Abstract
Long-range migrating progenitor cells generate hypaxial muscle, for instance the muscle of the limbs, hypoglossal cord, and diaphragm. We show here that migrating muscle progenitors express the chemokine receptor CXCR4. The corresponding ligand, SDF1, is expressed in limb and branchial arch mesenchyme; i.e., along the routes and at the targets of the migratory cells. Ectopic application of SDF1 in the chick limb attracts muscle progenitor cells. In CXCR4 mutant mice, the number of muscle progenitors that colonize the anlage of the tongue and the dorsal limb was reduced. Changes in the distribution of the muscle progenitor cells were accompanied by increased apoptosis, indicating that CXCR4 signals provide not only attractive cues but also control survival. Gab1 encodes an adaptor protein that transduces signals elicited by tyrosine kinase receptors, for instance the c-Met receptor, and plays a role in the migration of muscle progenitor cells. We found that CXCR4 and Gab1 interact genetically. For instance, muscle progenitors do not reach the anlage of the tongue in CXCR4;Gab1 double mutants; this target is colonized in either of the single mutants. Our analysis reveals a role of SDF1/CXCR4 signaling in the development of migrating muscle progenitors and shows that a threshold number of progenitor cells is required to generate muscle of appropriate size.
Keywords: CXCR4, Gab1, hypaxial muscle, migration
Skeletal muscle of vertebrate embryos originates from the dermomyotome, a derivative of the somite. On particular axial levels, cells of the ventral dermomyotomal lip lose their epithelial morphology, delaminate, and migrate as single cells over long distances using stereotypic routes. These migrating progenitor cells generate hypaxial muscles of the extremities, tongue, and diaphragm (Chevallier et al. 1977; Christ et al. 1977; Christ and Ordahl 1995).
Various genes are essential for the development of migrating hypaxial progenitors; these function in the specification of muscle progenitor cells, their delamination from the dermomyotome, and their subsequent migration, survival, proliferation, and differentiation. Pax3 is required for the correct establishment of the progenitor pool in the ventral dermomyotome. Consequently, the development of all hypaxial muscle is impaired in Pax3 mutant mice (Franz et al. 1993; Bober et al. 1994; Tajbakhsh et al. 1997). The tyrosine kinase receptor c-Met and its ligand, scatter factor/hepatocyte growth factor (SF/HGF), are essential for the delamination of the progenitors that are destined to migrate, and all muscle groups that derive from migrating progenitors are absent in c-Met or SF/HGF mutant mice (Bladt et al. 1995; Dietrich et al. 1999). Six1 controls the proliferation of muscle progenitor cells and forms a complex with Eya1 and Dach. This complex permits the expression of c-myc, a key molecule in the control of proliferation. Specific limb muscle groups are reduced in size in Six1 mutant mice, whereas Six1;Eya1 double mutants show a complete absence of limb musculature (Li et al. 2003). Migration of muscle progenitor cells is a complex process and requires signals that allow the cells to remain motile and find their targets. Gab1 encodes an adaptor molecule that transmits c-Met signals and its mutation impairs but does not completely abolish delamination of muscle progenitors (Sachs et al. 2000). A detailed analysis of Gab1 mutant mice indicates that c-Met signals mediated by Gab1 are essential not only for delamination, but also for migration and survival of muscle progenitor cells (Sachs et al. 2000; M. Strehle and C. Birchmeier, unpubl.). Lbx1 encodes a homeodomain transcription factor that is expressed exclusively in migrating progenitor cells that will form hypaxial muscle. Inactivation of Lbx1 severely impairs migration of those progenitors that move to the limbs while other populations of migrating muscle progenitors find their targets (Schafer and Braun 1999; Brohmann et al. 2000; Gross et al. 2000). This indicates that different subpopulations of migrating muscle progenitors encounter and respond to distinct guidance cues during migration. In the limbs of chick embryos, muscle progenitors that express the EphA4 receptor are repelled from ectopically applied ephrinA5, suggesting that EphA4/ephrin-A5 signals prevent their entry into inappropriate domains (Swartz et al. 2001). Progenitor cells that reach their targets continue to proliferate and start to express myogenic regulatory factors like MyoD, Myf5, myogenin, and MRF4 that determine their terminal differentiation program (Arnold and Braun 2000; Perry and Rudnick 2000; Buckingham 2001).
Chemokine receptors and their ligands regulate migration of cells in the developing and adult organism. This role of chemokine receptors first became apparent in the hematopoietic system, and the analysis of mutant mice demonstrated that various migration events of lymphoid cells are controlled by chemokine receptors (for review, see Müller et al. 2003). SDF1, the ligand of the CXCR4 receptor, acts as a potent chemoattractant for cultured B lymphocytes, monocytes, and CD34-positive hematopoietic progenitor cells (Nagasawa et al. 1994; Bleul et al. 1996). The analysis of CXCR4 or SDF1 mutant mice has, however, revealed numerous developmental functions in cell lineages other than hematopoietic cells (Nagasawa et al. 1996; Ma et al. 1998; Zou et al. 1998). Mutations of CXCR4 or SDF1 affect the migration of cerebellar granule cells and of hippocampal and cortical neuronal progenitors (for review, see Lazarini et al. 2003). Furthermore, CXCR4/SDF1 signaling is important for the migration of primordial germ cells, a function that is conserved in fish, birds, and mammals (Doitsidou et al. 2002; Knaut et al. 2003; Molyneaux et al. 2003; Stebler et al. 2004). Finally, chemokine signals also control the migration of malignant cancer cells. Metastasis of human breast cancer cells to particular preferred sites correlates with the expression of CXCR4 in tumor cells, and with the expression of SDF1 in the organ invaded by the metastatic cells (Müller et al. 2001). In addition to the regulation of various migration processes, CXCR4/SDF1 also controls growth and survival of different cells types (Zou et al. 1998; Bagri et al. 2002; Molyneaux et al. 2003). CXCR4 is expressed in cell lines derived from muscle satellite cells, the stem cells of the adult skeletal muscle. The ability of SDF1 to attract and stimulate the activation of cultured satellite cells suggested possible functions of CXCR4/SDF1 in muscle cells (Ratajczak et al. 2003).
We used microarray technology to determine the expression profile of genes in migrating muscle progenitors. Among the expressed genes, we identified the chemokine receptor CXCR4. SDF1, which encodes the corresponding ligand, is expressed in the mesenchyme of the limb and the first branchial arch, which represent targets of the migrating cells. Application of SDF1 into the limb of chick embryos directs the muscle progenitor cells toward the ectopic source of the factor and inhibits their differentiation. Analysis of CXCR4 mutant mice demonstrates changes in the distribution of migrating muscle progenitors. Furthermore, we observe a genetic interaction between CXCR4 and Gab1. For example, the anlage of the tongue is reached in CXCR4 and Gab1 mutant mice, but migrating muscle progenitor cells do not arrive at this site in the double-mutant mice. This genetic interaction might reflect a cross-talk between signaling cascades employed by G-protein-coupled receptors (GPCRs) and tyrosine kinases.
Results
The Lbx1GFP allele allows the sorting of muscle progenitor cells for gene expression profiling
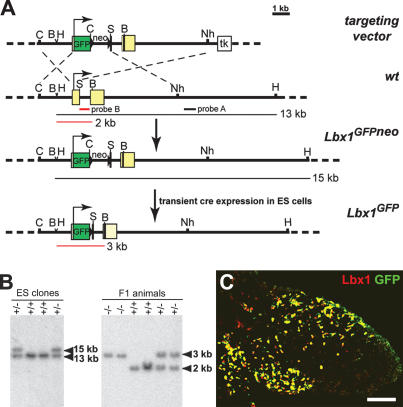
Previous studies showed that Lbx1 is expressed during development of long-range migrating hypaxial cells, but not in other types of muscle progenitor cells (Jagla et al. 1995). To allow the isolation of muscle progenitor cells by cell sorting, we generated a mutant Lbx1 allele, in which GFP was fused to the initiation codon by homologous recombination in ES cells (Fig. 1A). The mutant ES cells were then used to generate an Lbx1GFP mouse strain (Fig. 1B; for additional information, see Materials and Methods). GFP protein was produced from the Lbx1GFP allele and was located in migrating muscle progenitors of the dorsal and ventral limb in a pattern similar to the endogenous Lbx1 protein (Fig. 1C).
Figure 1.
