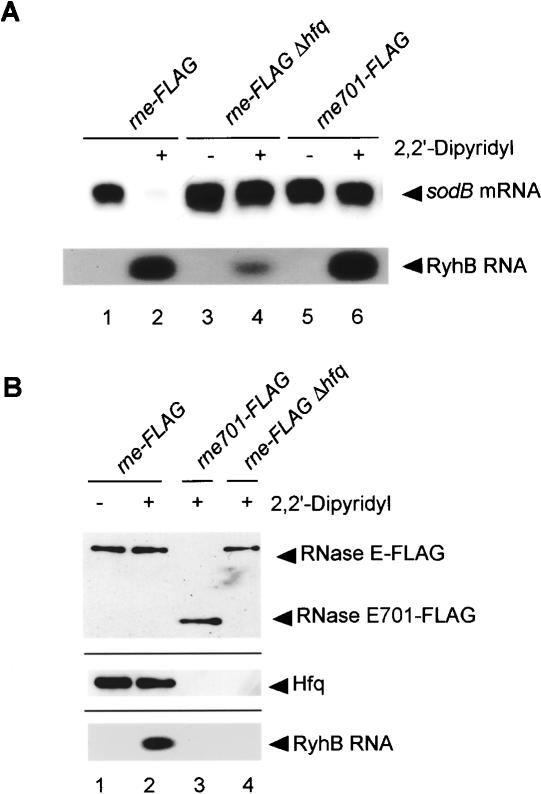
To examine whether other Hfq-binding small RNAs associate with RNase E through Hfq, similar experiments were carried out focusing on RyhB RNA and its target sodB mRNA encoding superoxide dismutase (Masse and Gottesman 2002; Masse et al. 2003). It is known that the addition of the Fe chelator 2, 2′-dipyridyl, to wild-type cells rapidly induced RyhB RNA along with the destabilization of sodB mRNA encoding superoxide dismutase (Masse and Gottesman 2002; Masse et al. 2003). We examined the effect of the Fe depletion on RyhB RNA induction and on sodB mRNA destabilization in our rne-Flag strain. As shown in Figure 7A, upon the addition of 2,2′-dipyridyl, RyhB RNA was induced, resulting in the destabilization of sodB mRNA. The degradation of sodB mRNA was essentially suppressed along with a significant reduction of RyhB in the Δhfq strain or in the rne701-Flag strain (Fig. 7A, lanes 3-6).These results are consistent with the previous data and suggest that RyhB destabilizes its target mRNA by the same mechanism as SgrS does.
Figure 7.
Analysis of the interaction between RNase E and RyhB RNA. (A) Effects of the C-terminal truncation of RNase E and hfq deletion on the expression of RyhB RNA and sodB mRNA. TM338 (rne-Flag-cat), TM618 (rne-Flag-cat Δhfq), and TM528 (rne701-Flag-cat) cells were grown in LB medium. At A600 = 0.6, the culture was split, 250 μM 2,2′-didipyridyl was added to one culture, and incubation was continued for 15 min. Total RNAs were prepared and 10 μg of each RNA sample was subjected to Northern blot analysis using sodB or ryhB probes. (B) Physical interaction between RNase E and RyhB RNA. TM338, TM618, and TM528 cells were grown in LB medium. At A600 = 0.6, the culture was split, 250 μM 2,2′-didipyridyl was added to one culture, and incubation was continued for 15 min. For analysis of proteins associated with RNase E, crude extracts were prepared and subjected to the pull-down assay using anti-Flag agarose. Proteins bound to the beads were analyzed by Western blotting in which 1 μL of each bound fraction was subjected to the gel electrophoresis. Anti-Flag and anti-Hfq antibodies were used to detect RNase E (RNase E701) and Hfq, respectively. For analysis of RNAs associated with RNase E analysis, 5 μL of deproteinized bound fraction (see Materials and Methods) was subjected to Northern blotting using the ryhB probe.
