Abstract
The histone-like nucleoid structuring (H-NS) protein is a global transcriptional regulator that is known to regulate stress response pathways and virulence genes in bacteria. It has also been implicated in the regulation of bacterial transposition systems, including Tn10. We demonstrate here that H-NS promotes Tn10 transposition by binding directly to the transposition complex (or transpososome). We present evidence that, upon binding, H-NS induces the unfolding of the Tn10 transpososome and helps to maintain the transpososome in an unfolded state. This ensures that intermolecular (as opposed to self-destructive intramolecular) transposition events are favored. We present evidence that H-NS binding to the flanking donor DNA of the transpososome is the initiating event in the unfolding process. We propose that by recruiting H-NS as a modulator of transposition, Tn10 has evolved a means of sensing changes in host physiology, as the amount of H-NS in the cell, as well its activity, are responsive to changes in environmental conditions. Sensing of environmental changes through H-NS would allow transposition to occur when it is most opportune for both the transposon and the host.
Keywords: Tn10, H-NS, DNA transposition, transposase, transpososome, nucleoid-associated proteins
Transposable elements constitute a major component of almost all genomes and are major contributors to genetic instability. Host organisms have developed a wide range of defenses that serve to limit transposition levels, and therefore the damage transposons cause. RNA interference and CpG methylation likely evolved to limit transposition events in some eukaryotes by reducing the expression of transposon-encoded proteins that are necessary to promote transposon mobility (Yoder et al. 1997; Plasterk 2002). In prokaryotes the expression of “transposition proteins” can also be limited by DNA methylation, as well as targeted degradation of RNA transcripts (for review, see Kleckner 1989). In the face of potent negative regulators of transposition, transposons have co-opted host proteins to promote transposition (for review, see Nagy and Chandler 2004). Such modulators may be critical in ensuring that transposition levels remain high enough to maintain a given transposon in a population. Interestingly, in the case of the bacterial transposon CTnDOT, stimulation of transposition is linked to an environmental signal. When Bacteroides species containing CTnDOT are exposed to tetracycline, excision and transfer of CTnDOT increases two to three orders of magnitude (Shoemaker and Salyers 1988). This response is mediated by transposon-encoded proteins, rteA, rteB, and rteC, which control the expression of excision and transfer genes (Wang et al. 2005).
Several members of the bacterial nucleoid-associated family of proteins, including integration host factor (IHF), HU, and histone-like nucleoid structuring protein (H-NS) have been shown to modulate selected transposition systems (Craigie et al. 1985; Morisato and Kleckner 1987; Surette and Chaconas 1989; Signon and Kleckner 1995; Shiga et al. 2001; Rouquette et al. 2004). It is particularly intriguing that H-NS should act in this fashion, because one of its main roles is to facilitate adaptation to environmental stresses (Dorman 2004). Thus, by being linked to H-NS function, selected transposons are well positioned to respond to changes in cellular physiology. In the current work we provide direct evidence that H-NS promotes Tn10 transposition by a novel mechanism, namely by unfolding the Tn10 transpososome.
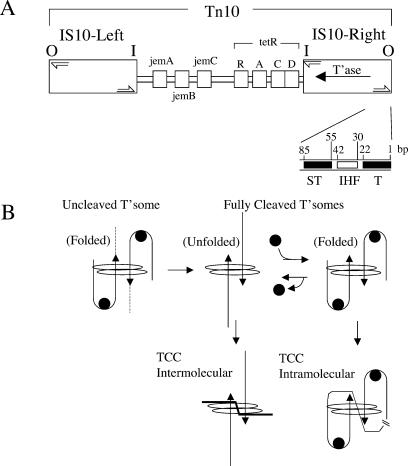
Tn10 is a bacterial transposon that confers resistance to tetracycline. Mobilization of Tn10 occurs through a “cut and paste” mechanism, wherein the transposon is first fully separated from flanking donor DNA. The excised transposon then integrates into a new location through a strand transfer reaction (for review, see Haniford 2002). All of the chemical steps in Tn10 transposition take place in the context of a higher-order protein-DNA complex called a transpososome, in which the termini of the transposon (referred to as the transposon ends) are brought together (Haniford et al. 1991; Sakai et al. 1995). IHF plays a key role in the assembly of the Tn10 transpososome; it binds to the outside end (OE) of Tn10 immediately adjacent to the primary transposase-binding site and bends the DNA (Fig. 1; Sakai et al. 1995). This bending permits transposase to contact the subterminal region of the OE, and in so doing, stabilizes the transposase-OE interaction—a critical step in transpososome formation (Sakai et al. 2000; Crellin and Chalmers 2001). While precise details of how the two ends are brought together are not known, once formed, the Tn10 transpososome is quite stable and does not require the continued presence of IHF. In fact, release of IHF subsequent to transpososome assembly, and the concomitant unfolding of the transposon arm that is linked to IHF release, is thought to drive the transposon excision reaction to completion (Crellin et al. 2004).