Generation of the Lbx1GFP mutant allele. (A) Schematic representation of the targeting vector, the wild-type Lbx1 locus, and the mutated Lbx1 allele before and after removal of the neomycin (neo) cassette. The Lbx1 gene has two exons (yellow boxes); the first was interrupted by the insertion of a Gap43-GFP cassette (green box). In addition, a frame-shift mutation (indicated by a black line) was introduced into the BglII site of exon 2 that encodes the Lbx1 homeodomain. Neomycin (neo) and thymidine kinase (tk) cassettes present in the targeting vector were used for positive and negative selection. The positions of the probes, A and B, used for Southern analysis are shown by black and red bars, respectively. The predicted fragment sizes obtained after HindIII (H) and BamHI (B) digestion of genomic DNA are indicated. In addition, the following restriction enzyme sites are indicated: ClaI (C), Sse8387I (S), and NheI (Nh). (B) Southern blot analysis of HindIII-digested genomic DNA from wild-type and Lbx1GFP/+ ES cells using probe A for hybridization (left) and BamHI-digested genomic DNA from wild-type, Lbx1GFP/+, and Lbx1GFP/GFP F1 animals using probe B for hybridization (right). (C) Immunohistological analysis of a forelimb section of an Lbx1GFP/+ embryo at E10 stained with anti-Lbx1 (red) and anti-GFP (green) antibodies. Bar, 125 μm.
GFP+ muscle progenitors were isolated from the embryonic limb bud. For this, the forelimbs of embryonic day 10.5 (E10.5) embryos were dissected, cells were dissociated, and GFP-positive cells were isolated by flow cytometry. Probes were generated from RNA obtained from these cells and used for hybridization on MG U74A/B/Cv2 Affymetrix GeneChips. Genes encoding cell surface molecules that might control cell migration were identified, and in situ hybridization was used to analyze their expression pattern in the embryo. Among the genes identified (Table 1) was the chemokine receptor CXCR4. CXCR4 and its ligand SDF1 are critical for migration, proliferation, and survival of various cell types (Cyster 2003; Lazarini et al. 2003; Raz 2003).
Table 1.
Selected genes encoding integral membrane proteins that are expressed in Lbx1+ muscle progenitor cells, as determined by microarray analysis
| Affy ID | GenBank ID | Gene | Detection p-value |
|---|---|---|---|
| 98169_s_at | AU020229 | Fzd3 | 0.0005 |
| 139426_r_at | AW228933 | Fgfr1 | 0.003 |
| 93090_at | M23362 | Fgfr2 | 0.006 |
| 108468_at | AW121015 | Bmpr1a | 0.002 |
| 95117_at | U04710 | Igf2r | 0.004 |
| 106644_at | AW047110 | Tgfbr1 | 0.0004 |
| 104188_at | AI853703 | Notch2 | 0.0004 |
| 102250_at | AF053005 | Il27ra | 0.002 |
| 102794_at | Z80112 | CXCR4 | 0.002 |
| 93430_at | AF000236 | Cmkor1 | 0.006 |
| 114749_at | AW107659 | Gpr23 | 0.0003 |
| 104673_at | X65138 | Epha4 | 0.02 |
| 98446_s_at | U06834 | Ephb4 | 0.0002 |
| 160857_at | U30244 | Efnb2 | 0.0004 |
| 95387_f_at | AA266467 | Sema4b | 0.0004 |
| 117151_at | AI838057 | c-Met | 0.0007 |
| 160480_at | X82288 | Ptprs | 0.0002 |
| 160760_at | L10106 | Ptprk | 0.006 |
| 97750_at | X06406 | Lamr1 | 0.0002 |
| 101585_at | AF042491 | Pgrmc1 | 0.0002 |
| 98094_f_at | AI843627 | Amfr | 0.002 |
| 103958_g_at | X57349 | Trfr | 0.002 |
| 103783_at | AI648965 | Xpr1 | 0.004 |
| 102852_at | M31131 | Cdh2 | 0.0002 |
| 100006_at | D21253 | Cdh11 | 0.0008 |
| 129896_at | AW125163 | Pcdh17 | 0.014 |
| 166351_f_at | AV245394 | Pcdh18 | 0.001 |
| 100124_r_at | X15202 | Itgb1 | 0.0002 |
| 95292_at | AA189389 | Itga4 | 0.010 |
| 94117_f_at | AF026465 | Punc | 0.0002 |
| 100153_at | X15052 | Ncam | 0.0002 |
| 100977_at | AA691492 | Glycam1 | 0.001 |
| 92558_at | M84487 | Vcam1 | 0.002 |
| 93604_f_at | AF061260 | Igsf4 | 0.0002 |
| 92270_at | AI847616 | Maged3 | 0.002 |
| 93389_at | AF039663 | Prom | 0.0006 |
Genes were annotated using the Affymetric Netaffy, Ensemble, and NCBI databases. The average detection p-value as calculated by Affymetrix MAS 5.0 software is given. Displayed are genes with a p-value for expression ≤0.02. Genes whose expression in migrating muscle progenitor cells was previously reported or verified by in situ hybridization are indicated in bold.
Expression of CXCR4 and SDF1 in mouse and chick embryos
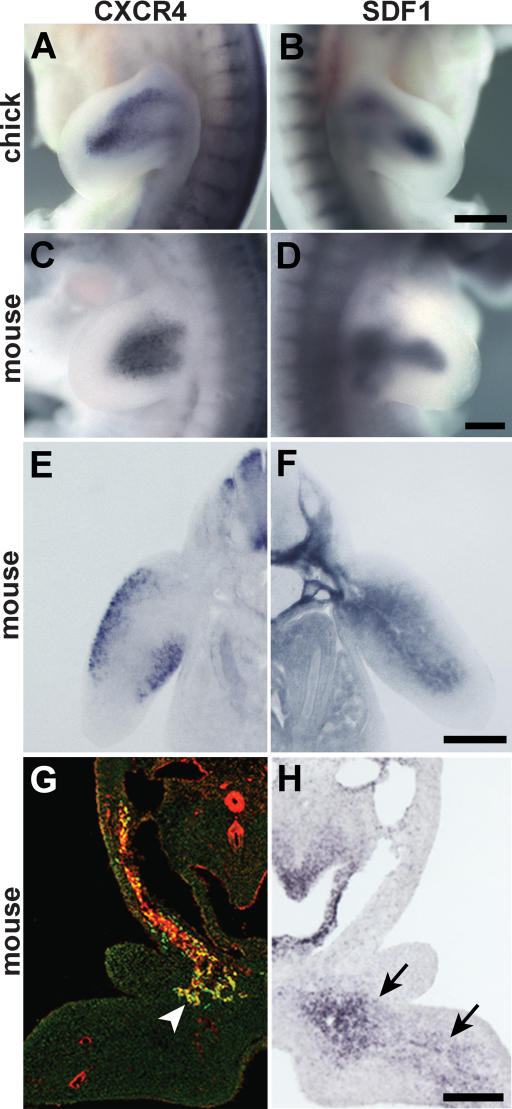
We examined expression of CXCR4 and SDF1 in the limbs of mouse and chick embryos. Indeed, CXCR4 is expressed in the limb buds. The distribution of CXCR4-expressing cells resembles, but is not entirely identical to, that of Pax3- or Lbx1-expressing muscle progenitors (Figs. 2A,C,E, 3; Supplementary Fig. 1). A stream of CXCR4+ cells is also observed along the hypoglossal chord and in the first branchial arch; i.e., along the route and at the target of those migrating muscle progenitor cells that will generate the intrinsic tongue muscle (Fig. 2G). However, CXCR4 expression in muscle progenitors that migrate to the anlage of the diaphragm is very low (data not shown). SDF1 expression can be detected in the limb bud mesenchyme, and the pattern of expression is dynamic during development (Fig. 2B,D,F; Supplementary Fig. 1). At the time muscle progenitor cells delaminate, SDF1 is expressed in the proximal limb bud (Supplementary Fig. 1A). After the cells enter the limb, SDF1 expression is also observed further distally in the mesenchyme (Fig. 2B,D,F; Supplementary Fig. 1B,C). Hence, migrating muscle progenitor cells in the limb are found in the vicinity of SDF1-expressing cells. Few muscle progenitors reach the limb of Lbx1-/- embryos (Schafer and Braun 1999; Brohmann et al. 2000; Gross et al. 2000). A comparison of the control and Lbx1-/- embryos demonstrates very similar expression patterns of SDF1, indicating that muscle progenitors do not express SDF1 (Supplementary Fig. 1C,D). SDF1 transcripts are also present in the floor of the first branchial arch (Fig. 2H). High expression of SDF1 is thus observed at the target of the migrating cells that move to the anlage of the tongue.
Figure 2.
