Abstract
The eukaryotic translation initiation factors 4A (eIF4A) and 4G (eIF4G) are crucial for the assembly of the translationally active ribosome. Together with eIF4E, they form the eIF4F complex, which recruits the 40S subunit to the 5′ cap of mRNA. The two-domain RNA helicase eIF4A is a very weak helicase by itself, but the activity is enhanced upon interaction with the scaffolding protein eIF4G. Here we show that, albeit both eIF4A domains play a role in binding the middle domain of eIF4G (eIF4G-m, amino acids 745-1003), the main interaction surface is located on the C-terminal domain. We use NMR spectroscopy to define the binding site and find that the contact surface is adjacent to the RNA-, ATP-, and eIF4A-NTD-interacting regions. Mutations of interface residues abrogated binding, confirmed the interface, and showed that the N-terminal end of eIF4G-m interacts with the C-terminal domain of eIF4A. The data suggest that eIF4G-m forms a soft clamp to stabilize the closed interdomain orientation of eIF4A. This model can explain the cooperativity between all binding partners of eIF4A (eIF4G, RNA, ATP) and stimulation of eIF4A activity in the eIF4F complex.
Keywords: Translation, protein-protein interaction, eIF4A, eIF4G, helicase, NMR
Expression of eukaryotic genes is highly regulated at the level of translation. Control of translation is crucial for cellular proliferation, development, differentiation, and death, and deficiencies in translational regulation have been correlated with cellular transformation. Recruitment of the small ribosomal subunit to the mRNA, scanning to the start codon, unwinding of the 5′ untranslated regions of RNA (5′UTRs), and many more tasks are carried out by a large set of proteins termed eukaryotic initiation factors (eIFs) (for reviews, see Sonenberg et al. 2000; Preiss and Hentze 2003; Sonenberg and Dever 2003; Kapp and Lorsch 2004; Marintchev and Wagner 2005). One of the key components in the assembly of the translationally active ribosome is eIF4F, a complex comprised of the ATP-dependent RNA helicase eIF4A, the adaptor protein eIF4G, and the 5′-cap-binding protein eIF4E. The eIF4G-eIF4A interaction is responsible for recruitment of the 43S subunit to capped (Morino et al. 2000) as well as some uncapped mRNAs (Pestova et al. 1996; Lomakin et al. 2000), and the eIF4G-mediated association of eIF4A with the preinitation complex supports scanning (Pestova and Kolupaeva 2002). The helicase activity of free eIF4A is weak but is enhanced when the protein is bound to eIF4G within the eIF4F complex, and when eIF4B and/or eIF4H are associated (see reviews quoted above and Abramson et al. 1988; Rogers et al. 1999, 2001).
Human eIF4G is the master scaffold protein for the assembly of the multiprotein complex that recruits the ribosome to mRNA. The N-terminal ∼600-residue region contains interaction sites for polyA-binding proteins (PABP) and eIF4E (for reviews, see Hershey et al. 1999; Sonenberg et al. 2000; Marintchev and Wagner 2005). The structure of the complex of the eIF4E-binding domain of yeast eIF4G with eIF4E has been solved (Gross et al. 2003). The crystal structure of the conserved middle domain of eIF4GII (eIF4G-m, amino acids 745-1003), which binds eIF4A and RNA, exhibits a crescent-like shape with five antiparallel α-helical hairpins (HEAT-repeats) arranged in a helical stack (Marcotrigiano et al. 2001). The C-terminal region (eIF4G-c, amino acids ∼1200-1600) contains another eIF4A-binding domain in its N-terminal half and a binding region for Mnk kinases in its C-terminal half (for reviews, see Prevot et al. 2003; Marintchev and Wagner 2005). The crystal structure of the C-terminal region of eIF4GI was also solved recently and shown to consist of two HEAT domains (Protein Data Bank [PDB] code 1UG3) (S.K. Burley, unpubl.). Therefore, the C-terminal two-thirds of human eIF4G consist of three consecutive HEAT domains, the first two of which interact with eIF4A. Both isoforms of yeast eIF4G (NP_011678, NP_011466) lack a region corresponding to the C-terminal region of human eIF4G and thus the second eIF4A-binding domain. Interestingly, the middle region of human eIF4G (amino acids ∼700-1200), comprised of the first HEAT domain (eIF4G-m) and the linker region important for eIF3 binding, is sufficient for the recruitment of capped and uncapped mRNA and scanning, whereas the eIF4E-binding region and the second eIF4A-binding domain of eIF4G are dispensable (Morino et al. 2000; Ali and Jackson 2001; Ali et al. 2001; Pestova and Kolupaeva 2002). This suggests a regulatory or accessory role for the second eIF4A-binding domain of eIF4G.
eIF4A is a prototypic member of the DEAD-box family of proteins, a subgroup in the large superfamily 2 (SF2) of helicases (Linder et al. 1989). The ∼400-residue protein consists of two α/β domains with a RecA-like (Story et al. 1992) topology. Crystal structures of helicases show different snapshots of possible domain orientations, ranging from an open conformation with two completely independent domains in a dumbbell-like arrangement to a tightly closed conformation of the active enzyme (Kim et al. 1998; Benz et al. 1999; Caruthers et al. 2000; Story et al. 2001; Shi et al. 2004; Zhao et al. 2004).
Currently, three DEAD box RNA helicase structures that contain both core domains are known. These are eIF4A from yeast (Benz et al. 1999; Caruthers et al. 2000), the archaeal eIF4A homolog MjDEAD (Story et al. 2001), and human UAP56 (Shi et al. 2004; Zhao et al. 2004). The N- and C-terminal domains are structural homologs, but the orientations of the N- and C-terminal domains to each other are strikingly different. The crystal structure of yeast eIF4A exhibits a completely open conformation, whereas closed conformations were observed for MjDEAD and UAP56. The closed form is considered the active enzyme, where the ATP-binding cleft is formed upon contact between the two domains.
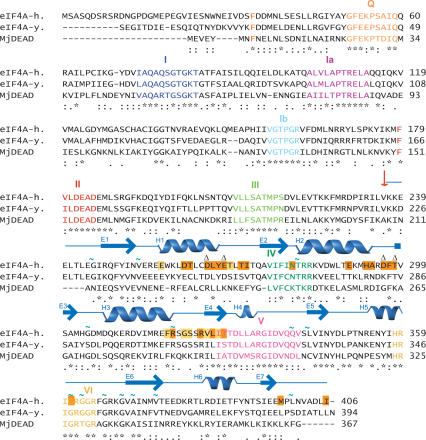
DEAD box helicases are characterized by a set of conserved motifs: Q, I (Walker A), Ia, Ib, II (Walker B), III, IV, V, and VI (Gorbalenya and Koonin 1993; Tanner et al. 2003). The helicase motifs are dispersed throughout the entire protein sequence, but most of them are clustered in close spatial proximity facing the cleft between the two domains in the closed conformation. The individual roles of each motif have been proposed based on analysis of crystal structures, genetic data, and biochemical experiments (for reviews, see Tanner and Linder 2001; Delagoutte and von Hippel 2002; Tanner et al. 2003): Motifs I, II (Walker motif A and B), and probably also motifs Q and VI, are involved in ATP binding and hydrolysis. Motifs Ia, Ib, IV, and V are proposed to be involved in RNA binding and unwinding (see Fig. 3A, below). Motif III (and possibly motif VI) has been suggested to link ATP hydrolysis with helicase activity (for reviews, see Tanner and Linder 2001; Delagoutte and von Hippel 2002; Rogers et al. 2002; Rocak and Linder 2004). Binding to RNA and ATP are coupled, and the helicase undergoes conformational rearrangements upon nucleotide binding, hydrolysis, and RNA unwinding (Lorsch and Herschlag 1998a,b). The structural mechanisms by which eIF4G modulates helicase and RNA binding activity are not yet known.
Figure 3.