Figure 1.
Tn10 and conformational changes in the Tn10 transpososome. (A) The structure of Tn10. The outside end (OE) contains the terminal-binding site (T) and subterminal-binding site (ST) for transposase and an IHF-binding site. The inside end (I) does not contain an IHF site. Half arrows are the terminal inverted repeats. Proteins encoded by Tn10 are indicated. (B) The Tn10 transpososome is shown to undergo several structural changes (see text) over the course of the transposition reaction. (Solid line with arrow) Duplex OE DNA; (dashed line) flanking donor DNA; (filled circle) IHF; (oval) transposase; (TCC) target capture complex.
The presence of IHF in the Tn10 transpososome also affects the types of transposition products formed (Fig. 1B). After full excision, IHF can rebind to the transpososome and constrain the ends in such a way that intratransposon insertion events are heavily favored. In contrast, when IHF is not bound to the cleaved transpososome, the above constraint is not imposed and intermolecular insertion events are favored. Thus, in addition to promoting Tn10 transposition by facilitating transpososome assembly and full excision, IHF can also inhibit intermolecular transposition events (Chalmers et al. 1998).
More recently, H-NS was shown to play a role in Tn10 transposition in vivo (Swingle et al. 2004). In an hns deletion strain, the frequency of Tn10 transposition is significantly reduced. Under conditions where cells gradually become nutrient-starved, Tn10 transposition exhibited almost a complete dependence on H-NS; similar results were observed for IS903 and Tn552 transposition. The hns mutant strain was shown to accumulate a Tn10 excision intermediate, an observation that is consistent with H-NS acting directly on the transpososome as opposed to affecting transposase expression. In contrast, H-NS influences bacteriophage Mu and IS1 transposition by influencing gene expression and protein stability, respectively (Falconi et al. 1991; Rouquette et al. 2004).
H-NS is the most abundant of the nucleoid-associated proteins, at ∼20,000 copies per cell (Spassky et al. 1984), although its level in the cell does fluctuate as a function of growth phase (Ali Azam et al. 1999). In addition to its role in structuring the nucleoid, H-NS either directly or indirectly regulates the expression of ∼5% of the genes in Escherichia coli, including genes involved in stress response and virulence pathways (Hommais et al. 2001). H-NS forms dimers in solution (Atlung and Ingmer 1997) and binds preferentially to curved DNA (Yamada et al. 1991). Unlike IHF, H-NS does not bind DNA in a sequence-specific manner. H-NS dimers bound to different DNA segments are able to interact, forming higher-order oligomers. Such interactions presumably account for the nucleoid compaction that has been attributed to H-NS. In addition, the interaction of H-NS dimers bound to promoter sequences is thought to trap RNA polymerase in the promoter region, thereby causing transcriptional silencing (Rimsky et al. 2001).
In the present study we set out to define the mechanism through which H-NS promotes Tn10 transposition. We provide evidence that H-NS is capable of binding to at least two different sites in the Tn10 transpososome in vitro. Once bound, H-NS appears to facilitate transpososome unfolding. The major impact this has on the Tn10 transposition reaction is to favor intermolecular transposition events by allowing the cleaved transpososome to readily interact with target DNAs that are unlinked to the transposon. Results are also presented that are consistent with H-NS affecting the structure of the transpososome in vivo. This work defines a new mode of modulating DNA transposition, by a host protein known to play an important role in regulating gene expression in response to the cellular environment.