Expression of CXCR4 and SDF1 in mouse and chick embryos. (A-F,H) In situ hybridization of chick embryos at HH25 (A,B) and mouse embryos at E10.25 (C-F) using probes specific for CXCR4 (A,C,E) and SDF1 (B,D,F). (G,H) Consecutive sections, displayed as mirror images, through the first branchial arch of wild-type mouse embryos at E10.25 were stained with antibodies against CXCR4 (red) and Lbx1 (green) (G), or hybridized with an SDF1-specific probe (H). CXCR4 and Lbx1 were coexpressed in muscle progenitors migrating toward the tongue anlage (arrowhead in G), whereas SDF1 transcripts were detected in the mesenchyme of the first branchial arch (arrows in H). Bars: A,B, 500 μm; C-H, 250 μm.
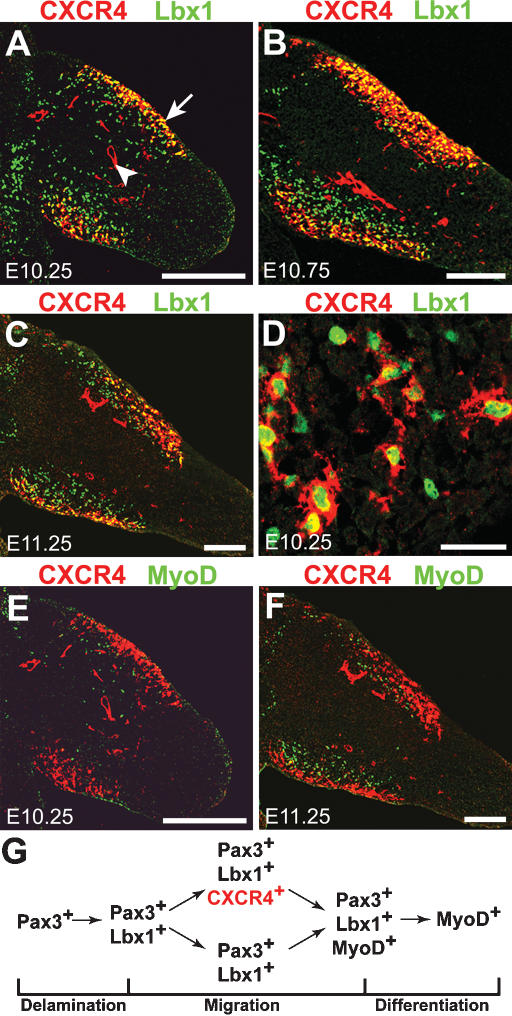
Figure 3.
Expression of CXCR4, Lbx1, and MyoD in limb muscle progenitors. (A-D) Sections of the forelimb of E10.25 (A,D), E10.75 (B), and E11.25 (C) mouse embryos stained with anti-CXCR4 (red) and anti-Lbx1 (green) antibodies. CXCR4+ and Lbx1+ cells are present in the muscle masses, and analysis at high magnification demonstrates that they are present in the same cells. (D) Note that not every Lbx1+ cell expressed CXCR4, but all CXCR4+ muscle progenitor cells were also positive for Lbx1. (A) In addition, CXCR4 was also present in limb endothelial cells (arrowhead). (E,F) Section of the forelimb of mouse embryo at E10.25 (E) and E11.25 (F) stained with anti-CXCR4 (red) and anti-MyoD (green) antibodies. CXCR4+/MyoD+ double-positive cells were very rare. (G) Schematic representation of gene expression in developing muscle progenitors. CXCR4 expression is induced after muscle progenitors have delaminated and have reached the limb, and is extinguished prior to their differentiation. Bars: A-C,E-F, 250 μm; D, 30 μm.
We analyzed the distribution of Lbx1 and CXCR4 by immunohistochemistry to identify if CXCR4 and Lbx1 are expressed in identical cell populations. We observed that all CXCR4+ cells in the premuscle masses of the limbs are also Lbx1+, demonstrating that CXCR4 is present in migrating muscle progenitors (Fig. 3A-D). However, not every Lbx1+ cell expresses CXCR4. Lbx1 appears in progenitors prior to their delamination from the ventrolateral dermomyotome, but CXCR4 protein is not detectable in the ventral dermomyotomal lip, in delaminating Lbx1+ cells, or in the myotome (Fig. 3A; data not shown). CXCR4 and Lbx1 are coexpressed only after the migrating progenitor cells have moved away from the dermomyotome. CXCR4 protein is present in cells that have already entered the forelimb at E10.0-E10.25, but present only in a subpopulation of the Lbx1+ cells that is located close to the ectoderm (Fig. 3A-C). Between E10.25 and E11.0, the number of CXCR4+ cells increases, but a significant number of Lbx1+ cells remain negative for CXCR4 (Fig. 3B,C). All Pax3+ cells in the limb coexpress Lbx1 (see also Gross et al. 2000; data not shown). Accordingly, Pax3+/CXCR4+ and Pax3+/CXCR4- cells exist in the limb. Similarly, the in situ hybridization patterns of CXCR4 and Pax3 are similar, but not identical, indicating that not all Pax3+ cells coexpress CXCR4 (Supplementary Fig. 1F,I,G,J). We also examined CXCR4 and Lbx1 expression in those muscle progenitors that migrate to the first branchial arch. Again, we observed that most, but not all, Lbx1+ cells coexpress CXCR4 (Fig. 2G).
Examination of CXCR4 and MyoD by immunohistology reveals that the CXCR4+ and the MyoD+ cell populations are distinct at E10.25, E10.5, and E11.25 (Fig. 3E,F; data not shown). In contrast, MyoD+/Lbx1+ or MyoD+/Pax3+ cells are frequently detected. In the developing limb, CXCR4+ cells locate closer to the ectoderm than MyoD+ cells. These data indicate that migrating muscle progenitors do not express CXCR4 at the time they are specified in dermomyotome. CXCR4+ cells coexpress Lbx1 and Pax3; i.e., proteins known to be expressed in migrating progenitors, but not the muscle differentiation factor MyoD. Thus, CXCR4 is down-regulated in muscle progenitors prior to their differentiation (for summary, see Fig. 3G).
Ectopic application of SDF1 in the chick limb attracts muscle progenitor cells and delays their differentiation
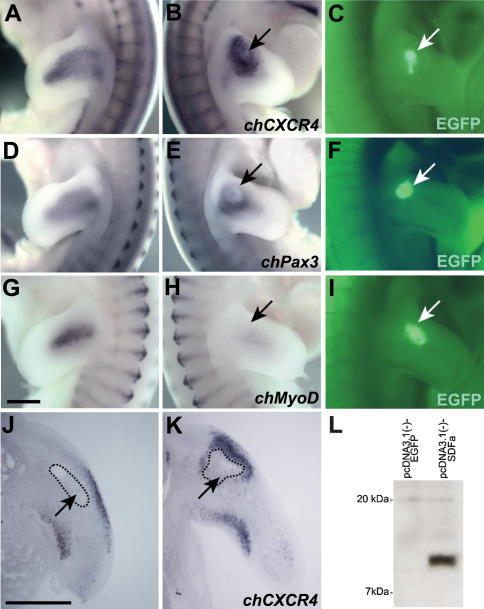
To determine if CXCR4+ muscle progenitor cells respond to SDF1, we applied SDF1 to ectopic positions within the chick limb. For this, COS1 cells were transiently transfected with an SDF1-expression construct or with a control construct. Western blotting was used to verify that SDF1 protein was secreted from transfected, but not from control, COS1 cells (Fig. 4L). The COS1 cells were harvested 36 h after transfection and implanted into the right forelimb bud of chick embryos (Hamburger-Hamilton stage 19-20; HH19-20); the untreated left forelimb served as a control. Embryos that had received an implant were examined by in situ hybridization (HH24-25). In the limb of those embryos that received an implant of SDF1-expressing COS1 cells, CXCR4-positive muscle progenitors accumulated at ectopic positions (16 out of 18 cases examined) (Fig. 4A-C,J,K). For instance, when the implant was located in the proximal portion of the dorsal limb, we observed that CXCR4+ cells assembled close to the implant (Fig. 4B,K). After implanting SDF1-expressing cells, a substantial redistribution of muscle progenitor cells was also observed by in situ hybridization with a Pax3 probe (11 out of 17 cases examined) (Fig. 4D-F). The change in the distribution of Pax3+ cells was less pronounced than the one observed for CXCR4+ cells, indicating that not all Pax3+ cells responded to SDF1. This is in accordance with the existence of a Pax3+/CXCR4- cell population. Dorsal or ventral CXCR4+ or Pax3+ cells assembled close to the implant, but such cells rarely moved around the implant into the central limb bud mesenyme. None of the embryos that received an implant of COS1 cells transfected with the control plasmid showed ectopically positioned muscle progenitors (n = 5). These data indicate that an ectopic source of SDF1 attracts CXCR4+ muscle progenitor cells.
Figure 4.