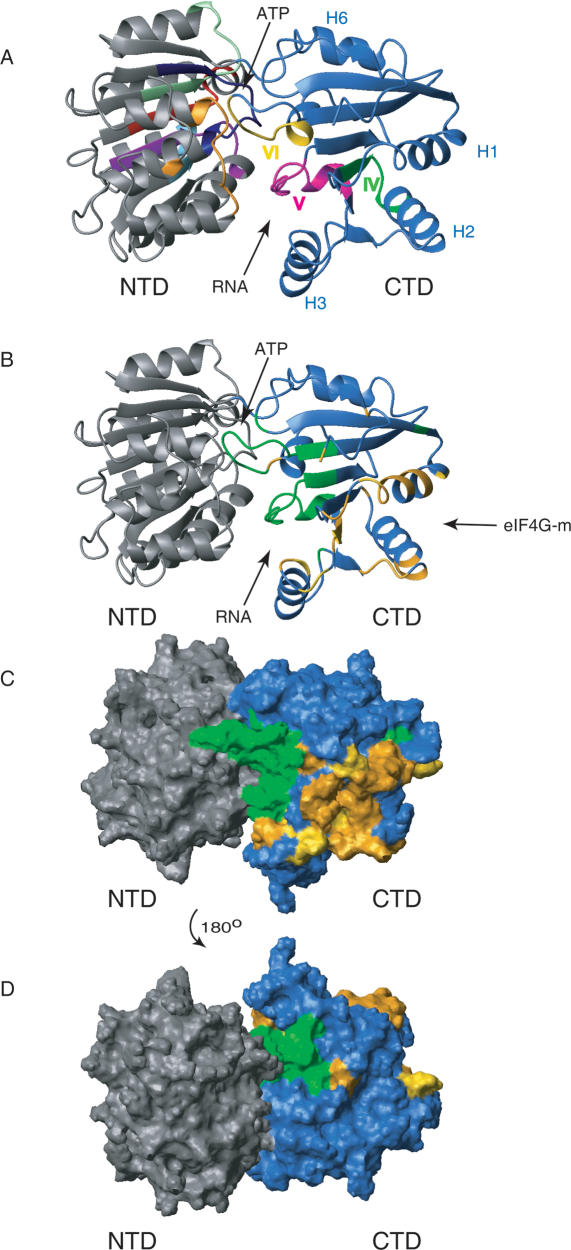
Structural model of full-length eIF4A and the eIF4G-m-binding regions on eIF4A-CTD. (A) Ribbon diagram of the homology model of full-length eIF4A. The N-terminal domain is colored gray; the C-terminal domain is colored blue. The conserved motifs are colored corresponding with Figure 2 (Q, orange; I, navy; Ia, purple; Ib, light blue; II, red; III, pale green; IV, dark green; V, magenta; VI, yellow). The position of motifs IV, V, and VI in eIF4A-CTD are highlighted with roman numerals. The faces of the interdomain clefts for substrate (RNA) and ATP binding are indicated with arrows. (B) Structural model of full-length eIF4A in the same orientation as in A. Residues involved in the binding to eIF4G-m are highlighted orange and gold; dynamic residues corresponding to very weak peaks and residues with exchange broadening are green (with the exceptions of R319 and G361, which are also affected by eIF4G-m binding and thus are colored orange). Surface representations in the same orientation (C) and after a 180° rotation (D) with the same color coding clearly show the distinct area involved in eIF4G-m binding.
Despite the great progress in solving structures of eIF4G domains involved in eIF4A binding, the architecture of the eIF4A/eIF4G complex has not yet been determined on a structural level, and also, the locations of the binding sites on either protein have remained elusive. As explained above, human eIF4G has two binding domains for eIF4A, whereas the second binding domain is missing in yeast. The stoichiometry of the interaction is subject of controversy and has been reported as 1:2 as well as 1:1 (Korneeva et al. 2001; Li et al. 2001). It also is unknown whether eIF4A uses the same or different surfaces for interaction with the two binding domains on eIF4G. Recent reports based on competition experiments and mutation analysis suggest different (but possibly overlapping) binding surfaces on eIF4A for the interactions with the middle and the C-terminal domains of eIF4G (Yang et al. 2003; Korneeva et al. 2005; Zakowicz et al. 2005), but again these questions remain to be answered on a structural level.
Here we analyze the mechanisms of the human eIF4AI/eIF4GII interaction. We find that the C-terminal domain of eIF4A is sufficient for binding eIF4G-m. However, the interaction of eIF4G-m with full-length eIF4A is stronger than that with eIF4A-CTD alone, indicating a role of eIF4A-NTD in the complex. We have identified the surface area of eIF4A-CTD that contacts eIF4G-m and find that it is adjacent to, but not overlapping with residues involved in the coupled mechanism of ATP and RNA binding, hydrolysis, and unwinding. Furthermore, our data suggest that the N-terminal region of eIF4G-m is responsible for interaction with eIF4A-CTD. The results presented here resolve recent conflicting reports on the eIF4A/eIF4G interaction (Korneeva et al. 2005; Zakowicz et al. 2005) and provide clues for the mechanisms by which eIF4G binding enhances eIF4A activity. The eIF4A/eIF4G interface revealed here also defines a potential target for discovery of compounds that specifically inhibit translation of mRNAs with long and structured 5′UTRs. Such mRNAs are often found for growth-promoting and oncogenic proteins (Kozak 1991), and inhibitors of their translation are hoped to have anti-tumor activity.
Results
The middle domain of eIF4GII binds the C-terminal domain of eIF4AI
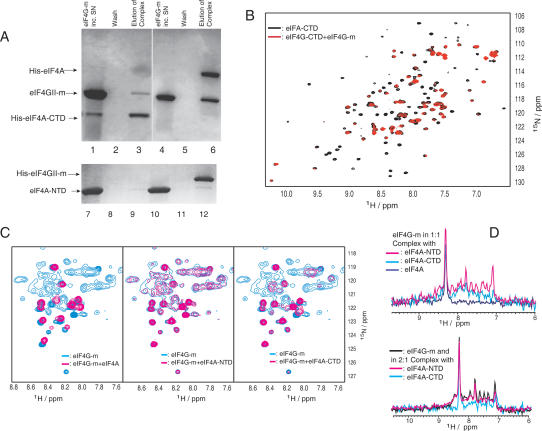
To confirm the interaction of eIF4AI with the middle domain of eIF4GII, and to narrow down the location of the interface, His-tagged full-length eIF4AI (eIF4A) and C-terminal domain (eIF4A-CTD, residues 237-406) were prepared and tested for interaction with eIF4GII (eIF4G-m, residues 745-1003). NMR and circular dichroism spectroscopy confirmed that the constructs were folded (data not shown). As expected, eIF4A was able to bind eIF4G-m effectively, and elution under native conditions resulted in bands corresponding to both proteins (Fig. 1A). Under the same experimental conditions, we also found that the C-terminal domain is sufficient for binding to eIF4G-m, although the interaction is weaker, as judged from the intensity of the Coomassie-stained bands (Fig. 1A). The binding between eIF4G-m and eIF4A-NTD was tested using His-tagged eIF4G-m and untagged eIF4A-NTD, because of the close similarity in molecular weight of the His-tagged N-terminal domain of eIF4A (eIF4A-NTD, residues 1-244) and eIF4G-m. eIF4A-NTD binding appeared only slightly above the background (Fig. 1A) but was visible above background at a lower salt concentration (data not shown).
Figure 1.
Binding of eIF4G-m to eIF4A detected by pull-down assays and NMR spectroscopy. (A) Lanes 1-6 depict the His-tag pull-down experiments when immobilized His8-eIF4A-CTD (lanes 1-3) and His6-eIF4A (lanes 4-6) were incubated with eIF4G-m (lanes 1,4). (Lanes 2,5) Unbound eIF4G-m was eluted completely by extensive washing steps. Bound eIF4G-m was eluted as a complex with eIF4A-CTD (lane 3) and eIF4A (lane 6). Lanes 7-12 show the reverse pull-down when untagged eIF4A-NTD was incubated on a control resin (lanes 7-9) and a resin with immobilized His6-eIF4G-m (lanes 10-12). After washing (lanes 8,11), bound proteins were eluted (lanes 9,12). (B) A characteristic overlay of 1H-15N-HSQC spectra of 15N-labeled eIF4A-CTD (500 μM) in free form (black) and complexed with the middle domain eIF4G-m (600 μM, red). (C) Overlays of 1H-15N-HSQC spectra of free 15N-labeled eIF4G-m (330 μM, blue) and 1:1 complexes of 15N-labeled eIF4G-m (130 μM, magenta) with equimolar amounts of full-length eIF4A (left), eIF4A-NTD (middle), and eIF4A-CTD (right) in buffer containing 300 mM NaCl. All spectra were acquired with the same number of scans. The intensities in the spectra of free 15N-labeled eIF4G-m were scaled to account for the difference in concentration. (D) 1D cross-sections of two-dimensional (2D) 1H-15N-HSQC spectra of free and complexed eIF4G-m taken at 119 ppm. The top and bottom panels show 1D slices as observed during titrations with buffer containing 300 and 100 mM NaCl, respectively. The different colors correspond to free eIF4G-m (black) and to eIF4G-m in complex with eIF4A-NTD (magenta), with eIF4A-CTD (cyan), and with eIF4A (navy).