Results
H-NS stimulates the Tn10 strand transfer reaction
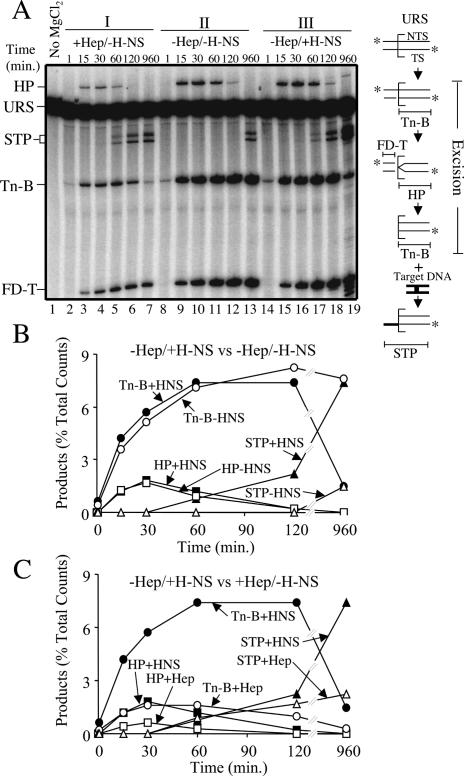
We examined the impact of H-NS on Tn10 transposition by adding purified H-NS to a complete in vitro transposition reaction (see Materials and Methods for reaction conditions). Reaction chemistry was initiated by adding MgCl2. Aliquots removed, at various time points after MgCl2 addition, were treated with phenol and the purified DNA was analyzed on a high-resolution denaturing gel (Fig. 2A). Comparison of panels II and III (Fig. 2A) reveals that addition of H-NS increases the amount of strand transfer products (STPs) formed. STPs are first clearly detected at 60 min in the reaction with H-NS (Fig. 2A, panel III), whereas in the absence of H-NS, STPs are not detected even after 120 min of incubation (Fig. 2A, panel II). At the end point of the experiment, STPs are detected in the reaction without H-NS; however, they are present at a fivefold lower level relative to the reaction with H-NS. Thus, the presence of H-NS increases the amount of strand transfer by at least fivefold.
Figure 2.
Kinetic analysis of in vitro transposition with and without H-NS. (A) Transposition reactions were set up as described in Materials and Methods and analyzed following electrophoresis through an 8% denaturing polyacrylamide gel. Illustrations of the substrate and products are shown to the right of the gel. The square bracket defines the position of the transposon-donor junction. (★) 5′ 32P labels; (TS) “transferred” strand (i.e., the transposon strand joined to the target site by transposase); (NTS) “nontransferred” strand (i.e., the transposon strand joined to the target site by host repair proteins); (URS) unreacted substrate. The strand transfer product (STP) results from joining Tn-B (defined in diagram) to either the top or bottom strand of the HisG1 target site. HP is the hairpin intermediate, and its formation coincides with the formation of FD-T. Different sized STPs are generated because the HisG1 target is asymmetrically positioned within the target DNA fragment (see diagram in Fig. 6). (Lane 19) Note that prolonged incubation with H-NS (960 min) resulted in some nucleolytic degradation of the OE DNA. In B and C, product levels are plotted as a function of time for the indicated reactions.
Importantly, there is no significant effect of H-NS addition on any of the steps in transposon excision. Prior to 60 min, the product profiles are very similar in reactions with and without H-NS. For example, the curves for hairpin formation (HP in Fig. 2B) are almost superimposable with each other. Similarly, the curves for the cleaved “bottom” strand of the transposon (Tn-B in Fig. 2B), which arises from first-strand nicking and later from hairpin resolution (Kennedy et al. 1998), are very similar up to 60 min. The Tn-B curves diverge beyond this point because Tn-B is the precursor for strand transfer and significantly more strand transfer occurs in the presence versus the absence of H-NS (7.5% of total substrate vs. 1.5%, respectively). Furthermore, H-NS did not influence transpososome formation in an experiment in which transpososome formation was monitored under noncatalytic conditions (data not shown). We therefore conclude that H-NS stimulates the in vitro transposition reaction by facilitating the conversion of excised transposon to intermolecular STPs, and not by enhancing transpososome formation, or early catalytic steps.