Muscle progenitors are attracted by an ectopic source of SDF1. COS1 cells cotransfected with SDF1 and GFP expression plasmids were implanted into the right wing bud of chick embryos at HH19-20. The distribution of muscle progenitor cells was analyzed at HH25 in the untreated (A,D,G) and treated (B,E,H) contra-lateral limb by in situ hybridization using chCXCR4-specific (A,B), chPax3-specific (D,E), and chMyoD-specific (G,H) probes. (C,F,I) The positions of the GFP positive implants are shown and are also indicated by arrows. Note the aberrant position of the CXCR4+ and Pax3+ progenitor cells and the reduction of the MyoD signal in the limb implanted with SDF1-expressing cells. (J,K) COS1 cells transfected with GFP expression plasmid only (J) or COS1 cells cotransfected with SDF1 and GFP expression plasmids (K) were implanted into the limb bud, and the distribution of muscle progenitors was analyzed on sections after in situ hybridization using chCXCR4. The position of the implant is indicated. (L) Western blot analysis of supernatant from COS1 cells transfected with a plasmid encoding SDF1 (right lane); as a control, a plasmid encoding GFP was transfected (left lane). Bars, 500 μm.
To test the effect of ectopic SDF1 on differentiation, chicken embryos with implants were hybridized with MyoD and Myf5 probes. Hybridization signals for MyoD (6 out of 6 cases examined) or Myf5 (5 of 5 cases examined) were lower in limbs that had received an implant of SDF1-producing cells than in the untreated, contra-lateral limbs (Fig. 4G-I; data not shown). In contrast, no change in MyoD and Myf5 expression was observed in embryos containing the control implant (5 out of 5 cases examined; data not shown). These data show that ectopic SDF1 not only attracts muscle progenitor cells, but also suppresses their differentiation.
Changes in the distribution and number of muscle progenitor cells in CXCR4-/- embryos
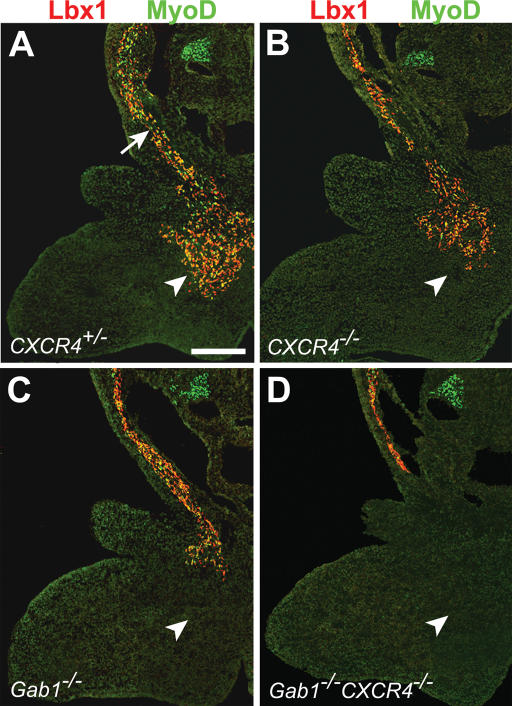
We examined the distribution of hypaxial muscle progenitor cells in mouse embryos that carry a mutation in the CXCR4 gene. The CXCR4 mutant allele was described previously (Ma et al. 1998). Antibodies directed against Lbx1 and MyoD were used to visualize the muscle progenitor cell population that moves to the anlage of the tongue in the first branchial arch of CXCR4+/- and CXCR4-/- embryos. In control embryos at E10.75, a stream of muscle progenitors along the hypoglossal cord could be observed, and a large number of muscle progenitor cells had reached the floor of the first branchial arch. The Lbx1+ or MyoD+ cell population that had reached the floor of the first branchial arch was reduced in number in CXCR4-/- embryos compared with controls (Fig. 5A,B). We also analyzed the effect of the CXCR4 mutation in a Gab1-/- genetic background. In the Gab1 mutant embryos, the number of Lbx1+ or MyoD+ cells in the first branchial arch was lower than in control mice (Fig. 5A,C). In CXCR4-/-Gab1-/- embryos, the migrating muscle progenitor cells did not reach this target, and Lbx1+ or MyoD+ cells were not detectable in the floor of the first branchial arch (Fig. 5D). Also at E11.5, we found no Lbx1+ or MyoD+ cells in the branchial arch of CXCR4-/-Gab1-/- embryos (data not shown). Thus, in the CXCR4;Gab1 double mutants, the migrating cells are not only delayed, but they do not reach this target. The complex morphology made it difficult to count the cells that migrate along the hypoglossal cord or reach the branchial arch. Therefore, quantification was performed for cells migrating into the limb.
Figure 5.
Migration of muscle progenitors along the hypoglossal cord. Sections of the first branchial arch of CXCR4+/- (A), CXCR4-/- (B), Gab1-/- (C), and CXCR4-/-Gab1-/- (D) embryos at E10.75 were analyzed with anti-Lbx1 (red) and anti-MyoD (green) antibodies to identify muscle progenitor cells. In control embryos, muscle progenitors were observed along the hypoglossal cord (arrow) and colonized the mesenchyme of the first branchial arch, the target (arrowhead). Note the reduction in the numbers of muscle progenitors in the first branchial arch of CXCR4-/- and Gab1-/- embryos, and their absence in CXCR4-/-Gab1-/- embryos. Bar, 250 μm.
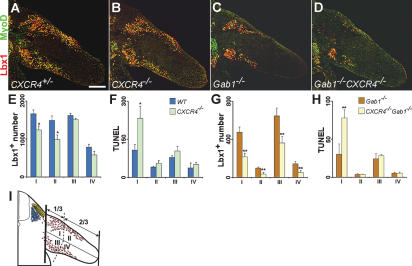
We examined the distribution and determined absolute numbers of muscle progenitor cells in the limbs of CXCR4-/- and control mice. For this, the limb was divided into four domains, dorsal proximal, dorsal distal, ventral proximal, and ventral distal, and the number of Lbx1+ cells was determined in each domain (Fig. 6I). In CXCR4-/- mice, we observed changes in the distribution of progenitor cells in the dorsal limb; i.e., reduction in the numbers of Lbx1+ cells at E10.75 (Fig. 6A,B,E, domains I and II). The reduction in cell numbers was more pronounced in the distal (35%) than in the proximal (25%) domain of the dorsal limb (Fig. 6E). Similarly, the number of MyoD+ cells was reduced in the dorsal limb, and the distal domain was more strongly affected than the proximal one (data not shown). However, the differentiation rate (number of MyoD+ cells/number of Lbx1+ cells) was not markedly altered (control: 59 ± 3%; CXCR4-/-: 50 ± 4%, p value = 0.04). Proliferation of muscle progenitors was analyzed using BrdU labeling showing no significant change in the proliferation rate of progenitors located in the dorsal or ventral limbs of CXCR4-/- and control embryos at E10.75 (control: 58 ± 5%; CXCR4-/-: 61 ± 5%, p value = 0.18). Cell death was analyzed by TUNEL staining and was rarely observed in Lbx1+ nuclei, indicating that the nuclear proteins are not properly maintained in apoptotic nuclei. We therefore counted all apoptotic nuclei in the area occupied by muscle progenitor cells and observed a significant increase in apoptosis in the proximal domain of the dorsal limb in CXCR4-/- compared with control embryos (Fig. 6F; for further detail, see Materials and Methods). In conclusion, these data show that muscle progenitor cells in the dorsal limb of CXCR4-/- mutant embryos are not correctly distributed and their survival is impaired.
Figure 6.
Distribution of muscle progenitor cells in the limb. (A-D) Sections of forelimbs of CXCR4+/- (A), CXCR4-/- (B), Gab1-/- (C), and CXCR4-/-Gab1-/- (D) embryos at E10.75 were analyzed with anti-Lbx1 (red) and anti-MyoD (green) antibodies. (E-H) Quantification of the numbers of Lbx1+ and TUNEL+ cells located in distinct domains of forelimbs of embryos with the genotypes CXCR4+/- (blue bars) and CXCR4-/- (green bars) (E,F); and Gab1-/- (orange bars) and CXCR4-/-Gab1-/- (yellow bars) (G,H). For this, consecutive sections of E10.75 embryos were analyzed by immunohistochemistry, and the Lbx1+ or TUNEL+ cell numbers were counted on every third section in the different limb domains; (★) p value < 0.007, n = 5; (★★) p value < 0.005, n = 4 (see also Material and Methods for further details). (I) Schematic drawing of a developing limb and the four domains defined therein: dorsal proximal (I), dorsal distal (II), ventral proximal (III), and ventral distal (IV). Bar, 250 μm.