NMR is an excellent method for studying weak protein-protein interactions. We recorded 1H-15N-HSQC spectra of uniformly 15N-labeled eIF4G-m in the absence or presence of these binding partners. In agreement with our pull-down experiments, we could observe a dramatic loss of signal intensities when eIF4A was added to eIF4G-m, indicative of the presence of a high-molecular-weight complex (Fig. 1C [left], D [top]). In contrast, addition of eIF4A-NTD introduced only minor changes in the peak intensities of the 1H-15N-HSQC spectrum of eIF4G-m, which indicates only a very weak interaction (Fig. 1C [middle], D). The leading role of eIF4A-CTD in binding of the middle domain of eIF4G was confirmed with the significant decrease of the intensities of the peaks in the 1H-15N-HSQC spectrum of labeled eIF4G-m (Fig. 1C [right], D). As a further control, we performed the reverse titration and observed only very few changes in the peaks of the 1H-15N-HSQC spectrum of 15N-labeled eIF4A-NTD upon addition of eIF4G-m (Supplementary Fig. S1).
The interaction of eIF4A-CTD with eIF4G-m is salt dependent. NMR titration experiments were carried out at 100 mM NaCl and at 300 mM NaCl concentration (Fig. 1D). Higher salt concentration increases the solubility and stability of eIF4A-CTD but reduces the affinity to eIF4G-m. Lowering the salt concentration resulted in more drastic decreases of signal intensities of eIF4G-m when adding substoichiometric concentrations of eIF4A-CTD. This indicates that charged regions of the protein surface are involved in the complex formation. Only minor changes were observed in the spectrum of 15N-labeled eIF4G-m upon addition of eIF4A-NTD, at both salt concentrations (Fig. 1D). Based on these results, we focused on the interaction between eIF4A-CTD and eIF4G-m. Using 15N-labeled eIF4A-CTD and addition of small aliquots of unlabeled eIF4G-m resulted in the predominant broadening and vanishing of a distinct set of peaks (Fig. 1B). In order to preserve the overall quality of the spectra, we performed most of the analysis at substoichiometric concentration of the unlabeled binding partner.
The broadening of the resonances in the titration experiments indicates that the eIF4G-m/eIF4A-CTD complex is in intermediate exchange on the NMR time scale. The line shapes in the 1H-15N-HSQC spectra are sensitive to the off-rates in the complex. Thus, the intermediate exchange behavior of the eIF4G-m/eIF4A-CTD complex suggests KD values in the low micromolar range (KD ∼1-10 μM), assuming diffusion-controlled on-rates. If full-length eIF4A is titrated to labeled eIF4G-m, it appears that some peaks change their position upon titration, indicating that some residues of this complex are in the fast exchange limit. These strong peaks most likely correspond to residues in flexible tail and loop regions on the periphery of the main protein-protein interface and thus display different dynamic properties. Although peaks in the intermediate exchange regime should reappear in new positions in excess of the unlabeled binding partner, it was difficult to unambiguously detect such peaks due to the high molecular weights of the complexes of eIF4G-m with eIF4A-CTD (∼50 kDa) and full-length eIF4A (∼80 kDa), despite the use of deuteration and TROSY-techniques (Pervushin et al. 1997).
Resonance assignments and model building of eIF4A-CTD
To be able to identify the eIF4G-m binding surface on eIF4A-CTD with NMR chemical shift mapping, we pursued the assignments of the peaks in the 1H-15N-HSQC spectra of free eIF4A-CTD to the corresponding amide groups in the protein. Using NMR triple-resonance experiments (for review, see Ferentz and Wagner 2000), backbone and partial side-chain resonance assignments were obtained for 156 (93%) residues of eIF4A-CTD. Since there is no structure available for human eIF4A, we built a model for full-length eIF4A based on the closed structures of homologous helicases (Figs. 2, 3). The modeled structure of eIF4A-CTD contains a twisted, seven-stranded parallel β-sheet (E1-E7) and six α-helices (H1-H6) connected by short loops and turns (Fig. 3). The placement of secondary structure elements in eIF4A-CTD was validated against our NMR data, including short- and medium-range NOEs and chemical-shift data analyzed with the TALOS program (Fig. 2; Cornilescu et al. 1999). The model is consistent with the experimental data, except that some secondary structure elements are slightly shorter or longer by one residue.
Figure 2.
Sequence alignments of human eIF4A-1 (accession no. P60842), yeast eIF4A (accession no. P10081), and MjDEAD (PDB entry 1HV8), the homologous protein from the hyperthermophile Methanococcus jannaschii. The conserved motifs of the DEAD-box helicase are labeled and depicted in different colors (Q, orange; I, navy; Ia, purple; Ib, light blue; II, red; III, pale green; IV, dark green; V, magenta; VI, yellow) and labeled with roman numerals. Identical amino acids are indicated with (★); residues with conserved and semi-conserved substitutions are indicated with (:) and (.), respectively. The start site of the C-terminal construct of eIF4A-CTD is indicated with a red arrow. Secondary structure elements of yeast eIF4A are blue (arrow, β-strand; helix, α-helix). The nomenclature for the secondary structure elements (E1-E7) as used throughout the text is printed in blue at the beginning of the respective sheets and helices. Residues for which a decrease in the signal is observed after addition of eIF4G-m at eIF4A-CTD/eIF4G-m ratio of 3:1 are highlighted with orange boxes. Yellow boxes indicate additionally affected residues if the entire titration range (up to eIF4A-CTD/eIF4G-m ratio of 1:1.5) is taken into account. The sites of the mutations D265R/E268K and D296A/T298K are highlighted with (Δ). Residues with relaxation contribution resulting from conformational exchange are depicted with (∼).
Identification of mobile regions of eIF4A-CTD in the putative NTD-interaction face
During the assignment process, it became evident that signals of some protein regions exhibited extensive line-broadening. We mapped these regions (colored green in Fig. 3B-D) onto the homology model of the eIF4A structure and noticed that these stretches corresponded to regions comprising the short helices H4 and H5, which are at the proposed interface with the N-terminal domain. For a qualitative assessment of this phenomenon, we measured the 15N relaxation parameters, R2, off-resonance R1ρ, and steady-state [1H]-15N NOEs, which are sensitive to internal mobility (Palmer et al. 2001).
Motions with fast subnanosecond time constants can be monitored with heteronuclear [1H]-15N NOE experiments and are typically used to identify fast-moving loops and tails. The profile for [1H]-15N NOE values (Supplementary Fig. S2, top) shows an increase of heteronuclear NOE values, starting from negative values for the very N-terminal residues, followed by a plateau region (average [1H]-15N NOE value 0.80 ± 0.07) and a decrease for the C-terminal residues. In addition, a region of lower NOE values can also be observed for residues G363 to G370 in the glycine-rich motif VI, which form a loop between helix H5 and strand E6 (average [1H]-15N NOE value 0.67 ± 0.11). Thus, in addition to the expected flexibilities of N- and C-terminal tails, the middle portion of the conserved glycine-rich motif VI exhibited fast mobility on the subnanosecond time scale. This motif is believed to be involved in the binding of the phosphates of ATP. Thus, differences in the dynamic behavior of these residues between the uncomplexed and complexed form are likely. Flexible residues (except N- and C-terminal tails) and residues with exchange contribution are colored green in Fig. 3B-D).