The above experiment was also performed with heparin in place of H-NS (panel I in Fig. 2A). Heparin perturbs the transpososome folding-unfolding equilibrium by binding IHF and preventing it from associating with the transpososome. This leads to transpososome unfolding (Junop and Haniford 1997; Crellin and Chalmers 2001). It is apparent that heparin addition increases the rate of strand transfer relative to the no treatment control (Fig. 2A, cf. lanes 5,6 and 11,12). The increase in the amount of STPs at early time points occurs despite the fact that fewer substrate molecules undergo excision relative to the reactions in panels II and III of Figure 2A. This is apparent from the reduced levels of HP, Tn-B and cleaved top strand (FD-T) in panel I relative to the panel II or III (Fig. 2A) samples. The reduced level of excision can be accounted for entirely by the fact that heparin inhibits transpososome formation by preventing IHF binding to the OE (data not shown).
Notably, of the population of substrate molecules that are assembled into transpososomes in the presence of heparin, the excision product profile is very similar to that observed for transpososomes formed in the absence of heparin. For example, the HP intermediate appears and disappears with very similar kinetics in reactions with and without H-NS (cf. the hairpin curves in Fig. 2C). Similarly, the curves for Tn-B have a comparable shape for the aforementioned reactions over the first 30 min. This leads us to conclude that heparin is promoting strand transfer by acting exclusively at the transition point between full excision and strand transfer. Furthermore, since heparin is known to unfold the fully cleaved transpososome (Junop and Haniford 1997), it is likely that heparin stimulates the in vitro transposition reaction by unfolding the excision complex.
Taken together, the experiments in this section indicate that H-NS promotes Tn10 transposition by at least fivefold. It does so by acting at the transition point between full transposon excision and strand transfer. Unfolding of the full excision complex is a critical and perhaps rate-limiting step at this transition point, and this raises the possibility that H-NS promotes Tn10 transposition by facilitating transpososome unfolding. Evidence is provided below supporting this mechanism of H-NS action.
H-NS binds directly to the uncleaved transpososome
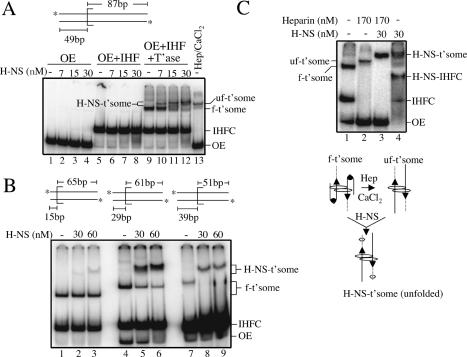
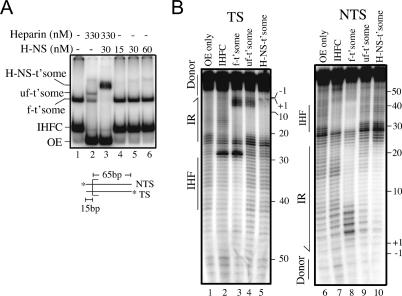
In principle, H-NS could exert its affect on the strand transfer reaction by interacting with the transpososome relatively early after its formation, or at a later stage in transpososome development. Alternatively, H-NS might bind IHF in solution. We show, using a “band-shift” assay, that H-NS binds preferentially to a transpososome containing >15 bp of flanking donor DNA.
We added H-NS to assembly reactions containing OE DNA, OE DNA and IHF, or OE DNA, IHF, and transposase. The OE DNA contained 49 bp of flanking donor DNA. Reactions were analyzed on a 5% native polyacrylamide gel (Fig. 3A). With H-NS present at up to a fivefold molar excess relative to OE DNA, H-NS specifically reduced the mobility of the complete transpososome complex (Fig. 3A, lanes 10-12). At the highest concentration of H-NS used (30 nM), ∼70% of the transpososome has bound H-NS.
Figure 3.
Electrophoretic band-shift assays with H-NS. (A) H-NS band-shift with OE substrate containing 49 bp of flanking donor DNA. (H-NS-t'some) H-NS-bound transpososome; (uf-t'some) unfolded transpososome; (f-t'some) folded transpososome; (IHFC) IHF complex; (OE) outside end DNA. In lane 13, heparin and CaCl2 were added after transpososome formation to unfold the transpososome. (B) H-NS band-shift with transpososomes formed with OE DNA containing different lengths of flanking donor DNA. Note that H-NS also failed to bind a transpososome formed with no flanking donor DNA (data not shown). (C) Effect of H-NS on transpososome mobility when H-NS is added to folded and unfolded transpososomes. As depicted, heparin and CaCl2 were added to unfold the transpososome prior to H-NS addition; based on data from Figure 4, H-NS (hexagon) is shown bound to the flanking donor DNA. Note that in C, OE DNA, IHF, and transposase were present at five, three-, and threefold lower concentrations, respectively, relative to A and B.