We then analyzed the effect of the CXCR4 mutation in the Gab1-/- background. For this analysis, CXCR4-/-Gab1-/- and CXCR4+/+Gab1-/- embryos were compared (Fig. 6C,D). In the dorsal and ventral limbs, the numbers of Lbx1+ and MyoD+ cells were further reduced in CXCR4-/-Gab1-/- compared with CXCR4+/+Gab1-/- embryos (Fig. 6G). The number of apoptotic cells in the limb areas occupied by muscle progenitor cells was increased in CXCR4-/-Gab1-/- mutants compared with CXCR4+/+Gab1-/- mice (Fig. 6H). We detected no significant differences in proliferation of Lbx1+ muscle progenitor cells in the limb of control, CXCR4+/+Gab1-/-, or CXCR4-/-Gab1-/- mice (proliferation rate: control, 58 ± 5%; CXCR4+/+Gab1-/-, 56 ± 4%; CXCR4-/-Gab1-/-, 53 ± 5%). A small reduction in the differentiation rate was observed in CXCR4+/+Gab1-/- and CXCR4-/-Gab1-/- mice (control: 59 ± 3%; CXCR4+/+Gab1-/-: 50 ± 8%; CXCR4-/-Gab1-/-: 44 ± 5%).
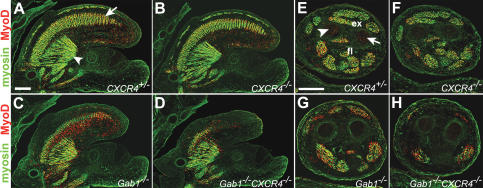
To assess whether the changes in progenitor numbers affected the generation of differentiated skeletal muscle, we compared tongue and limb muscle in control and mutant E13.5 embryos using anti-myosin and anti-MyoD antibodies (Fig. 7). In the tongue, several muscle groups are present at this stage. The extrinsic tongue muscle and proximal component of intrinsic tongue muscle derive from the head mesenchyme, whereas the distal component of the intrinsic tongue muscle is formed mainly by long-range migrating cells that derive from occipital somites (Huang et al. 1999; in Fig. 7A, intrinsic and extrinsic muscle are indicated by an arrow and arrowhead, respectively). No major difference was observable when the tongue muscles of CXCR4-/- and control mice were compared (Fig. 7A,B). However, when we compared the tongue muscle in CXCR4-/-Gab1-/- and CXCR4+/+Gab1-/- animals, major differences were noted (Fig. 7C,D). In CXCR4+/+Gab1-/- mice, the intrinsic tongue muscle was small but muscle tissue was observable in the proximal and distal tongue. In CXCR4-/-Gab1-/- mice, only a fragment of the intrinsic tongue muscle was present in the proximal tongue, and the overall size of the tongue was very small (Fig. 7C,D). The appearance of the tongue muscle of CXCR4-/-Gab1-/- embryos is comparable to that observed in c-Met-/- or SF/HGF-/- mutants; i.e., comparable to the morphology of a muscle that completely lacks the contribution of migrating muscle progenitor cells (data not shown). Comparison of the size and distribution of muscle groups in the fore- or hind-limbs revealed occasionally minor, but no reproducible, differences between CXCR4+/- and CXCR4-/- embryos at E13.5 (Fig. 7E,F). Thus, the moderate reduction in the number of progenitors at early developmental stages was subsequently compensated for and did not affect the final muscle size. The limb muscle of CXCR4+/+Gab1-/- mutant mice was, however, affected (Fig. 7G). Thus, the reduction in the number of progenitor cells observed at earlier stages of Gab1 mutants was not compensated. When we compared the limb muscle groups in CXCR4+/+Gab1-/- and CXCR4-/-Gab1-/- mice, additional major differences were observed. In the proximal portion of the lower forelimb, particular extensor muscles of the dorsal limb (for instance extensor carpi radialis, extensor digitorum communis, extensor digitorum lateralis) were absent in CXCR4-/-Gab1-/- mice (Fig. 7H). In CXCR4+/+Gab1-/- mice, these were observable but were reduced in size compared with control mice (Fig. 7G). Flexor muscles in the ventral limb were still present, but significantly smaller in CXCR4-/-Gab1-/- than in CXCR4+/+Gab1-/- mice (Fig. 7G,H). This indicates that compensatory mechanisms operate only if a threshold number of muscle progenitor cells reaches the limb.
Figure 7.
Differentiated muscle groups of the limb and tongue. (A-D) Sections of the tongue of CXCR4+/- (A), CXCR4-/- (B), Gab1-/- (C), and CXCR4-/-Gab1-/- (D) embryos at E13.5 stained with antibodies to myosin (green) and MyoD (red). (A) The intrinsic and extrinsic tongue muscles are indicated by an arrow and arrowhead, respectively. (E-H) Transverse sections through the proximal part of the lower forelimb of CXCR4+/- (E), CXCR4-/- (F), Gab1-/- (G), and CXCR4-/-Gab1-/- (H) embryos at E13.5 stained with antibodies to myosin (green) and MyoD (red). (E) Indicated are extensor (ex) and flexor (fl) muscles; arrowhead and arrow point toward the radius and ulna, respectively. Bars, 250 μm.
Discussion
Cell sorting allowed us to determine the gene expression profile of embryonic muscle progenitor cells obtained from limb buds. Among the expressed genes, we identified the chemokine receptor CXCR4. The corresponding ligand, SDF1, is expressed in the mesenchyme of the limbs and the first branchial arch. In CXCR4-/- mice, a reduction in the number of muscle progenitor cells was observed at particular target sites of migration. Furthermore, we observed a genetic interaction between CXCR4 and Gab1. Thus, the chemokine receptor CXCR4 that uses G-proteins for signaling and Gab1 that transduces signals of tyrosine kinases, for instance of the c-Met receptor, elicit similar cellular responses in vivo, which might reflect cross-talk between such signaling systems. We conclude that the signals provided by CXCR4 and Gab1 control migration and survival of muscle progenitor cells.
CXCR4 is transiently expressed during migration of muscle progenitor cells
We show here that the chemokine receptor CXCR4 is expressed only transiently in migrating muscle progenitor cells of chick and mouse. CXCR4 protein in muscle progenitors is present only after delamination from the dermomyotome and is observed on forelimb levels only in those muscle progenitor cells that had already entered the limb. CXCR4 expression in progenitor cells is thus observed after the onset of Pax3 or Lbx1 expression. In Lbx1 mutant mice, CXCR4 was not expressed in the muscle progenitor cells on limb levels; CXCR4 expression was, however, observed in the progenitors of the hypoglossal stream of Lbx1 mutant mice (E. Vasyutina and C. Birchmeier, unpubl.). Thus, Lbx1 might participate in controlling CXCR4 expression. Pax3+ or Lbx1+ muscle progenitor cells in the limb are heterogeneous with respect to CXCR4 expression. However, CXCR4 is exclusively expressed in MyoD-negative cells, and thus only in undifferentiated progenitor cells. Therefore, CXCR4 is expressed during migration of the muscle progenitor cells. Endogenous SDF1 expression is observed in the mesenchyme of the limb close to the positions occupied by progenitor cells. Along the route of muscle progenitor cells that migrate into the head, expression of SDF1 is found at the migration target; i.e., in the floor of the first branchial arch. The distribution of the endogenous SDF1 indicates that migrating muscle progenitors can encounter the factor during migration.
Ectopic application of SDF1 demonstrated that Pax3+ and CXCR4+ muscle progenitor cells migrate toward the SDF1 source. The redistribution of CXCR4+ cells was more pronounced than the redistribution of Pax3+ cells, in accordance with the existence of two Pax3+ cell populations, one population that expresses CXCR4 and responds to SDF1, and a second population that is CXCR4-negative and nonresponsive. These gain-of-function experiments indicate that SDF1 can provide directional cues for migrating muscle progenitor cells. Moreover, a loss-of-function mutation of CXCR4 in mice causes a change in the distribution of muscle progenitor cells in the dorsal forelimbs at E10.75. A reduction in numbers of progenitor cells is most pronounced in the distal limb of CXCR4 mutant mice. Increased apoptosis of limb cells was not detected in this distal domain, but was observed proximally. Thus, CXCR4 provides signals that control distribution and survival of migrating muscle progenitor cells. However, the highest concentration of SDF1 mRNA is observed in the central and distal limb bud at E11. The distribution of the protein is unknown, as antibodies that allow the detection of SDF1 within the tissue are not available. Muscle progenitors migrate distally, but not into the central domain. This indicates that the distribution of SDF1 cannot account for all aspects of the migratory behavior of the cells, raising the possibility that more than a single factor guides the migration. Such additional factors, for example, members of the ephrin family, could repulse muscle progenitor cells from the center of the limb (Swartz et al. 2001). Despite the significant reduction in the number of muscle progenitor cells of CXCR4 mutant mice at E10.75, reproducible changes in the size of muscle groups were not observed at E13.5, indicating that changes in progenitor numbers can be compensated at subsequent developmental stages.