Cell extracts were prepared from the rne-Flag, rne701-Flag, and rne-Flag Δhfq strains grown in the presence or absence of 2,2′-dipyridyl and incubated with anti-Flag M2-agarose beads. Proteins bound to the agarose beads were analyzed by Western blotting using anti-Flag and anti-Hfq antibodies. Hfq copurified with RNase E-Flag, indicating that Fe depletion also does not affect the RNase E-Hfq interaction (Fig. 7B, lanes 1,2). The affinity-purified RNase E-Flag was treated with phenol and subjected to Northern blotting. A significant amount of RyhB RNA was detected in the affinity-purified RNase E-Flag sample prepared from cells grown in the presence but not absence of 2,2′-dipyridyl (Fig. 7B, lanes 1,2). RyhB RNA was not detected in the RNase E701-Flag sample prepared from cells grown in the presence of 2,2′-dipyridyl (Fig. 7B, lane 3) or in the RNase E-Flag sample prepared from Δhfq cells grown in the presence of 2,2′-dipyridyl (Fig. 7B, lane 4). We conclude that RyhB RNA also forms a ribonucleoprotein complex with RNase E through Hfq.
Discussion
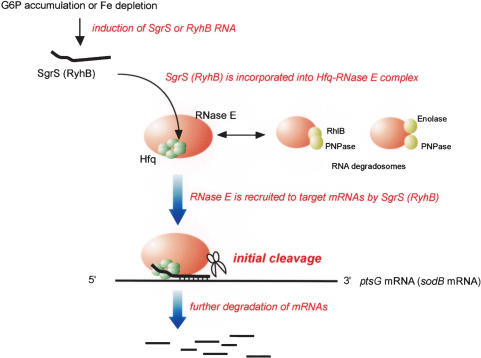
A salient common feature regarding the functions of SgrS and RyhB is to stimulate the degradation of their target mRNAs in an RNase E-dependent fashion. However, the mechanisms by which these small RNAs lead to the destabilization of target mRNAs have remained unclear. The major question addressed in the present study is how small RNAs and Hfq facilitate the degradation of target mRNAs by RNase E. There are two possible ways to explain the functional cooperation between RNase E and small RNAs/Hfq. First, it can be explained if the mRNA target itself becomes more sensitive to RNase E after base-pairing with the cognate small RNA. Alternatively, it could occur if RNase E is recruited near the target mRNAs by the small RNA and/or Hfq. We have discovered that Hfq is stably associated with the C-terminal scaffold region of RNase E. More importantly, we have discovered that SgrS and RyhB associate with RNase E through Hfq when they are expressed. These findings have led us to conclude that RNase E forms ribonucleoprotein complexes with Hfq along with small antisense RNAs to act as specialized RNA decay machines. The physical association of Hfq/small RNAs with RNase E nicely explains how the functional cooperation of small RNAs/Hfq and RNase E is achieved. It also explains how only specific mRNAs are destabilized under a specific stress condition in which a specific cognate small RNA is expressed. We would like to propose the following model for the combined action of small RNAs/Hfq and RNase E (Fig. 8). Under a particular stress condition such as phosphosugar stress and Fe depletion, a specific small RNA is highly expressed and associates with RNase E through Hfq to form a specialized ribonucleoprotein complex. The small RNA/Hfq appears to be able to associate with RNase E only when its C-terminal scaffold region is not occupied with other degradosome components because Hfq and major components of the degradosome do not bind to RNase E simultaneously. Alternatively, the association of small RNA/Hfq with RNase E may reduce the affinity between RNase E and other degradosome components. In either case, each specific complex containing a small antisense RNA, Hfq, and RNase E, which is clearly distinct from the degradosome itself, could act only on the cognate target mRNAs as a specialized RNA decay machine. The role of each small RNA in mRNA destabilization is to guide RNase E near target mRNAs through Hfq while Hfq acts as an adaptor between RNase E and small RNAs. This model also explains why a small regulatory RNA and its mRNA targets are rapidly degraded by RNase E in a concerted manner (Masse and Gottesman 2002; Masse et al. 2003).
Figure 8.