Conformational mobility on the microsecond-to-millisecond time scale can be identified by comparing R2 and R1ρ relaxation rates. Whereas relaxation due to exchange is suppressed in the R1ρ experiment, it is not, or is only partially, suppressed in the R2 experiment. Experimental R2 relaxation rates for eIF4A-CTD increase for the first N-terminal residues until they plateau at 19.0 (±2.4) sec-1 for most of the residues. The rates decrease again toward the very C terminus (data not shown). Several residues—including T242, G245, V253, T281, D305, R319, V325, Q340, L343, G361, G363, and A272—display substantial R2 relaxation rates above average, which could arise from conformational exchange contributions. Comparing R2 with R1ρ relaxation rates (Supplementary Fig. S2, bottom) we found that the glycine-rich loop G361-G370 and the first residues in strand 6 (E6) (V371, A372) undergo conformational exchange on this time scale. This type of motion is also indicated for residues in the loop adjacent to helix H4 (Q340, L343), which is proposed to form interdomain contacts. However, signal overlap and line-broadening of these peaks prevented a more detailed analysis. Other residues that show exchange contributions are G245, V253, T281, G304, and R319. Residues with exchange contribution are annotated with ∼ in Figure 2. Almost all of these are located in loops and turns connecting the helical and extended segments in the interface with the NTD (Fig. 3B-D).
The eIF4G-m contact surface of eIF4A-CTD is on the periphery of the RNA-, ATP-, or NTD-interacting surfaces but is distinct from the conserved motifs
To elucidate the eIF4G-m-binding surface on eIF4A-CTD, we mapped the changes in the 1H-15N-HSQC of labeled eIF4A-CTD upon titration with unlabeled eIF4G-m onto the modeled structure of eIF4A (colored orange and gold in Figs. 2, 3). The binding interface is a contiguous surface area, including the C termini of helices H1, H2, H3, their adjacent loop regions, and strand E4 (Fig. 3B-D). Viewing eIF4A from the “top” as in the orientation of Figure 3A-C, the eIF4G-m interaction surface (colored gold and orange) is located at the bottom right. The interacting residues are adjacent to, but generally not overlapping with, the conserved helicase motifs IV, V, and VI. Motifs IV and V are thought to be involved in RNA binding, and motif VI is believed to be engaged in ATP binding and coupling of nucleotide hydrolysis to RNA binding (for review, see Tanner and Linder 2001; Rocak and Linder 2004). The conserved RNA-binding motifs IV and V (colored green and magenta in Fig. 3A) cluster midway to beneath the center of the molecule in this orientation (cf. Fig. 5, below). They are in close proximity to the residues affected by eIF4G-m binding. Residues I327 and T328 in the strand E4 are on the periphery of the conserved motif V and are affected by binding to eIF4G-m. The mapped interaction region between eIF4A-CTD and eIF4G-m leaves the putative interdomain surface accessible to interact with eIF4A-NTD and thus allows the formation of the ATP-binding pocket. Likewise, rearrangements of the domains upon ATP binding, hydrolysis, and RNA unwinding would not be impeded.
Figure 5.
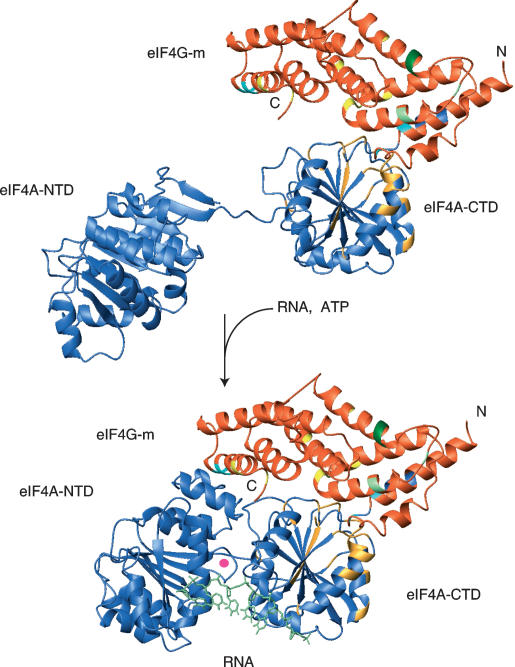
Model of the eIF4G-m as a soft clamp of eIF4A. We propose that eIF4G-m (orange) binds to the C-terminal domain of eIF4A (eIF4A-CTD, blue) and stabilizes the closed conformation of the helicase. Accessibility of the RNA- and ATP-binding surfaces of the helicase is retained. Residues contributing to the eIF4G-m-binding surface of eIF4A-CTD are colored gold. Surface mutations in eIF4G-m abolishing/drastically reducing binding of eIF4A and RNA are colored blue and green, respectively. Surface mutations that showed no effect are yellow. The open and closed conformations in our model correspond to the interdomain orientation as observed in the crystal structures of yeast eIF4A and MjDEAD (Caruthers et al. 2000; Story et al. 2001). To model the binding site of the oligonucleotide, human eIF4A-CTD from our homology model was superimposed to the structure of HCV NS3 in complex with bound oligonucleotide (Kim et al. 1998). For the sake of clarity, the structure of HCV NS3 is omitted and only the orientation of the oligonucleotide is shown. The SO4- ion found in the loop of the ATP-binding motif I is indicated by a filled circle.
The types of amino acids located in the interface indicate that electrostatic interaction contributes to the binding. Of the 30 residues that are identified as affected upon binding of eIF4G-m, five (17%) residues are threonines; six (20%) and two (7%) are negatively and positively charged, respectively. Out of the nine (30%) hydrophobic residues (ILVF), three are located in the buried strand E2. This pattern is consistent with the observed salt dependence of the strength of the eIF4A-CTD/eIF4G-m interaction and complements the positively charged surface residues on eIF4G-m found to be important for binding to eIF4A (Marcotrigiano et al. 2001).
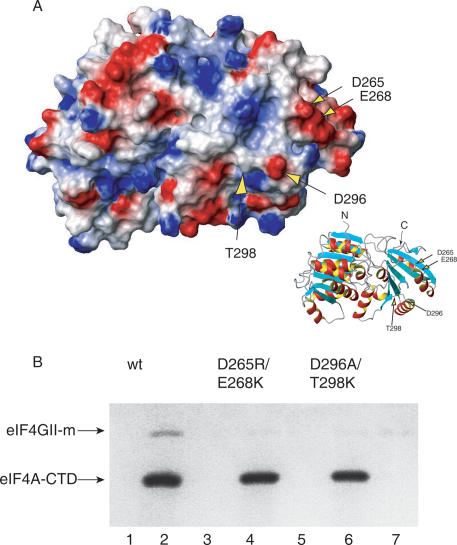
eIF4A-CTD mutants D265R/E268K and D296A/T298K lose their ability to bind eIF4G-m
In order to validate the eIF4G-m-interacting surface by a method independent from NMR spectroscopy, we introduced double mutations of adjacent solvent-exposed residues (Fig. 4) in the proposed interaction site. Mutation D265R/E268K harbors a double mutation of two negative residues that are conserved in eukaryotes and exchanged them to basic residues as found in archaea. In a similar manner, the mutation D296A/T298K was designed to resemble the sequence of the homolog from archaea more closely to ensure a change of surface properties without disruption of the fold. In addition, one-dimensional (1D) NMR spectra of unlabeled mutant proteins were recorded and showed that the resonance dispersion is typical for folded proteins and the qualitative appearances of the spectra are conserved (data not shown). Both mutants, D265R/E268K and D296A/T298K, were tested in His-tagged pull-down assays and were unable to bind eIF4G-m at the detection limit of the assay (Fig. 4). These results are in excellent agreement with the predicted disruption or weakening of the interaction based on the binding surface on eIF4A-CTD as identified by NMR spectroscopy.
Figure 4.