Using substrates with progressively shorter amounts of donor DNA, we show in Figure 3B that efficient H-NS binding to a transpososome requires >15 bp of donor DNA. Greater than 90% of the folded transpososome formed with OE substrates containing 29 or more base pairs of flanking donor DNA is shifted to an H-NS-bound form (Fig. 3B, cf. lanes 4 and 5,6, and lanes 7 and 8,9). In contrast, <5% of the folded transpososome formed with the OE substrate containing 15 bp of flanking donor DNA is shifted to an H-NS-bound form (Fig. 3B, cf. lanes 1 and 2,3). Importantly, the reduction in the H-NS-bound form of the transpososome for the latter substrate did correlate with a loss of H-NS stimulation of strand transfer. When we used the OE with only 15 bp of flanking donor as a substrate in a time-course experiment analogous to that shown in Figure 2A, we failed to see a stimulation in strand transfer (data not shown). Thus, an H-NS interaction with the flanking donor DNA plays a critical role in the stimulation of the in vitro transposition reaction. Direct evidence for H-NS binding to the flanking donor DNA is presented below. Taken together, the results in Figure 3A and B show that H-NS preferentially binds to the transpososome and that this interaction is aided by the presence of flanking donor DNA.
Insight into how H-NS promotes the in vitro transposition reaction comes from a comparison of the mobility of the H-NS-bound transpososome when it is generated from folded and unfolded transpososomes; the unfolded transpososome was generated by treatment of the folded transpososome with heparin and CaCl2. In the experiment in Figure 3C we added equal amounts of H-NS to folded and unfolded transpososomes and compared the extent of the mobility shift. The results show that the unfolded transpososome is capable of binding H-NS (Fig. 3C, lane 3), and that the resulting complex has roughly the same mobility as the H-NS-bound transpososome generated from the folded form in Figure 3C, lane 4. We note that in the absence of H-NS, folded and unfolded transpososomes have different mobilities. We can think of two likely explanations to account for the similar mobilities of the H-NS complexes. If the folded transpososome was to bind significantly more H-NS than the unfolded transpososome, then the extra mass could account for the H-NS-bound folded transpososome having the same mobility as the H-NS-bound unfolded transpososome. Alternatively, if H-NS were to unfold the transpososome, the H-NS-bound forms of the folded and unfolded transpososomes would be expected to have the same mobility. We provide more direct evidence below that H-NS does unfold Tn10 transpososomes.
H-NS binds to donor DNA and induces transpososome unfolding
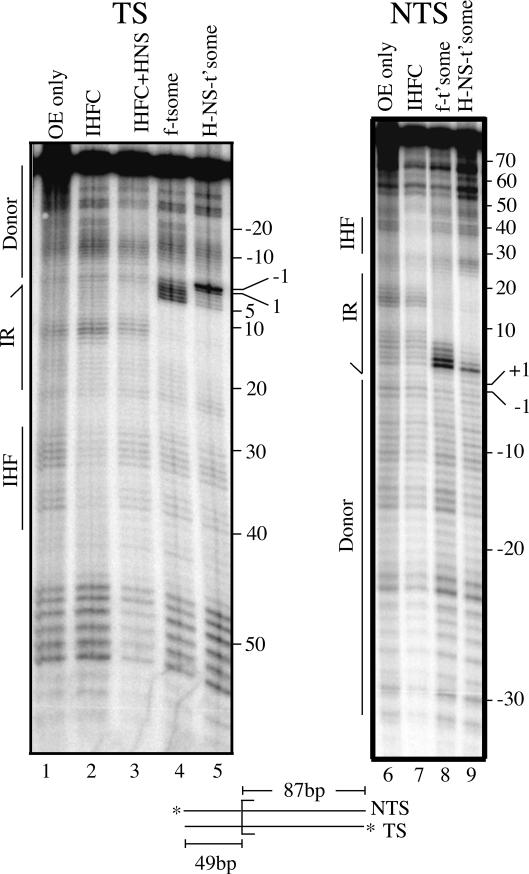
We used 1,10 phenanthroline copper (OP-Cu), a minor groove-specific footprinting reagent (Sigman et al. 1991), to localize H-NS within the transpososome. Transferred strand and nontransferred strand footprints for the folded transpososome and the H-NS-bound transpososome are shown in Figure 4. In the folded transpososome, there is a broad region of protection against OP-Cu cleavage in the terminal inverted repeat region that is due to transposase binding. On the transferred strand this encompasses nucleotides 6-18 (Fig. 4, cf. lanes 2,4), and on the nontransferred strand nucleotides 8 to at least 19 (Fig. 4, cf. lanes 7,8). Enhanced OP-Cu cleavage is also apparent at nucleotides 2-5 of the transferred strand and nucleotides 3-6 of the nontransferred strand. There is no clear region of protection over the IHF site of the folded transpososome, probably because IHF is bound tightly to only one OE (Sewitz et al. 2003).