Model for the concerted action of a small RNA, Hfq, and RNase E on the target mRNA. Hfq associates with the C-terminal scaffold region of RNase E lacking PNPase, Rhl B helicase, and enolase. Under a stress condition such as the accumulation of G6P (αMG6P) or the Fe depletion, the cognate small RNA is rapidly synthesized and incorporated into the RNase E/Hfq complex. The RNase E containing Hfq and the small RNA is recruited to the target mRNA through base-pairing between the small RNA and the target mRNA. This RNase E efficiently initiates endonucleolytic cleavage of the targeted mRNA. Following the initial cleavage of mRNA by the RNase E/Hfq/small RNA complex, the RNA degradosome and other ribonucleases would be also involved in further degradation of the mRNA.
It should be noted that the partially purified RNase E was shown to contain tightly bound RNA in an early study (Miczak et al. 1996). Subsequent studies revealed that the degradosome contained a significant amount of heterogeneous RNAs in addition to various protein components (Bessarab et al. 1998; Lin-Chao et al. 1999). The RNAs in the degradosome were shown to consist of primarily ribosomal RNAs and their fragments. These RNAs were thought to be substrates for RNase E and/or degradation intermediates rather than the functional components of the degradosome. Our study has established that RNase E forms true ribonucleoprotein complexes with functional small RNAs through Hfq. Thus, RNase E-based complexes are much more versatile than originally thought. Concerning the versatility of the degradosome, it has been shown recently that a specialized degradosome containing CsdA, one of the DEAD-box RNA helicases, is formed at a low temperature (Prud'homme-Genereux et al. 2004). This “cold shock” degradosome has been shown to be required for efficient decay of a model mRNA at low temperature.
More than 50 small noncoding RNAs are expressed in E. coli cells (Gottesman 2004; Storz et al. 2004). Many of these RNAs appear to act as an antisense regulatory RNA to regulate the translation and/or stability of target mRNAs through base-pairing. So far, the mRNA destabilization effect has been shown clearly only for SgrS and RyhB. Although we have shown that these two Hfq-binding RNAs are able to associate with the RNase E/Hfq complex to destabilize selectively the target mRNAs, there is no reason that the association with RNase E is restricted to only SgrS and RyhB. It is reasonable to speculate that other Hfq-binding small RNAs involved in translational inhibition could also destabilize the target mRNAs by associating with RNase E. In fact, we observed in preliminary experiments that MicF induced by paraquat (Chen et al. 2004) seems to destabilize the target ompF mRNA, suggesting that MicF might also associate with RNase E. The destabilization of target mRNAs mediated by small RNAs has been found in the Vibrio harveyi and Vibrio cholerae quorum sensing system (Lenz et al. 2004). It is plausible that the small RNAs and Hfq may also act by associating with RNase E in these cases. On the other hand, it is unlikely that small RNAs such as DsrA (Majdalani et al. 1998, 2002) destabilize the target mRNAs when they act to stimulate their translation. It is certainly interesting to test whether Hfq-binding small RNAs involved in translational activation associate with RNase E.
Post-transcriptional gene regulation mediated by small antisense RNAs is widespread in both prokaryotes and eukaryotes. The functional similarity between bacterial small RNAs and eukaryotic counterparts such as miRNAs and siRNAs is rather clear because both RNAs act to down-regulate the target genes at post-transcriptional levels by decreasing translation and/or mRNA stability. However, the mechanistic commonality between two systems was not clear except that these small RNAs may act by base-pairing with target mRNAs in general. Eukaryotic small RNAs are assembled into large ribonucleoprotein complexes known as the RNA-induced silencing complex (RISC) prior to acting on the target mRNAs (Bartel 2004). The RISC consists of a variety of proteins such as the RNA-binding protein, RNA helicase, and nuclease, although the precise components and their roles remain unclear. The discovery that bacterial small RNAs and Hfq form multiple ribonucleoprotein complexes with RNase E has raised the possibility that the pathway and mechanism of action of small RNAs in two systems may also resemble each other at least with certain aspects.