(A) Electrostatic surface of eIF4A with the locations of the four mutated residues in the double mutants D265R/E268K and D296A/T298K indicated by arrows. The orientation is shown in the insert on the right. (B) eIF4G-m binding to wild-type eIF4A-CTD and to the double mutants D265R/E268K and D296A/T298K detected by pull-down assays. Wild-type and mutant eIF4A-CTD protein was immobilized on a His-resin and incubated with eIF4G-m. (Lanes 1,3,5) Unbound eIF4G-m was eluted completely by extensive washing steps. Bound eIF4G-m was eluted as a complex with eIF4A-CTD wild type (wt) (lane 2), but no binding to eIF4G-m could be detected in the elution fractions of eIF4A-CTD D265R/E268K (lane 4) and eIF4A-CTD D296A/T298K (lane 6).
The N-terminal helical hairpins of eIF4G-m face the C-terminal domain of eIF4A
In order to determine the orientation of the rod-shaped eIF4G-m relative to eIF4A-CTD, we performed NMR titration experiments with eIF4G-m mutants 756 and 814 (Marcotrigiano et al. 2001). The eIF4G-m mutant 756 has amino acid substitutions within the first N-terminal helical hairpin, whereas the eIF4G-m mutant 814 has amino acid substitutions in the second helical hairpin and the adjacent loop. Numerous peaks are broadened or disappear in eIF4A-CTD upon addition of wild-type eIF4G-m, which is consistent with tight binding and slow to intermediate exchange binding kinetics. In contrast, only very small chemical shift changes are seen on few cross-peaks upon addition of the mutants eIF4G-m 756 (Supplementary Fig. S4) and eIF4G-m 814 (data not shown), which is consistent with fast exchange on the NMR time scale and very low affinity. Residues that show minor changes map to the binding site on eIF4A-CTD as identified above. These data strongly suggest that residues within the first N-terminal helical hairpins of the HEAT domain are responsible for the interaction with the C-terminal domain of eIF4A.
Discussion
A model for the structure of human eIF4A
While crystal structures have been reported for yeast (Benz et al. 1999; Caruthers et al. 2000) and archaeal (Story et al. 2001) homologs, no experimental structure for human eIF4A is available yet, and the mode of interaction with eIF4G is still unclear. We pursued homology modeling and NMR spectroscopy to elucidate the structure of human eIF4A and the mechanism of its interaction with eIF4G. Both domains of eIF4A have been shown to be involved in ATP binding, hydrolysis, and RNA binding; it is very likely that they can rearrange relative to each other, which may be necessary for interaction with different ligands (Lorsch and Herschlag 1998a,b). Based on these observations, we used homology modeling to predict the structures of the two domains of human eIF4A, and consulted biological data and the closed conformation observed in the structure of the archaeal eIF4A homolog (Story et al. 2001) to suggest the relevant domain orientation. This model provides an allocation of ATP and RNA-binding regions. 15N relaxation properties within eIF4A-CTD show that the region that forms the putative interface with eIF4A-NTD has enhanced mobility on the microsecond to millisecond time scale. This is indicative of slow conformational exchange of a surface that has the plasticity for accommodating ligands. This result is in agreement with reports that motifs V and VI, which are part of this interface, are disordered and/or show different conformations in different crystal structures of the DEAD box helicase UAP56 (Shi et al. 2004; Zhao et al. 2004). Such regions with conformational exchange properties are often associated with “hinge” segments or surfaces interacting with ligands or other proteins.
The contact surface of eIF4G-m on eIF4A-CTD is adjacent to the conserved DEAD box motifs
Mapping the residues of eIF4A that are affected upon titration with eIF4G-m (Fig. 1) onto the homology-modeled structure of eIF4A-CTD revealed a contiguous surface as the binding site for eIF4G-m. It extends over a surface area defined by residues in loop regions and the C termini of helices H1, H2, and H3 (Figs. 2, 3). The vast majority of the residues that are affected by binding of eIF4G-m do not coincide with residues in the conserved motifs responsible for RNA binding, ATP binding, and hydrolysis. Thus, the binding site for eIF4G-m is adjacent to but distinct from the DEAD-box motifs and the RNA binding site. This is consistent with the notion that mRNA and eIF4G can bind simultaneously, and eIF4G binding enhances helicase activity.
Binding of eIF4G-m causes some allosteric effects in eIF4A-CTD. The chemical shift changes observed in strand E4 are associated with interior residues and cannot be due to direct interaction with eIF4G-m. Conformational rearrangement of these residues at the periphery of the conserved RNA-binding motif could be the cause of the observed changes. Such an arrangement with the eIF4G-m contact surface adjacent to the proposed RNA-binding region is consistent with a mechanism where eIF4G-m stimulates mRNA binding of eIF4A through a cooperative three-way interaction of eIF4G, eIF4A, and mRNA. The eIF4G-m contact surface is also adjacent to residues in motif VI. As discussed above, the peaks corresponding to motif VI are very weak and difficult to analyze unambiguously due to conformational mobility. Therefore, it cannot be ruled out completely that the eIF4G-m contact surface does not extend into this area of eIF4A-CTD, and motif VI may indeed be partially involved in eIF4G-m binding.
The mapped interaction surface on eIF4A-CTD was further corroborated with two double mutants (D265R/E268K and D296A/T298K) (Fig. 4). The mutants were deficient in binding eIF4G-m, confirming the interaction surface as mapped by NMR.
eIF4G-m may act as a soft clamp for eIF4A stabilization
Surface mutations of eIF4G-m that abrogate or weaken eIF4A binding can be mapped onto the crystal structure of eIF4G-m (Marcotrigiano et al. 2001), which provides further insights into the mechanisms of eIF4A/eIF4G interaction. These data suggest the involvement of an extended surface region of the rod-shaped eIF4G-m in binding eIF4A. Mutations within the first helical hairpin (helix 1a and 1b) abolish or drastically reduce eIF4A binding (Imataka and Sonenberg 1997; Lomakin et al. 2000; Marcotrigiano et al. 2001). It should be noted, however, that some of the mutations described by Imataka and Sonenberg (1997) are within the hydrophobic core of the protein and could exhibit a loss of function more likely because of the disruption of the native tertiary structure of eIF4G. Of the mutated residues shown in Figure 5 and Supplementary Figure S3, only one is buried (L762) from mutant M1 (Imataka and Sonenberg 1997; Lomakin et al. 2000; Marcotrigiano et al. 2001), whereas the rest are solvent exposed. Mutations within helix 5b, at the C-terminal end of the helical stack, have a similar effect (Fig. 5; Supplementary Fig. S3). This indicates a second eIF4A-binding surface on eIF4G-m. Possibly, the entire contiguous surface of the molecule, stretching between the two sites as identified with mutations, is involved in complex formation. Comparing the size and the shape of the crystal structure of eIF4G-m (Fig. 5) with structures of eIF4A one realizes that the distance between these mutations on eIF4G-m is larger than the diameter of a single domain of eIF4A. Assuming that all those mutated residues interact directly and simultaneously with eIF4A, it is tempting to speculate that the eIF4G-m spans across the two domains of eIF4A in a bridging mechanism (Fig. 5). We show, that the mutants in the N-terminal end of the helical stack have dramatically reduced binding affinity to eIF4A-CTD compared with wild-type eIF4G-m (Supplementary Fig. S4), suggesting the principal orientation of eIF4G-m bound on eIF4A-CTD. We propose that the main interaction resides within eIF4A-CTD, which thereby functions as the primary anchoring domain for the N-terminal hairpins of eIF4G-m. Importantly, the binding of eIF4G-m to eIF4A-CTD does not occlude the eIF4A-NTD-interacting surface on eIF4A-CTD, which is a prerequisite to form the ATP-binding pocket. When eIF4A is in a closed conformation, additional interaction of eIF4G-m with eIF4A-NTD can be established with eIF4G-m spanning across the interdomain groove. Coupled ATP binding and hydrolysis could take place in such a closed conformation. Due to the weaker binding affinity of eIF4G-m to eIF4A-NTD, reported conformational changes required for helicase activity, sliding along RNA, displacement of additional protein factors, and shuttling of eIF4A in and out of eIF4F would still be enabled. Our proposed mechanism renders eIF4G-m as a soft clamp stabilizing the relative orientation of the two helicase domains and favoring the closed “active” conformation of eIF4A (Fig. 5).