We observed a genetic interaction between CXCR4 and Gab1. Gab1 is an important c-Met signal transduction molecule, which binds and recruits other signal transduction components to activated c-Met (for review, see Birchmeier et al. 2003). In Gab1 mutant mice, muscle progenitor cells reach the first branchial arch and the forelimb in reduced numbers. The deficit in progenitor numbers cannot be compensated at later developmental stages and is reflected in a change in size (or even the absence of) particular muscle groups (this study; Sachs et al. 2000). In CXCR4-/-Gab1-/- mice, a further decrease in the number of progenitor cells that reach the forelimb is observed that is more pronounced than in either of the single-mutant mice. The deficit is not compensated for and causes substantial changes in the size and distribution of muscle groups in CXCR4-/-Gab1-/- mice compared with CXCR4+/+Gab1-/- mice. Furthermore, migrating muscle progenitor cells do not reach the floor of the first branchial arch in CXCR4-/-Gab1-/- double-mutant mice; this target is colonized in either of the single-mutant mice. The consequence is a severe deficit in development of the intrinsic tongue muscle in the double-mutant mice. Increased apoptosis is observed in the hypoglossal cord and limbs, indicating that impaired cell migration and survival contribute to the changes in the distribution of progenitor cells.
The reduction in the numbers of distal muscle progenitor cells in the dorsal limb of CXCR4 mutants is compensated for at subsequent stages, since we did not observe reproducible changes in the size of differentiated limb muscles. In contrast, the reduction in the numbers of dorsal progenitors in Gab1-/- or CXCR4-/-Gab1-/- double-mutant mice was more pronounced and resulted in a smaller size or even an absence of limb muscles. In none of these mutant strains did we observe a change in the proliferation rate of muscle progenitors at E10.75; i.e., at the developmental stage at which the progenitor numbers were counted. Compensatory mechanisms might prolong the proliferative phase of the progenitors at subsequent stages. Our data indicate that compensation is only possible if a threshold number of progenitor cells reaches the limb. We attempted to estimate a critical number of progenitor cells, which ensures the generation of muscle of normal size. For this, the reduction in progenitor cell numbers caused by mutation of CXCR4 or Gab1 in particular limb domains (see Fig. 6I) were compared with resulting abnormalities of differentiated muscle groups observed at later development stages (E13.5). A 35% reduction in progenitors of the dorsal distal limb domain of CXCR4-/- mice did not affect the size of extensor muscle. The 60% reduction in the progenitor numbers in the ventral proximal limb domain of Gab1-/- mice results in only a small size reduction of flexor muscles. This indicates that the critical number corresponds to about half of the number of progenitors present in the limb of wild-type animals. A balance between proliferation and differentiation controls the muscle progenitor pool and the size of the differentiated muscle (Amthor et al. 1999). Compensatory proliferation of muscle progenitors might be only possible during a limited time period in which a favorable environment is provided in the developing limb.
A genetic interaction of Gab1 and CXCR4
Tyrosine kinase receptors and chemokine receptors are two important classes of molecules implicated in the regulation of cell migration, and can also affect proliferation and cell death. Gab1 is an adaptor molecule that functions in the signal transduction of c-Met and of other tyrosine kinases (Gu and Neel 2003). The phenotypes observed in the developing muscle of the Gab1 and CXCR4 mutations are similar: Both mutations affect the distribution and the survival of muscle progenitor cells, albeit to a different extent. In addition, a genetic interaction between the two loci can be observed: The muscle lineage of CXCR4;Gab1 double-mutant mice is more strongly affected than in single CXCR4 or Gab1 mutants.
Cell motility relies on the ordered disruption and reformation of cell adhesion sites, as well as on cytoskeletal dynamics (Ridley et al. 2004). Gab1 and CXCR4 signaling might have distinct endpoints, for instance one in the control of cell matrix attachment and the other in the control of actin polymerization, but might nevertheless cooperate to regulate cell migration. Alternatively, the genetic cooperation of Gab1 and CXCR4 might reflect the fact that both employ similar signaling cascades. Gab1 contains multiple docking sites for SH2 domain containing proteins like Shp2, the p85 subunit of PI3kinase, Src, Crk, and PLCγ (for review, see Birchmeier et al. 2003; Gu and Neel 2003). Tyrosine phosphorylation of Gab1 results in the activation of Ras/MAPK, PI3K/Akt, and PLCγ/PKC signaling pathways that regulate cell motility, proliferation, and survival. Chemokine receptors such as CXCR4 use G-proteins to transmit signals in the cytoplasm. Classical GPCR signaling involves activation of second messenger-regulated serine/threonine kinases or ion channels (for review, see Mellado et al. 2001). More recently, GPCRs were also found to stimulate tyrosine phosphorylation cascades (Daub et al. 1996). The molecular mechanisms responsible for this depend on the exact cell type. Signals provided by GPCR can induce the activity of metalloproteinases, which cause cell surface shedding of ligands of tyrosine kinase receptors like HB-EGF, and thus activate the signaling cascades downstream of the corresponding tyrosine kinase receptors (Prenzel et al. 1999; Yan et al. 2002). G-αi, which is used by CXCR4 to transmit signals, directly binds and activates the c-Src tyrosine kinase, which in turn activates the Ras/MAPK and PI3K/Akt pathways (Ma et al. 2000). Experiments performed on cultured cells indicate that signaling molecules shared by tyrosine kinase and chemokine receptors regulate cell migration. For instance, SF/HGF and SDF1 cooperate to elicit chemotaxis in cultured rhabdomyosarcoma cells, and PI3K/Akt activities are required for this response (Jankowski et al. 2003). c-Kit and CXCR4 signals attract hematopoietic progenitor cells, act synergistically, and both receptors rely on MAPK activity to transduce their chemotactic signals (Dutt et al. 1998). The genetic interaction that we observe for Gab1 and CXCR4 might reflect the fact that the signal transduction cascades used by these molecules converge on identical effectors to control migration and survival of muscle progenitor cells.
Materials and methods
Generation of an Lbx1GFP mutant allele
The Lbx1 targeting vector was assembled using the pTV vector (Riethmacher et al. 1995), which contains a neomycin gene flanked by loxP sites as well as a thymidine kinase gene to allow positive and negative selection, respectively. Lbx1 genomic sequences were isolated from the 129Sv library. The Gap43-GFP cassette (provided by U. Mueller, Friedrich Miescher Institute, Basel, Switzerland) was fused to the initiation codon of Lbx1; a frameshift mutation was introduced into the BglII site in the second exon of Lbx1. In addition, the targeting vector contains 3.9 kb upstream and 6.1 kb downstream homologous sequences of the Lbx1 locus. The linearized targeting vector was introduced into E14.1 embryonic stem (ES) cells by electroporation. ES colonies were selected with G418 and gancyclovir and screened for homologous recombination events using Southern blot hybridization. The neomycin resistance cassette was removed by transient expression of Cre in ES cells. Lbx1GFP ES cells were injected into C57B6 blastocysts to generate a mouse strain that carries the Lbx1GFP allele. CXCR4 mutant mice were obtained from the Jackson Laboratory and are described in Ma et al. (1998). The generation of the Gab1 mutant mice was detailed previously (Sachs et al. 2000).
FACS sorting, cRNA probe preparation and hybridization of Affymetrix GeneChips
The forelimbs of Lbx1+/GFP embryos were dissected, the cells were dissociated by treatment with 0.02% Trypsin (PAN Biotech GmbH) for 10 min, and the GFP-positive cells were isolated using fluorescence-activated cell sorting (FACS). Cell sorting was performed on a FACSVantage SE (Becton-Dickinson) using an argon ion laser (488 nm) for excitation. Total RNA from sorted cells was used for preparation of biotinylated cRNA probes according to the Affymetrix protocol. The probes of three independent cell pools were hybridized on MG U74A/B/Cv2 Affymetrix GeneChips. The results on the expression of a selected set of genes that encode surface molecules are indicated in Table 1. Four probes for SDF1 are present on the MG U74A/B/Cv2 Affymetrix GeneChips, 162234_f_at, 100112_at, 105704_at, and 160511_at. The hybridization signals of three of these, 162234_f_at, 105704_at, and 160511_at were called absent; signals of 100112_at were called present, but the intensity of the signal was low (p value = 0.006; hybridization signal intensity = 330 ± 65).