Figure 4.
OP-Cu footprinting of transpososomes formed with and without H-NS. Transferred (TS) and nontransferred strand (NTS) OP-Cu footprints are shown. The +1 position is the first base pair of the transposon, and the -1 position is the first base pair of the flanking donor DNA. Boundaries for the terminal inverted repeat (IR), IHF-binding site, and flanking donor DNA are indicated. (IHFC + H-NS) IHF complex that was supershifted by H-NS.
H-NS addition to the folded transpososome has a major impact on the OP-Cu footprint in two ways. First, enhanced OP-Cu cleavage is shifted from nucleotides 2-5 to nucleotides 1 and -1 on the transferred strand (Fig. 4, cf. lanes 4,5), and on the nontransferred strand there is almost a complete suppression of OP-Cu hypersensitivity, with only nucleotide 3 remaining moderately hypersensitive (Fig. 4, cf. lanes 8,9). The shift in hypersensitivity to nucleotides 1 and -1 is particularly important because it has been shown that transpososome unfolding causes nucleotides 1 and -1 of the transferred strand to become hypersensitive to cleavage by chemical nucleases (Crellin and Chalmers 2001; Allingham and Haniford 2002). Thus, our footprinting data provide strong evidence that H-NS has unfolded the transpososome.
Second, a new zone of protection is evident that spans nucleotides -16 to -20 on the transferred strand and nucleotides -15 to -21 on the nontransferred strand. Transposase contacts do not extend out this far into the flanking donor DNA (Crellin and Chalmers 2001), and thus, we conclude that this protection is due to H-NS binding. H-NS can also bind to this site in the IHF complex (Fig. 4, lane 3); however, this requires fivefold more H-NS, and the presence of transposase in the reaction.
H-NS binding to the terminal inverted repeat stabilizes transpososomes in the unfolded conformation
We have presented evidence that H-NS induces the unfolding of the initial transpososome (Figs. 3C, 4) and shown that H-NS increases the efficiency of conversion of the excised transposon to intermolecular STPs (Fig. 2). However, the H-NS effect on strand transfer is dependent on H-NS binding to the flanking donor DNA, and this DNA is absent in the final excision product. For locking a transpososome lacking flanking donor DNA in the unfolded form we would predict that H-NS is able to bind at least one site within the transpososome. We show below that H-NS is able to bind within the terminal inverted repeat of a transpososome, and that this interaction alone is sufficient to lock a transpososome in the unfolded form.
H-NS does not bind with an appreciable affinity to a folded transpososome constructed with 15 bp of flanking donor DNA (Fig. 3B). However, we show in Figure 5A that if we transiently unfold this transpososome, by treating it with heparin, H-NS now binds efficiently to this transpososome. Treatment with heparin alone resulted in ∼50% conversion of folded to unfolded transpososome (Fig. 5A, lane 2). Strikingly, all of the transpososome in the heparin-treated sample is converted to an H-NS-bound form when H-NS is added after heparin treatment (Fig. 5A, lane 3). In contrast, when H-NS is added without prior heparin treatment very little H-NS-bound transpososome is formed (Fig. 5A, lanes 4-6).
Figure 5.
H-NS binding to heparin-treated transpososomes formed with a short donor flank. (A) H-NS was added to a transpososome (formed with the substrate containing 15 bp of flanking donor DNA) that was either treated (lane 3) or not treated (lanes 4-6) with heparin. (Lane 2) Note that heparin treatment of the folded transpososome alone resulted in only ∼50% conversion to unfolded transpososome. (B) OP-Cu footprinting was carried out on the indicated species generated in A and a parallel experiment in which the opposite strand was 5′-end-labeled.