This clamp model also provides an explanation for the cooperativity between all binding partners of eIF4A (eIF4G, RNA, ATP). Binding of eIF4A to the EMCV IRES is strongly enhanced by the central domain of eIF4G (Pestova et al. 1996). Consequently, binding of eIF4G to eIF4A should not impair but assist interactions that are necessary for the functionality of the enzyme. In our model, eIF4G can stimulate RNA binding to eIF4A by simultaneously binding to RNA and eIF4A-CTD (Fig. 5). RNA bound to eIF4G-m would be positioned in close proximity of eIF4A upon binding and in turn would increase the RNA affinity of eIF4A. Mutations in eIF4G-m that were shown to reduce affinity for eIF4A and RNA (Marcotrigiano et al. 2001) are compatible with this model. eIF4G-m stimulates RNA cross-linking to eIF4A (Pestova et al. 1996) and to the dominant-negative eIF4A mutant R362Q (Korneeva et al. 2005). This effect is consistent with our model suggesting that eIF4G-m binds to a site distinct from the active site.
In other helicases, additional domains are often involved in substrate binding, such as domain 3 in the case of HCV NS3 (Kim et al. 1998). It is possible that these other domains define the relative orientation of the two RecA-like domains. Human eIF4A lacks such additional domains, and their role may be adopted by binding proteins, such as eIF4G, eIF4B, or eIF4H. This could explain the low intrinsic unwinding activity of the free eIF4A in the absence of other factors.
The direct identification of the eIF4G-eIF4A interface presented here resolves a controversy on the location of the binding sites. We identified the binding region of eIF4G-m on eIF4A-CTD by NMR chemical shift mapping and confirmed the contact surface with site-directed mutagenesis. Previously, the binding region of eIF4G on eIF4A has been a subject of conflicting reports. In a recent paper, Korneeva et al. (2005) proposed the N-terminal domain of eIF4A to be the primary binding site for the middle domain of eIF4G-I, whereas Zakowicz et al. (2005) proposed the perimeters of the hinge region between the eIF4A-NTD and the eIF4A-CTD to be the interaction region for eIF4G. The conclusions from both studies are at variance with each other and are inconsistent with the results presented here.
These discrepancies could in part be due to different experimental conditions. Korneeva et al. (2005) used isoform eIF4G-I for their studies, whereas we performed our experiments with the middle domain of eIF4G-II. However, it seems likely that both isoforms of the middle domain of eIF4G-m bind to the same surface on eIF4A based on sequence conservation (Gradi et al. 1998; Marcotrigiano et al. 2001) and functional data (Li et al. 1999). Most likely, the conclusion in the report by Korneeva et al. (2005) is due to the method used, namely, a pull-down assay with a mix of fragments of eIF4A obtained by tryptic digest. The strongest band corresponds to the entire C-terminal domain plus an additional 70 or 100 residues from the N-terminal domain, which could be interpreted as consistent with our data. The authors did not try to confirm their results by testing whether isolated eIF4A-NTD or eIF4A-CTD bind eIF4G-m.
The point mutations reported in Zakowicz et al. (2005) targeted the conserved helicase motifs of both eIF4A domains. They inhibited translation but also abolished binding to eIF4G-m in vivo. Nevertheless, these mutations did not prevent (or even increased) binding of these eIF4A mutants to endogenous eIF4F. The results of Zakowicz et al. (2005) were perplexing in several aspects: (1) Some of the mutations in eIF4A that eliminated eIF4G-m binding affect conserved-motif residues that are known or proposed to directly contact ATP. However, eIF4G-m does not compete with ATP interaction but stimulates ATP binding and hydrolysis instead. (2) Several of the mutations abolished binding to both eIF4G-m and Pdcd4 even though the two proteins do not compete with each other for eIF4A binding (Yang et al. 2003). (3) Some of the eIF4A motif mutants were defective in binding to both eIF4G-m and eIF4G-c but nevertheless were incorporated in eIF4F at least as efficiently as wild-type eIF4A. Since eIF4G is the only known binding partner of eIF4A in eIF4F, it would be expected that the eIF4A mutants that are defective in binding to both binding sites in eIF4G would not be efficiently incorporated in eIF4F. However, previous studies have shown that these eIF4A mutants are dominant negative and are efficiently incorporated in eIF4F in vitro and in vivo (Pause and Sonenberg 1992; Pause et al. 1994; Svitkin et al. 2001). It was proposed that the mutants are trapped on mRNA as part of eIF4F due to their defect in ATP hydrolysis and RNA unwinding. In this paper we show that the main contact surface with eIF4G-m is distinct from the conserved motifs. Therefore, our results support the previous interpretation and data that these dominant-negative mutants are deficient in ATP hydrolysis and are trapped in the context of the eIF4F complex. Thus we suggest that the eIF4G-m-binding defects reported for these mutants by Zakowicz et al. (2005) are indirect.
In summary, our model of eIF4A-CTD interaction with eIF4G-m is consistent with previous biochemical reports and provides a plausible explanation for the increase of helicase activity upon binding of eIF4G. More structural studies will allow a consensus on binding stoichiometry, answer the question whether the same binding surfaces of eIF4A are used for the interaction with eIF4G-m and eIF4G-c, and eventually enable insight at the molecular detail of eIF4F.
Materials and methods
Vectors, protein expression, and purification
The expression vectors for full-length human eIF4A (isoform 1), the N-terminal domain of eIF4A (amino acids 1-244), the middle domain of eIF4GII (745-1003, eIF4G-m), and the eIF4G-m mutants 756 and 814 (Marcotrigiano et al. 2001) were a generous gift from Tatyana Pestova (State University of New York, Brooklyn, NY). The expression vectors and purification procedures for human eIF4A and eIF4G-m have been described previously (Pestova et al. 1996; Marcotrigiano et al. 2001). The C-terminal domain of eIF4A (amino acids 237-406, eIF4A-CTD) (Fig. 2) was subcloned from the vector expressing full-length eIF4A. An N-terminal 8xHis-tag was introduced into the expression vector pDEST14 using a Gateway kit (Invitrogen) according to the manufacturer's instruction. The eIF4A mutants D265R/E268K and D296A/T298K were obtained using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The mutations were verified by restriction analysis and sequencing. Unlabeled and labeled eIF4A-CTD were expressed for 6 h at 37°C in the Escherichia coli strain BL21 STAR (DE3), lysed by sonication, and purified on a Talon Co2+ affinity column (Clontech) in buffer containing 20 mM Tris-HCl (pH 7.0), 500 mM NaCl, 7 mM β-mercaptoethanol, and 0.2 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF). Unlabeled eIF4A-NTD was expressed for 6 h at 37°C in the E. coli strain BL21 (DE3) and purified following the same protocol as eIF4A-CTD. The N-terminal His-tag of eIF4A-CTD was removed by digestion with enterokinase (NEB). The His-tag was removed by affinity chromatography, and the untagged flow-through was collected and concentrated. All proteins were >95% pure as estimated from Coomassie-stained SDS-PAGE gels. For 13Cor 15N-isotope labeling in minimal medium, [13C6]-D-glucose or 15NH4Cl (Cambridge Isotope Laboratories) was used as a sole carbon or nitrogen source, respectively. Amino acid-specific 15N-labeled samples were prepared for Ile, Leu, Val, and Lys. Reverse labeled samples (uniformly 15N-labeled residues and unlabeled target amino acids) were prepared for Arg residues for help in the assignment process.
His-tag pull-down assays
One hundred micrograms of His-eIF4A, His-eIF4A-CTD, and the His-eIF4A-CTD mutants D265R/E268K and D296A/T298K were incubated with nutation with 100 μL pre-equilibrated Talon resin (Clontech) for 20 min at room temperature in buffer containing 20 mM Tris-HCl buffer (pH 7.0), 200 mM NaCl, and 7 mM β-mercaptoethanol. The resin was washed several times with 400 μL binding buffer before 150 μg of untagged eIF4G-m was added. After further incubation for 15 min, the unbound proteins were eluted with several washes with buffer. The bound complex was eluted in the same buffer containing 200 mM imidazole. The pull-down assay with eIF4A-NTD was performed in a similar way, with the exception that His-tagged eIF4G-m was immobilized on the resin and then incubated with untagged eIF4A-NTD. Binding above background level became detectable when the incubation was carried out with the buffer containing 150 mM NaCl; 150 μg albumin was added to the binding reactions to inhibit nonspecific binding to the resin. Incubation supernatants, wash, and elution fractions were analyzed using Coomassie-stained SDS-PAGE gels.