Materials and methods
Media and growth conditions
Cells were grown aerobically at 37°C in LB medium or M9 minimum medium supplemented with indicated sugars. Antibiotics were used at the following concentrations when needed: ampicillin (50 μg/mL) and chloramphenicol (15 μg/mL). Bacterial growth was monitored by determining the optical density at 600 nm.
Strains
The E. coli K12 strains used in this study are listed in Table 1. IT1568 (W3110 mlc) was used as a parent wild-type strain. TM338 (rne-Flag-cat), TM528 (rne701-Flag-cat), TM433 (pnp-HA rhlB-HA rne-Flag-cat), TM388 (Δpnp::cat), TM390 (ΔrhlB::cat), TM447 (cat-PBAD-eno), TM461 (Δpgi Pbla-ptsG), and TM509 (Δpgi Pbla-ptsG PBAD-eno) were described previously (Morita et al. 2004). TM587 (Δhfq::cat) was constructed from IT1568 by one-step gene inactivation protocol (Datsenko and Wanner 2000). TM589 (Δhfq), TM522 (rne-Flag), TM611 (rne701-Flag), and TM450 (pnp-HA rhlB-HA rne-Flag) were constructed from TM587, TM338, TM528, and TM433, respectively, by eliminating the cat marker by using an FLP expression plasmid pCP20 (Datsenko and Wanner 2000). The rne-Flag-cat allele of TM338 was moved to TM589 by P1 transduction to construct TM618. The Δpnp::cat or ΔrhlB::cat allele of TM388 or TM390 was moved to TM450 by P1 transduction to construct TM581 (rhlB-HA rne-Flag Δpnp::cat) or TM583 (pnp-HA rne-Flag ΔrhlB::cat), respectively. The cat-PBAD-eno allele of TM447 was moved to TM522 by P1 transduction to construct TM452 (rne-Flag cat-PBAD-eno). TM615 (hfq-Flag-cat) was constructed from IT1568 according to the modified Datsenko-Wanner protocol using pSU313 harboring the Flag-tag sequence (Uzzau et al. 2001). TM619 was constructed from TM615 by eliminating the cat marker. TM641 (rne-HA-cat) and TM642 (rne701-HA-cat) were constructed from IT1568 according to the modified Datsenko-Wanner protocol using pSU313 harboring the HA-tag sequence (Uzzau et al. 2001). The rne-HA-cat and rne701-HA-cat alleles of TM641 and TM642 were moved to TM619 by P1 transduction to construct TM645 (hfq-Flag rne-HA-cat) and TM646 (hfq-Flag rne701-HA-cat), respectively. TM651 (hfq-Flag rne-HA) and TM652 (hfq-Flag rne701-HA) were constructed from TM645 and TM646 by eliminating the cat marker. TM640 (rhlB-Flag-cat) was constructed from IT1568 according to the modified Datsenko-Wanner protocol using pSU313 harboring the Flag-tag sequence (Uzzau et al. 2001). TM647 (rhlB-Flag) was constructed from TM640 by eliminating the cat marker.
Table 1.