We further characterized the H-NS-bound transpososome that was formed after heparin treatment by subjecting it to OP-Cu footprinting (Fig. 5B). This experiment demonstrates that occupancy of IHF sites is low in this transpososome, which is indicative of an unfolded transpososome (Sakai et al. 2000; Crellin and Chalmers 2001). With this substrate IHF binding is revealed by hypersensitivity to OP-Cu at nucleotides 27 and 28, and by OP-Cu protection in the region of nucleotides 31-45 on the transferred strand (Fig. 5B, cf. lanes 2,3 and 1). Both the hypersensitivity to OP-Cu and the protection against cleavage are lost in the unfolded transpososome and in the H-NS-bound transpososome (Fig. 5B, cf. lanes 4,5 and 3). The lack of protection (28-44) on the nontransferred strand further confirms that IHF is absent from both the unfolded transpososome and the H-NS-bound transpososome (Fig. 5B, cf. lanes 6,7 and 8,10). Thus, with regard to IHF occupancy, the H-NS-bound transpososome behaves precisely the same as the unfolded transpososome. In addition, nucleotides +1 and -1 of the transferred strand are mildly hypersensitive to OP-Cu cleavage in the H-NS-bound transpososome and the unfolded transpososome (Fig. 5B, cf. lanes 3,5). The above similarities in the footprints, and data from Figure 3, lead us to conclude that the H-NS-bound transpososome is locked in the unfolded form.
A simple scenario to explain the unfolded state of the H-NS-bound transpososome was that H-NS occupied the IHF site and prevented IHF rebinding and folding the arms. However, there is no compelling evidence in the footprinting experiment that H-NS is binding to the IHF site. Instead, H-NS addition does suppress the OP-Cu hypersensitivity that extends from nucleotides 2 to 4 on the transferred strand of the folded transpososome, and this suppression is not observed in the unfolded transpososome (Fig. 5B, lanes 3-5). Thus, this difference cannot be attributed to transpososome unfolding per se. Since H-NS clearly binds this transpososome, we conclude that at least one H-NS-binding site is adjacent to transposase in the proximal region of the terminal inverted repeat. Furthermore, we suggest that H-NS binding at this location is responsible for locking the transpososome in the unfolded state. Presumably, this binding site becomes accessible only when the transpososome is transiently unfolded.
H-NS stimulates intermolecular strand transfer in a staged reaction
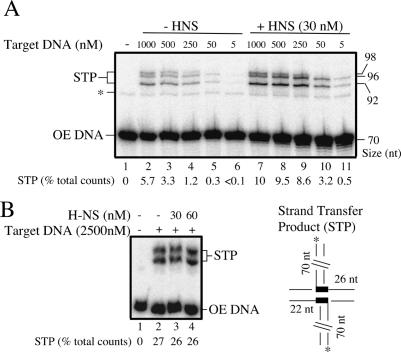
If H-NS helps to trap the transpososome in an unfolded form, then it should directly stimulate intermolecular strand transfer in a staged reaction with a folded cleaved transpososome. If, however, H-NS affects the strand transfer chemistry directly, then stimulation of strand transfer would still be observed if the cleaved transpososome was unfolded by heparin treatment prior to H-NS addition.
In previous work we showed that addition of heparin and a target DNA containing the HisG1 hotspot for Tn10 insertion allows efficient conversion of the folded cleaved transpososome to STPs (Junop and Haniford 1997). Both heparin and the target DNA act to sequester dissociated IHF from the folded cleaved transpososome, thereby allowing this species to unfold and interact with the target DNA. We show in Figure 6A that in the absence of heparin treatment, H-NS addition increases the frequency of strand transfer relative to reactions where H-NS is not added. Furthermore, H-NS addition has the greatest impact at the lowest concentrations of target DNA. For example, at 50 nM target DNA there is a 10-fold increase in STPs when H-NS is present, compared with a less than twofold increase at 1000 nM target DNA (Fig. 6A, cf. lanes 5,10). This inverse relationship is expected if H-NS and target DNA both perturb the equilibrium between folded and unfolded forms of the cleaved transpososome in the direction of unfolding.
Figure 6.