Cell transfection and implantation in ovo
Coding sequences of chicken SDF1 (Stebler et al. 2004) or EGFP (BD Biosciences Clontech) were inserted into the pcDNA3.1(-) expression vector (Invitrogen). COS1 cells were cotransfected with 10 μg of pcDNA-SDF1 and/or pcDNA-EGFP, using Lipofectamine2000 (GIBCO-BRL). Transfection efficiency was estimated by GFP expression. Thirty-six hours after transfection, cell aggregates were implanted into the right limb of chick embryos (HH19-20). At HH24-25, embryos in which we detected GFP-expressing cells in the limb were analyzed.
We analyzed the secreted SDF1 protein from transfected COS1 cells by Western blot. For cotransfection, 10 μg of each plasmid (pcDNA-SDF1 and pcDNA-EGFP) was used; for a control, 10 μg pcDNA-EGFP was transfected. Cells were grown for 36 h in a serum-containing medium, which was followed by a 24 h culture in a serum-depleted medium. The supernatants were then collected, precipitated with 20% (w/v) trichloroacetic acid, and dissolved in SDS-containing loading buffer. The samples were analyzed for the presence of SDF1 by Western blot analysis using goat anti-human SDF1 antibody (Sigma).
In situ hybridization and immunohistochemistry
Whole-mount in situ hybridization analysis of mouse and chick embryos was performed as described (Wilkinson 1992). The following RNA probes were used for in situ hybridization: A 945-bp fragment encoding mouse CXCR4; a fragment (660 bp) that encompasses the 3′ coding sequence and 3′ UTR of the mouse SDF1 gene; chicken Pax3 (Goulding et al. 1994); chicken MyoD (Lin et al. 1989); chicken Myf5 (Dechesne et al. 1994); chicken CXCR4 and SDF1 (Stebler et al. 2004). RNA transcripts were synthesized using a digoxigenin (DIG)-labeling kit (Roche Mannheim) according to the manufacturer's protocol.
Immunofluorescence staining was performed on 12-μm cryosections of embryonic tissue that had been fixed with 4% paraformaldehyde for 2 h. The following antibodies were used: monoclonal mouse anti-skeletal fast myosin (Sigma); rabbit anti-mouse MyoD (Santa Cruz); rabbit anti-GFP (Molecular Probes); guinea pig anti-mouse Lbx1 (Müller et al. 2002); rabbit anti-mouse CXCR4 (Stumm et al. 2002); and secondary antibodies conjugated with Cy2, Cy3, or Cy5 (Jackson ImmunoResearch Laboratories). To visualize CXCR4 protein, the signal obtained after incubation with a biotinylated goat anti-rabbit antibody (KPL) was amplified using the Cy3-TSA Fluorescence System (PerkinElmer Life Siences).
To assess the proliferation rate, BrdU (75 μg/g of body weight; Sigma) was injected intraperitoneally into the dams 2 h prior to the dissection of the embryos; incorporated BrdU was detected using mouse anti-BrdU antibodies (Sigma). Cell death was determined by TUNEL staining using an Apop-Tag fluorescein in situ apoptosis detection kit (Intergen). To determine the number of Lbx1+ cells in the limb bud, consecutive sections of stage and size-matched embryos (E10.75) were analyzed by immunohistochemistry, and the number of Lbx1+, BrdU+/Lbx1+ double-positive, and MyoD+ cells were counted on every third section in different limb domains. Additionally, TUNEL+ cells that were also Lbx1+ or positioned close to Lbx1+ cells were counted. The proliferation rate was determined as the number of BrdU+/Lbx1+ cells divided by the total number of Lbx1+ cells. Similarly, the differentiation rate was determined as the number of MyoD+ cells divided by the total number of Lbx1+ cells.
Acknowledgments
We thank Walter Birchmeier, Alistair Garratt, and Thomas Müller for critically reading the manuscript, and Henning Brohmann for valuable advice and discussion. We also thank Martyn Goulding (The Salk Institute, La Jolla, CA) for kindly providing Pax3 antibody. We gratefully acknowledge Alex Scheffold and Katharina Raba (Deutsches Rheuma-Forschung-szentrum, Berlin, Germany) for the FACS sorting, Stefan Britsch and Peter Westermann for help with the Affymetrix chip hybridization experiments, and Michael Stehle for help and advice with the bioinformatics analysis. C.B. is supported by grants from the DFG and BMBF.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.346205.
References
- Amthor H., Christ, B., and Patel, K. 1999. A molecular mechanism enabling continuous embryonic muscle growth—A balance between proliferation and differentiation. Development 126: 1041-1053. [DOI] [PubMed] [Google Scholar]
- Arnold H.H. and Braun, T. 2000. Genetics of muscle determination and development. Curr. Top. Dev. Biol. 48: 129-164. [DOI] [PubMed] [Google Scholar]
- Bagri A., Gurney, T., He, X., Zou, Y.R., Littman, D.R., Tessier-Lavigne, M., and Pleasure, S.J. 2002. The chemokine SDF1 regulates migration of dentate granule cells. Development 129: 4249-4260. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Birchmeier, W., Gherardi, E., and Vande Woude, G.F. 2003. Met, metastasis, motility and more. Nat. Rev. Mol. Cell. Biol. 4: 915-925. [DOI] [PubMed] [Google Scholar]
- Bladt F., Riethmacher, D., Isenmann, S., Aguzzi, A., and Birchmeier, C. 1995. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376: 768-771. [DOI] [PubMed] [Google Scholar]
- Bleul C.C., Fuhlbrigge, R.C., Casasnovas, J.M., Aiuti, A., and Springer, T.A. 1996. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1). J. Exp. Med. 184: 1101-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bober E., Franz, T., Arnold, H.H., Gruss, P., and Tremblay, P. 1994. Pax-3 is required for the development of limb muscles: A possible role for the migration of dermomyotomal muscle progenitor cells. Development 120: 603-612. [DOI] [PubMed] [Google Scholar]
- Brohmann H., Jagla, K., and Birchmeier, C. 2000. The role of Lbx1 in migration of muscle precursor cells. Development 127: 437-445. [DOI] [PubMed] [Google Scholar]
- Buckingham M. 2001. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 11: 440-448. [DOI] [PubMed] [Google Scholar]
- Chevallier A., Kieny, M., and Mauger, A. 1977. Limb-somite relationship: Origin of the limb musculature. J. Embryol. Exp. Morphol. 41: 245-258. [PubMed] [Google Scholar]
- Christ B. and Ordahl, C.P. 1995. Early stages of chick somite development. Anat. Embryol. (Berl.) 191: 381-396. [DOI] [PubMed] [Google Scholar]
- Christ B., Jacob, H.J., and Jacob, M. 1977. Experimental analysis of the origin of the wing musculature in avian embryos. Anat. Embryol. (Berl.) 150: 171-186. [DOI] [PubMed] [Google Scholar]
- Cyster J.G. 2003. Homing of antibody secreting cells. Immunol. Rev. 194: 48-60. [DOI] [PubMed] [Google Scholar]
- Daub H., Weiss, F.U., Wallasch, C., and Ullrich, A. 1996. Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature 379: 557-560. [DOI] [PubMed] [Google Scholar]
- Dechesne C.A., Wei, Q., Eldridge, J., Gannoun-Zaki, L., Millasseau, P., Bougueleret, L., Caterina, D., and Paterson, B.M. 1994. E-box- and MEF-2-independent muscle-specific expression, positive autoregulation, and cross-activation of the chicken MyoD (CMD1) promoter reveal an indirect regulatory pathway. Mol. Cell. Biol. 14: 5474-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich S., Abou-Rebyeh, F., Brohmann, H., Bladt, F., Sonnenberg-Riethmacher, E., Yamaai, T., Lumsden, A., Brand-Saberi, B., and Birchmeier, C. 1999. The role of SF/HGF and c-Met in the development of skeletal muscle. Development 126: 1621-1629. [DOI] [PubMed] [Google Scholar]
- Doitsidou M., Reichman-Fried, M., Stebler, J., Koprunner, M., Dorries, J., Meyer, D., Esguerra, C.V., Leung, T., and Raz, E. 2002. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell 111: 647-659. [DOI] [PubMed] [Google Scholar]
- Dutt P., Wang, J.F., and Groopman, J.E. 1998. Stromal cell-derived factor-1 α and stem cell factor/kit ligand share signaling pathways in hemopoietic progenitors: A potential mechanism for cooperative induction of chemotaxis. J. Immunol. 161: 3652-3658. [PubMed] [Google Scholar]