Bacterial strains used in this study
| Strain | Relevant genotype and property | Source |
|---|---|---|
| IT1568 | W3110mlc | Wild type |
| TM587 | W3110mlc Δhfq::cat | This study |
| TM589 | W3110mlc Δhfq | This study |
| TM338 | W3110mlc rne-Flag-cat | Morita et al. 2004 |
| TM618 | W3110mlc Δhfq rne-Flag-cat | This study |
| TM528 | W3110mlc rne701-Flag-cat | Morita et al. 2004 |
| TM461 | W3110mlc Δpgi Pbla-ptsG | Morita et al. 2004 |
| TM509 | W3110mlc Δpgi Pbla-ptsG PBAD-eno | Morita et al. 2004 |
| TM433 | W3110mlc rhlB-HA pnp-HA rne-Flag-cat | Morita et al. 2004 |
| TM388 | W3110mlc Δpnp::cat | Morita et al. 2004 |
| TM390 | W3110mlc ΔrhlB::cat | Morita et al. 2004 |
| TM450 | W3110mlc rhlB-HA pnp-HA rne-Flag | This study |
| TM581 | W3110mlc rhlB-HA Δpnp::cat rne-Flag | This study |
| TM583 | W3110mlc ΔrhlB::cat pnp-HA rne-Flag | This study |
| TM447 | W3110mlc cat-PBAD-eno | Morita et al. 2004 |
| TM452 | W3110mlc rne-Flag cat-PBAD-eno | This study |
| TM522 | W3110mlc rne-Flag | This study |
| TM611 | W3110mlc rne701-Flag | This study |
| TM615 | W3110mlc hfq-Flag-cat | This study |
| TM619 | W3110mlc hfq-Flag | This study |
| TM641 | W3110mlc rne-HA-cat | This study |
| TM645 | W3110mlc hfq-Flag rne-HA-cat | This study |
| TM651 | W3110mlc hfq-Flag rne-HA | This study |
| TM642 | W3110mlc rne701-HA-cat | This study |
| TM646 | W3110mlc hfq-Flag rne701-HA-cat | This study |
| TM652 | W3110mlc hfq-Flag rne701-HA | This study |
| TM640 | W3110mlc rhlB-Flag-cat | This study |
| TM647 | W3110mlc rhlB-Flag | This study |
Northern blotting
Total RNAs were isolated from cells grown to mid-log phase as described (Aiba et al. 1981). For Northern blot analysis, indicated amounts of total RNAs were resolved by 1% (for the ptsG and sodB mRNAs) or 1.5% (for the SgrS and RyhB RNAs) agarose gel electrophoresis in the presence of formaldehyde and blotted onto Hybond-N+ membranes (Amersham Biosciences). The mRNAs were visualized using digoxigenin (DIG) reagents and kits for nonradioactive nucleic acid labeling and detection system (Roche Molecular Biochemicals) according to the procedure specified by the manufacturer. The following DIG-labeled DNA probes were prepared by PCR using DIG-dUTP: 305-bp fragment corresponding to the 5′-region of ptsG; 305-bp fragment corresponding to the 5′-region of sodB; 150-bp fragment corresponding to sgrS; 120-bp fragment corresponding to ryhB; 350-bp fragment corresponding to ssrA (tmRNA).
Western blotting
The sample was heated at 100°C for 5 min and subjected to a polyacrylamide-0.1% SDS gel electrophoresis and transferred to Immobilon membrane (Millipore). The 8% and 15% polyacrylamide gels were used to detect RNase E and Hfq, respectively, while the 12% gel was used for other proteins. The membranes were treated with anti-Flag monoclonal antibody (Sigma), anti-HA (Santa Cruz Biotechnology) and anti-enolase, anti-CRP, and anti-Hfq polyclonal antibodies. Signals were visualized by the ECL system (Amersham). Anti-enolase and anti-Hfq polyclonal antibodies were obtained by immunizing rabbits with purified enolase and Hfq proteins, respectively. Anti-CRP polyclonal antibody was described previously (Ishizuka et al. 1993).
Pull-down assay
Cells were grown in 200 mL of LB or M9 medium to A600 of 0.6, harvested, and washed with 10 mL of 10 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA. The cell pellets were suspended in 5 mL of ice-cold IP buffer 1 (20 mM Tris-HCl at pH 8.0, 0.1 M KCl, 5 mM MgCl2, 10% glycerol, 0.1% Tween 20) containing Complete Mini (Roche). The cell suspension was sonicated and centrifuged at 10,000 × g for 1 h at 4°C. The supernatant (crude extract) was incubated with 50 μL of anti-Flag M2-agarose suspension (Sigma) for 1 h at 4°C. When indicated, the supernatant was incubated with 40 units of micrococcal nuclease (Takara) in the presence of 2.5 mM CaCl2 for 30 min at 37°C prior to the incubation with the M2-agarose beads. The mixture was filtered by using a mini chromatography column (Bio-Rad). The filtrate was used as unbound fraction (UB). The proteins bound to the beads trapped on the column were eluted with 50 μL of IP buffer containing 1 mg/mL Flag peptide (Sigma) and used as bound fraction. The crude extract, unbound fraction, and bound fraction were analyzed by Western blotting. The amount of protein loaded was as follows: crude extract, 10 μL; unbound fraction, 10 μL; and bound fraction, 1 μL. To analyze RNAs, each of the crude extracts (100 μL), unbound fractions (100 μL), and the bound fractions (10 μL) was treated with phenol, precipitated, and washed with ethanol. The precipitate was dissolved in 10 μL of distilled water and 5 μL of RNA sample was subjected to Northern blotting.