NMR resonance assignments
NMR spectra were recorded at 298 K on a Varian Inova 750-MHz, a Bruker Avance 600-MHz, a Bruker Avance 500-MHz, or a Varian Inova 500-MHz spectrometer, most of them equipped with cryogenic probes. Samples for NMR measurements typically contained 0.4-1 mM protein in buffer containing 20 mM Tris-HCl (pH 7.0), 300 mM NaCl, 5 mM DTT, 1 mM EDTA, 0.01% NaN3, 0.2 mM AEBSF, and 10% D2O. The spectra were processed with NMRPipe and analyzed with Xeasy (Bartels et al. 1995; Delaglio et al. 1995). Sequential resonance assignments were obtained from standard triple-resonance NMR experiments [HNCA, HN(CO)CA, HN(CA)CB, HN(COCA)CB, HNCO, HN(CA)CO] on uniformly 15N-/13C-labeled samples with 70% deuteration. 15N-edited NOESY-HSQC and TOCSY-HSQC experiments were recorded on uniformly 15N-labeled samples. Backbone and partial side-chain resonance assignments were obtained for 93% of the residues. Secondary structure elements were derived by inspection of the amide backbone NOE pattern and from 13C chemical shifts using TALOS (Cornilescu et al. 1999).
Model building
Human eIF4A was modeled using the protein structure homology-modeling server Swiss-Model (Schwede et al. 2003). The sequence of human eIF4A was aligned to the sequences of yeast eIF4A and the archaeal eIF4A homolog MjDEAD, whose structures have been determined (Caruthers et al. 2000; Story et al. 2001) using ClustalW (Thompson et al. 1994). The structure of human eIF4A was first modeled using the yeast eIF4A structure as a template, resulting in an “open” conformation. To achieve the interdomain orientation for a more closed conformation, a second model was built based on the archaeal eIF4A structure. The C-terminal domain obtained with the yeast structure template was then ligated with the closed-conformation model, and the loop regions P55-A57 and G361-G370 (located within motifs Q and VI, respectively) were energy-minimized using InsightII (Accelrys) to avoid clashes between side-chains of these residues. NMR data obtained from sequential NOE peaks from 15N-edited NOESY-HSQC spectra and chemical shift information were used to identify secondary structure elements of eIF4A-CTD and evaluate the model.
NMR titrations
For the NMR titration experiments, the unlabeled binding partner (e.g., eIF4G-m) was added in small aliquots to uniformly 15N- or 13C-labeled protein (e.g., eIF4A-CTD). HSQC spectra were collected at every titration step to monitor the changes upon binding. Progressive decrease in intensity was observed for a subset of the peaks in the HSQC spectra of eIF4A-CTD. Some peaks already completely disappeared at titration points where the concentration of the unlabeled eIF4G-m was only 30% that of labeled eIF4A-CTD. This indicates that the eIF4A-CTD/eIF4G-m complex is in intermediate exchange on the NMR time scale, which is accompanied by line-broadening. When unlabeled eIF4G-m was added in excess, an overall strong decrease in the intensities of most peaks was observed due to the large size of the eIF4A-CTD/eIF4G-m complex (∼50 kDa). Only few peaks corresponding to flexible regions in the N-terminal linker of eIF4A-CTD remained significantly less affected by the binding. Therefore, the analysis of the effects of eIF4G-m binding was performed starting at titration steps of 15% unlabeled eIF4G-m, where the overall quality of the spectra was still good and the peaks affected by the interaction were still above the noise level. Peaks with significant decrease in intensity were identified in an iterative process: The peak intensities of the various titration points were normalized with respect to peaks that are not involved in the binding interface to account for dilution upon addition of the binding partner. Those reference peaks are strong peaks corresponding to N-terminal residues that are part of the linker region and unaffected in the presence of the binding partner. Then, the average change in peak intensities was calculated, and those peaks more than two standard deviations away from the average intensity change were identified and also omitted in the subsequent round of iteration. Significantly affected peaks were defined as those, for which the intensities decrease by more than two standard deviations after two iterative steps. An alternative approach for identifying peaks significantly affected by eIF4G-m binding, where decay rates were calculated by fitting a curve of peak intensity versus concentration of eIF4G-m for the different titration steps, produced the same set of peaks as the above iterative approach. The orientation of eIF4G-m on eIF4A-CTD was probed by titrating unlabelled eIF4G-m mutants 756 and 814 (Marcotrigiano et al. 2001) to 15N-labeled eIF4A-CTD. In contrast to the strong binding observed with eIF4G-m wild type, no significant changes in the overall peak intensities of eIF4A-CTD were observed, even in 2:1 excess of the mutant proteins. For very few residues, the NMR resonances shift in the presence of the eIF4G-m mutant, indicative of fast exchange on the NMR time scale and consistent with very low binding affinity. Remarkably, the few peaks that show minor changes are in the center of the binding site that was mapped as the interface for wild-type eIF4G-m.
Relaxation measurements
All relaxation experiments were recorded on uniformly 15N labeled sample at a concentration of ∼0.5 mM. Steady-state [1H]-15N NOEs were derived from 1H-15N-HSQC spectra recorded in an interleaved manner with and without 1H presaturation at a temperature of 298 K and a magnetic field strength of 11.7 T (500 MHz). The spectrum with presaturation had a prescan delay of 3 sec followed by presaturation for 2 sec. In the spectrum without presaturation, the prescan delay was 5 sec. The steady-state [1H]-15N NOE values were calculated as the ratio of Isat/Inosat, where Isat and Inosat are the peak intensities in the spectra recorded with and without presaturation. Transverse relaxation rates and estimates for R1ρ were obtained at 298 K and a magnetic field strength of 14.1 T. For R2 measurements, the spectra were recorded with relaxation delays of 0.010, 0.03, 0.05, 0.07, 0.130, and 0.170 sec. The interpulse delay for the 180° 15N pulse in the CPMG experiment was 625 μsec. For R1ρ measurements, the spectra were recorded with relaxation delays of 0.010, 0.020, 0.030, 0.050, 0.070, 0.130, and 0.170 sec. All spectra were acquired with a spinlock radio frequency pulse at the beginning of each pulse sequence to ensure average sample heating. Relaxation rates were calculated by fitting the experimental data to a two-parameter exponential decay function. Contribution of conformational exchange to relaxation rates was analyzed qualitatively using a R1ρ spectrum in addition to the R2 data. The R1ρ spectrum was assumed to be free of contributions from conformational exchanges, whereas conformational exchange can still contribute to the transverse relaxation rates obtained from the CPMG experiments. The rates extracted from the R1ρ experiment were subtracted from the experimentally derived R2 rates, which therefore identify regions with relaxation contribution from conformational exchange.
Acknowledgments
We thank T.V. Pestova (SUNY, Brooklyn) for providing plasmids used in this study. M.O. gratefully acknowledges funding by an Erwin-Schroedinger Fellowship from the Austrian Science Funds (FWF) and funding from the Max Kade Foundation. A.M. was funded by post-doctoral fellowship PF-02-117-01-GMC from the American Cancer Society. The work was supported by NIH grant CA68262 to G.W.
Supplementary material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1335305.