- Franz T., Kothary, R., Surani, M.A., Halata, Z., and Grim, M. 1993. The Splotch mutation interferes with muscle development in the limbs. Anat. Embryol. (Berl.) 187: 153-160. [DOI] [PubMed] [Google Scholar]
- Goulding M., Lumsden, A., and Paquette, A.J. 1994. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development 120: 957-971. [DOI] [PubMed] [Google Scholar]
- Gross M.K., Moran-Rivard, L., Velasquez, T., Nakatsu, M.N., Jagla, K., and Goulding, M. 2000. Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development 127: 413-424. [DOI] [PubMed] [Google Scholar]
- Gu H. and Neel, B.G. 2003. The `Gab' in signal transduction. Trends Cell Biol. 13: 122-130. [DOI] [PubMed] [Google Scholar]
- Huang R., Zhi, Q., Izpisua-Belmonte, J.C., Christ, B., and Patel, K. 1999. Origin and development of the avian tongue muscles. Anat. Embryol. (Berl.) 200: 137-152. [DOI] [PubMed] [Google Scholar]
- Jagla K., Dolle, P., Mattei, M.G., Jagla, T., Schuhbaur, B., Dretzen, G., Bellard, F., and Bellard, M. 1995. Mouse Lbx1 and human LBX1 define a novel mammalian homeobox gene family related to the Drosophila lady bird genes. Mech. Dev. 53: 345-356. [DOI] [PubMed] [Google Scholar]
- Jankowski K., Kucia, M., Wysoczynski, M., Reca, R., Zhao, D., Trzyna, E., Trent, J., Peiper, S., Zembala, M., Ratajczak, J., et al. 2003. Both hepatocyte growth factor (HGF) and stromal-derived factor-1 regulate the metastatic behavior of human rhabdomyosarcoma cells, but only HGF enhances their resistance to radiochemotherapy. Cancer Res. 63: 7926-7935. [PubMed] [Google Scholar]
- Knaut H., Werz, C., Geisler, R., and Nusslein-Volhard, C. 2003. A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature 421: 279-282. [DOI] [PubMed] [Google Scholar]
- Lazarini F., Tham, T.N., Casanova, P., Arenzana-Seisdedos, F., and Dubois-Dalcq, M. 2003. Role of the α-chemokine stromal cell-derived factor (SDF-1) in the developing and mature central nervous system. Glia 42: 139-148. [DOI] [PubMed] [Google Scholar]
- Li X., Oghi, K.A., Zhang, J., Krones, A., Bush, K.T., Glass, C.K., Nigam, S.K., Aggarwal, A.K., Maas, R., Rose, D.W., et al. 2003. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 426: 247-254. [DOI] [PubMed] [Google Scholar]
- Lin Z.Y., Dechesne, C.A., Eldridge, J., and Paterson, B.M. 1989. An avian muscle factor related to MyoD1 activates muscle-specific promoters in nonmuscle cells of different germ-layer origin and in BrdU-treated myoblasts. Genes & Dev. 3: 986-996. [DOI] [PubMed] [Google Scholar]
- Ma Q., Jones, D., Borghesani, P.R., Segal, R.A., Nagasawa, T., Kishimoto, T., Bronson, R.T., and Springer, T.A. 1998. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc. Natl. Acad. Sci. 95: 9448-9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.C., Huang, J., Ali, S., Lowry, W., and Huang, X.Y. 2000. Src tyrosine kinase is a novel direct effector of G proteins. Cell 102: 635-646. [DOI] [PubMed] [Google Scholar]
- Mellado M., Rodriguez-Frade, J.M., Manes, S., and Martinez, A.C. 2001. Chemokine signaling and functional responses: The role of receptor dimerization and TK pathway activation. Ann. Rev. Immunol. 19: 397-421. [DOI] [PubMed] [Google Scholar]
- Molyneaux K.A., Zinszner, H., Kunwar, P.S., Schaible, K., Stebler, J., Sunshine, M.J., O'Brien, W., Raz, E., Littman, D., Wylie, C., et al. 2003. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development 130: 4279-4286. [DOI] [PubMed] [Google Scholar]
- Müller A., Homey, B., Soto, H., Ge, N., Catron, D., Buchanan, M.E., McClanahan, T., Murphy, E., Yuan, W., Wagner, S.N., et al. 2001. Involvement of chemokine receptors in breast cancer metastasis. Nature 410: 50-56. [DOI] [PubMed] [Google Scholar]
- Müller T., Brohmann, H., Pierani, A., Heppenstall, P.A., Lewin, G.R., Jessell, T.M., and Birchmeier, C. 2002. The homeodomain factor Lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron 34: 551-562. [DOI] [PubMed] [Google Scholar]
- Müller G., Hopken, U.E., and Lipp, M. 2003. The impact of CCR7 and CXCR5 on lymphoid organ development and systemic immunity. Immunol. Rev. 195: 117-135. [DOI] [PubMed] [Google Scholar]
- Nagasawa T., Kikutani, H., and Kishimoto, T. 1994. Molecular cloning and structure of a pre-B-cell growth-stimulating factor. Proc. Natl. Acad. Sci. 91: 2305-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa T., Hirota, S., Tachibana, K., Takakura, N., Nishikawa, S., Kitamura, Y., Yoshida, N., Kikutani, H., and Kishimoto, T. 1996. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature 382: 635-638. [DOI] [PubMed] [Google Scholar]
- Perry R.L. and Rudnick, M.A. 2000. Molecular mechanisms regulating myogenic determination and differentiation. Front. Biosci. 5: D750-D767. [DOI] [PubMed] [Google Scholar]
- Prenzel N., Zwick, E., Daub, H., Leserer, M., Abraham, R., Wallasch, C., and Ullrich, A. 1999. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature 402: 884-888. [DOI] [PubMed] [Google Scholar]
- Ratajczak M.Z., Majka, M., Kucia, M., Drukala, J., Pietrzkowski, Z., Peiper, S., and Janowska-Wieczorek, A. 2003. Expression of functional CXCR4 by muscle satellite cells and secretion of SDF-1 by muscle-derived fibroblasts is associated with the presence of both muscle progenitors in bone marrow and hematopoietic stem/progenitor cells in muscles. Stem Cells 21: 363-371. [DOI] [PubMed] [Google Scholar]
- Raz E. 2003. Primordial germ-cell development: The zebrafish perspective. Nat. Rev. Genet. 4: 690-700. [DOI] [PubMed] [Google Scholar]
- Ridley A., Peckham, M., and Clark, P. 2004. Cell motility: From molecules to organisms. Wiley, Chichester, United Kingdom; Hoboken, NJ.
- Riethmacher D., Brinkmann, V., and Birchmeier, C. 1995. A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc. Natl. Acad. Sci. 92: 855-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs M., Brohmann, H., Zechner, D., Müller, T., Hülsken, J., Walther, I., Schaeper, U., Birchmeier, C., and Birchmeier, W. 2000. Essential role of Gab1 for signaling by the c-Met receptor in vivo. J. Cell Biol. 150: 1375-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer K. and Braun, T. 1999. Early specification of limb muscle precursor cells by the homeobox gene Lbx1h. Nat. Genet. 23: 213-216. [DOI] [PubMed] [Google Scholar]
- Stebler J., Spieler, D., Slanchev, K., Molyneaux, K.A., Richter, U., Cojocaru, V., Tarabykin, V., Wylie, C., Kessel, M., and Raz, E. 2004. Primordial germ cell migration in the chick and mouse embryo: The role of the chemokine SDF-1/CXCL12. Dev. Biol. 272: 351-361. [DOI] [PubMed] [Google Scholar]
- Stumm R.K., Rummel, J., Junker, V., Culmsee, C., Pfeiffer, M., Krieglstein, J., Hollt, V., and Schulz, S. 2002. A dual role for the SDF-1/CXCR4 chemokine receptor system in adult brain: Isoform-selective regulation of SDF-1 expression modulates CXCR4-dependent neuronal plasticity and cerebral leukocyte recruitment after focal ischemia. J. Neurosci. 22: 5865-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz M.E., Eberhart, J., Pasquale, E.B., and Krull, C.E.. 2001. EphA4/ephrin-A5 interactions in muscle precursor cell migration in the avian forelimb. Development 128: 4669-4680. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Rocancourt, D., Cossu, G., and Buckingham, M. 1997. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 89: 127-138. [DOI] [PubMed] [Google Scholar]
- Wilkinson D.G. 1992. In situ hybridization: A practical approach. Oxford University Press, Oxford, United Kingdom.
- Yan Y., Shirakabe, K., and Werb, Z. 2002. The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein-coupled receptors. J. Cell. Biol. 158: 221-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y.R., Kottmann, A.H., Kuroda, M., Taniuchi, I., and Littman, D.R. 1998. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature 393: 595-599. [DOI] [PubMed] [Google Scholar]