Affinity-purification of Flag-tagged proteins
Cells were grown in 1 L of LB to A600 of 0.6, harvested, and washed with 20 mL of 10 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM EDTA. The cell pellets were suspended in 10 mL of ice-cold IP buffer 2 (20 mM Tris-HCl at pH 8.0, 0.2 M KCl, 5 mM MgCl2, 10% glycerol, 0.1% Tween 20) containing Complete Mini. The cell suspension was sonicated and centrifuged at 10,000 × g for 1 h at 4°C. The supernatant (crude extract) was incubated with 100 μL of anti-Flag M2-agarose suspension (Sigma) for 1 h at 4°C. The mixture was filtered by using a mini chromatography column (Bio-Rad). The agarose beads were washed three times with 10 mL of IP buffer, and proteins bound to the beads were eluted with 100 μL of IP buffer containing 1 mg/mL Flag peptide (Sigma). The purified proteins were analyzed by electrophoresis using a 4%-12% polyacrylamide gradient gel, NuPAGE GEL (Bio-Rad), and the gel was stained with Coomassie Brilliant Blue (CBB).
Mass spectrometry
The protein bands stained with CBB were cut out from the gel, and a small piece of each band was treated with 125 ng of trypsin (Wako) in 10 μL of 20 mM ammonium bicarbonate for 12 h at 37°C. The digested peptides were eluted with 300 μL of 50% acetonitrile, 5% formic acid, and concentrated to 20 μL. Then, the sample was desalted with a zip-tip reverse-phase column, mixed with 1% α-CHCA (α-cyano-4-hydroxycinnamic acid) in 70% acetonitrile, and subjected to MALDI/TOF-MS.
Acknowledgments
We are grateful to Susan Gottesman for comments on the manuscript. This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by Ajinomoto Co., Inc. T.M. is grateful to Masami Oguchi for continuous encouragement during this work.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1330405.
References
- Aiba H., Adhya, S., and de Crombrugghe, B. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256: 11905-11910. [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [DOI] [PubMed] [Google Scholar]
- Bessarab D.A., Kaberdin, V.R., Wei, C.L., Liou, G.G., and Lin-Chao, S. 1998. RNA components of Escherichia coli degradosome: Evidence for rRNA decay. Proc. Natl. Acad. Sci. 95: 3157-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan A.J., Aurikko, J.P., Ilag, L.L., Gunter Grossmann, J., Chandran, V., Kuhnel, K., Poljak, L., Carpousis, A.J., Robinson, C.V., Symmons, M.F., et al. 2004. Studies of the RNA degradosome-organizing domain of the Escherichia coli ribonuclease RNase E. J. Mol. Biol. 340: 965-979. [DOI] [PubMed] [Google Scholar]
- Carpousis A.J. 2002. The Escherichia coli RNA degradosome: Structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 30: 150-155. [PubMed] [Google Scholar]
- Carpousis A.J., Van Houwe, G., Ehretsmann, C., and Krisch, H.M. 1994. Copurification of E. coli RNAase E and PNPase: Evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76: 889-900. [DOI] [PubMed] [Google Scholar]
- Chen S., Zhang, A., Blyn, L.B., and Storz, G. 2004. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 186: 6689-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn G.A., Miao, X., Briant, D.J., and Mackie, G.A. 1999. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes & Dev. 13: 2594-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K.A. and Wanner, B.L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97: 6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. 2004. The small RNA regulators of Escherichia coli: Roles and mechanisms. Annu. Rev. Microbiol. 58: 303-328. [DOI] [PubMed] [Google Scholar]
- Ishizuka H., Hanamura, A., Kunimura, T., and Aiba, H. 1993. A lowered concentration of cAMP receptor protein caused by glucose is an important determinant for catabolite repression in Escherichia coli. Mol. Microbiol. 10: 341-350. [DOI] [PubMed] [Google Scholar]