References
- Abramson R.D., Dever, T.E., and Merrick, W.C. 1988. Biochemical evidence supporting a mechanism for cap-independent and internal initiation of eukaryotic mRNA. J. Biol. Chem. 263: 6016-6019. [PubMed] [Google Scholar]
- Ali I.K. and Jackson, R.J. 2001. The translation of capped mRNAs has an absolute requirement for the central domain of eIF4G but not for the cap-binding initiation factor eIF4E. Cold Spring Harb. Symp. Quant. Biol. 66: 377-387. [DOI] [PubMed] [Google Scholar]
- Ali I.K., McKendrick, L., Morley, S.J., and Jackson, R.J. 2001. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 20: 4233-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C., Xia, T., Billeter, M., Güntert, P., and Wüthrich, K. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 5: 1-10. [DOI] [PubMed] [Google Scholar]
- Benz J., Trachsel, H., and Baumann, U. 1999. Crystal structure of the ATPase domain of translation initiation factor 4A from Saccharomyces cerevisiae: The prototype of the DEAD box protein family. Structure Fold. Des. 7: 671-679. [DOI] [PubMed] [Google Scholar]
- Caruthers J.M., Johnson, E.R., and McKay, D.B. 2000. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. 97: 13080-13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13: 289-302. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277-293. [DOI] [PubMed] [Google Scholar]
- Delagoutte E. and von Hippel, P.H. 2002. Helicase mechanisms and the coupling of helicases within macromolecular machines, part I: Structures and properties of isolated helicases. Q. Rev. Biophys. 35: 431-478. [DOI] [PubMed] [Google Scholar]
- Ferentz A.E. and Wagner, G. 2000. NMR spectroscopy: A multifaceted approach to macromolecular structure. Q. Rev. Biophys. 33: 29-65. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E. and Koonin, E.V. 1993. Helicases: Amino-acid-sequence comparisons and structure-function-relationships. Curr. Opin. Struct. Biol. 3: 419-429. [Google Scholar]
- Gradi A., Imataka, H., Svitkin, Y.V., Rom, E., Raught, B., Morino, S., and Sonenberg, N. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18: 334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J.D., Moerke, N.J., von der Haar, T., Lugovskoy, A.A., Sachs, A.B., McCarthy, J.E., and Wagner, G. 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115: 739-750. [DOI] [PubMed] [Google Scholar]
- Hershey P.E., McWhirter, S.M., Gross, J.D., Wagner, G., Alber, T., and Sachs, A.B. 1999. The Cap-binding protein eIF4E promotes folding of a functional domain of yeast translation initiation factor eIF4G1. J. Biol. Chem. 274: 21297-21304. [DOI] [PubMed] [Google Scholar]
- Imataka H. and Sonenberg, N. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 17: 6940-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp L.D. and Lorsch, J.R. 2004. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 73: 657-704. [DOI] [PubMed] [Google Scholar]
- Kim J.L., Morgenstern, K.A., Griffith, J.P., Dwyer, M.D., Thomson, J.A., Murcko, M.A., Lin, C., and Caron, P.R. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligo-nucleotide: The crystal structure provides insights into the mode of unwinding. Structure 6: 89-100. [DOI] [PubMed] [Google Scholar]
- Korneeva N.L., Lamphear, B.J., Hennigan, F.L., Merrick, W.C., and Rhoads, R.E. 2001. Characterization of the two eIF4A-binding sites on human eIF4G-1. J. Biol. Chem. 276: 2872-2879. [DOI] [PubMed] [Google Scholar]
- Korneeva N.L., First, E.A., Benoit, C.A., and Rhoads, R.E. 2005. Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J. Biol. Chem. 280: 1872-1881. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266: 19867-19870. [PubMed] [Google Scholar]
- Li Q., Imataka, H., Morino, S., Rogers Jr., G.W., Richter-Cook, N.J., Merrick, W.C., and Sonenberg, N. 1999. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol. Cell. Biol. 19: 7336-7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Belsham, G.J., and Proud, C.G. 2001. Eukaryotic initiation factors 4A (eIF4A) and 4G (eIF4G) mutually interact in a 1:1 ratio in vivo. J. Biol. Chem. 276: 29111-29115. [DOI] [PubMed] [Google Scholar]
- Linder P., Lasko, P.F., Ashburner, M., Leroy, P., Nielsen, P.J., Nishi, K., Schnier, J., and Slonimski, P.P. 1989. Birth of the D-E-A-D box. Nature 337: 121-122. [DOI] [PubMed] [Google Scholar]
- Lomakin I.B., Hellen, C.U., and Pestova, T.V. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20: 6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch J.R. and Herschlag, D. 1998a. The DEAD box protein eIF4A, 1: A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry 37: 2180-2193. [DOI] [PubMed] [Google Scholar]
- ____. 1998b. The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry 37: 2194-2206. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J., Lomakin, I.B., Sonenberg, N., Pestova, T.V., Hellen, C.U., and Burley, S.K. 2001. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell 7: 193-203. [DOI] [PubMed] [Google Scholar]
- Marintchev A. and Wagner, G. 2005. Translation initiation: Structures, mechanisms and evolution. Q. Rev. Biophys. 37: 1-88. [DOI] [PubMed] [Google Scholar]
- Morino S., Imataka, H., Svitkin, Y.V., Pestova, T.V., and Sonenberg, N. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20: 468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A.G., Kroenke, C.D., and Loria, J.P. 2001. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 339: 204-238. [DOI] [PubMed] [Google Scholar]
- Pause A. and Sonenberg, N. 1992. Mutational analysis of a DEAD box RNA helicase: The mammalian translation initiation factor eIF-4A. EMBO J. 11: 2643-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A., Methot, N., Svitkin, Y., Merrick, W.C., and Sonenberg, N. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13: 1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K., Riek, R., Wider, G., and Wuthrich, K. 1997. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. 94: 12366-12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V. and Kolupaeva, V.G. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 16: 2906-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Shatsky, I.N., and Hellen, C.U. 1996. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16: 6870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T. and Hentze, M.W. 2003. Starting the protein synthesis machine: Eukaryotic translation initiation. Bioessays 25: 1201-1211. [DOI] [PubMed] [Google Scholar]
- Prevot D., Darlix, J.L., and Ohlmann, T. 2003. Conducting the initiation of protein synthesis: The role of eIF4G. Biol. Cell. 95: 141-156. [DOI] [PubMed] [Google Scholar]
- Rocak S. and Linder, P. 2004. DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell. Biol. 5: 232-241. [DOI] [PubMed] [Google Scholar]
- Rogers G.W., Richter, N.J., and Merrick, W.C. 1999. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274: 12236-12244. [DOI] [PubMed] [Google Scholar]
- Rogers G.W., Richter, N.J., Lima, W.F., and Merrick, W.C. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276: 30914-30922. [DOI] [PubMed] [Google Scholar]
- Rogers G.W., Komar, A.A., and Merrick, W.C. 2002. eIF4A: The godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 72: 307-331. [DOI] [PubMed] [Google Scholar]
- Schwede T., Kopp, J., Guex, N., and Peitsch, M.C. 2003. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31: 3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Cordin, O., Minder, C.M., Linder, P., and Xu, R.M. 2004. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc. Natl. Acad. Sci. 101: 17628-17633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N. and Dever, T.E. 2003. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 13: 56-63. [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Hershey, J.W., and Mathews, M.B. 2000. Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Story R.M., Weber, I.T., and Steitz, T.A. 1992. The structure of the E. coli recA protein monomer and polymer. Nature 355: 318-325. [DOI] [PubMed] [Google Scholar]
- Story R.M., Li, H., and Abelson, J.N. 2001. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc. Natl. Acad. Sci. 98: 1465-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y.V., Pause, A., Haghighat, A., Pyronnet, S., Witherell, G., Belsham, G.J., and Sonenberg, N. 2001. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7: 382-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N.K. and Linder, P. 2001. DExD/H box RNA helicases: From generic motors to specific dissociation functions. Mol. Cell 8: 251-262. [DOI] [PubMed] [Google Scholar]
- Tanner N.K., Cordin, O., Banroques, J., Doere, M., and Linder, P. 2003. The Q motif: A newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell 11: 127-138. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.S., Jansen, A.P., Komar, A.A., Zheng, X., Merrick, W.C., Costes, S., Lockett, S.J., Sonenberg, N., and Colburn, N.H. 2003. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 23: 26-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakowicz H., Yang, H.S., Stark, C., Wlodawer, A., Laronde-Leblanc, N., and Colburn, N.H. 2005. Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI. RNA 11: 261-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Shen, J., Green, M.R., MacMorris, M., and Blumenthal, T. 2004. Crystal structure of UAP56, a DExD/H-box protein involved in pre-mRNA splicing and mRNA export. Structure (Camb.) 12: 1373-1381. [DOI] [PubMed] [Google Scholar]