- Kawamoto H., Morita, T., Shimizu, A., Inada, T., and Aiba, H. 2005. Implication of membrane localization of target mRNA in the action of a small RNA: Mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes & Dev. 19: 328-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemici V. and Carpousis, A.J. 2004. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 51: 777-790. [DOI] [PubMed] [Google Scholar]
- Kimata K., Tanaka, Y., Inada, T., and Aiba, H. 2001. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. EMBO J. 20: 3587-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz D.H., Mok, K.C., Lilley, B.N., Kulkarni, R.V., Wingreen, N.S., and Bassler, B.L. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118: 69-82. [DOI] [PubMed] [Google Scholar]
- Lin-Chao S., Wei, C.L., and Lin, Y.T. 1999. RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc. Natl. Acad. Sci. 96: 12406-12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N., Cunning, C., Sledjeski, D., Elliott, T., and Gottesman, S. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. 95: 12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N., Hernandez, D., and Gottesman, S. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46: 813-826. [DOI] [PubMed] [Google Scholar]
- Masse E. and Gottesman, S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. 99: 4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E., Escorcia, F.E., and Gottesman, S. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes & Dev. 17: 2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall K.J. and Cohen, S.N. 1996. The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA-binding site. J. Mol. Biol. 255: 349-355. [DOI] [PubMed] [Google Scholar]
- Miczak A., Kaberdin, V.R., Wei, C.L., and Lin-Chao, S. 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. 93: 3865-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B.K., Maples, V.F., and Kushner, S.R. 2004. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 54: 905-920. [DOI] [PubMed] [Google Scholar]
- Morita T., El-Kazzaz, W., Tanaka, Y., Inada, T., and Aiba, H. 2003. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J. Biol. Chem. 278: 15608-15614. [DOI] [PubMed] [Google Scholar]
- Morita T., Kawamoto, H., Mizota, T., Inada, T., and Aiba, H. 2004. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 54: 1063-1075. [DOI] [PubMed] [Google Scholar]
- Prud'homme-Genereux A., Beran, R.K., Iost, I., Ramey, C.S., Mackie, G.A., and Simons, R.W. 2004. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: Evidence for a `cold shock degradosome.' Mol. Microbiol. 54: 1409-1421. [DOI] [PubMed] [Google Scholar]
- Py B., Higgins, C.F., Krisch, H.M., and Carpousis, A.J. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381: 169-172. [DOI] [PubMed] [Google Scholar]
- Storz G., Opdyke, J.A., and Zhang, A. 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 7: 140-144. [DOI] [PubMed] [Google Scholar]
- Sukhodolets M.V. and Garges, S. 2003. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry 42: 8022-8034. [DOI] [PubMed] [Google Scholar]
- Uzzau S., Figueroa-Bossi, N., Rubino, S., and Bossi, L. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. 98: 15264-15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P., Eriksen, M., and Udesen, C. 2004. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol. Microbiol. 51: 1525-1533. [DOI] [PubMed] [Google Scholar]
- Vanderpool C.K. and Gottesman, S. 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 54: 1076-1089. [DOI] [PubMed] [Google Scholar]
- ____. 2005. Noncoding RNAs at the membrane. Nat. Struct. Mol. Biol. 12: 285-286. [DOI] [PubMed] [Google Scholar]
- Vanzo N.F., Li, Y.S., Py, B., Blum, E., Higgins, C.F., Raynal, L.C., Krisch, H.M., and Carpousis, A.J. 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes & Dev. 12: 2770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Wassarman, K.M., Rosenow, C., Tjaden, B.C., Storz, G., and Gottesman, S. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 50: 1111-1124. [DOI] [PubMed] [Google Scholar]