Effect of H-NS on strand transfer with a preformed, fully cleaved transpososome. A preformed, fully cleaved transpososome was either mixed with target DNA and then H-NS (A), or pretreated with target DNA and heparin before addition of H-NS (B), in the presence of MgCl2. Reactions were then analyzed as in Figure 2. The amount of STP formed is indicated for each reaction as a percentage of the total counts loaded. The expected STPs are shown in the diagram. The HisG1 hotspot is represented by thick lines within the target fragment.
In Figure 6B we show that if the folded cleaved transpososome is first converted to an unfolded cleaved transpososome, by treatment with both heparin and target DNA, subsequent addition of H-NS does not increase the level of STPs (cf. lanes 2 and 3,4). Thus, the results of the experiments in Figure 6 support the idea that H-NS stimulates intermolecular transposition by unfolding the transpososome and not by affecting the chemical steps in strand transfer directly, a possibility that could not be ruled out by the experiment in Figure 2.
H-NS affects transpososome structure in vivo
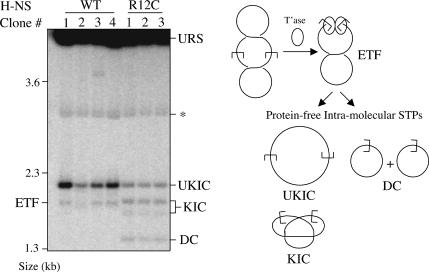
In a strain expressing the wild-type form of H-NS, transposition events from a mini-Tn10-containing plasmid partition between intermolecular insertions and intratransposon insertions (Swingle et al. 2004). The latter products are almost exclusively unknotted inversion circles (Benjamin and Kleckner 1989). These intratransposon events arise as a consequence of geometric and topological constraints imposed by DNA supercoiling and IHF binding to the outside ends of the transpososome (Chalmers et al. 1998). If H-NS does influence the structure of the transpososome in vivo, as we have provided evidence for in vitro, then some mutant forms of H-NS might produce a different distribution of STPs. We screened a small collection of H-NS mutant strains for possible affects on product partitioning by expressing transposase in the presence of a multicopy plasmid containing a mini-Tn10 element (see Materials and Methods). DNA was isolated from these strains, and intratransposon transposition products were detected by Southern blot analysis (Haniford and Kleckner 1994).
We show in Figure 7 that in the hns12 strain (encodes H-NS with an R to C substitution at amino acid 12) (Ueguchi et al. 1996), the intratransposon transposition products formed are dramatically different compared with the wild-type strain. In the wild-type strain two transposition products are detected, the unknotted inversion circle and excision product. Importantly, no knotted inversion circles or deletion circles are detected. In contrast, products from the hns12 strain include knotted inversion circles and deletion circles in addition to unknotted inversion circles, and the sum of the knotted inversion circles and deletion circles is roughly equivalent to the unknotted inversion circles. Also, the amount of unknotted inversion circles formed in hns12 isolates can be as much as 10-fold less than that formed in the wild-type strain (Fig. 7, cf. hns12 clones 1-3 and clone 1 from the wild-type strain). The qualitative and quantitative changes in the STP species are indicative of a transpososome structure in which the transposon ends are less tightly constrained relative to the folded transpososome (Chalmers et al. 1998). Furthermore, since these changes are dependent on a mutation in H-NS, the results support the idea that H-NS plays an important role in modulating the structure of the transpososome in vivo.
Figure 7.
Analysis of transposition products formed in vivo. A Southern blot with DNA isolated from isogenic E. coli strains encoding either wild-type H-NS (four separate clones) or mutant H-NS (R12C) (three separate clones) is shown. A fragment derived from the mini-Tn10 element present in each strain was used as the probe. The diagram shows the origin of the transposition products detected on the blot. (UKIC) Unkotted inversion circle; (KIC) knotted inversion circle; (DC) deletion circle; (ETF) excised transposon fragment; (URS) unreacted substrate plasmid; (★) transposase-encoding plasmid, which cross-hybridizes weakly with the probe.
Discussion
H-NS is an important regulatory protein in bacteria. It acts as a transcriptional repressor for a wide variety of genes, many of which encode proteins involved in stress response and virulence pathways (Dorman 2004). We have shown in this work that H-NS stimulates Tn10 transposition in vitro by acting at the transition point between transposon excision and integration. H-NS binds to the initial transpososome through an interaction with the flanking donor DNA. We provide evidence that H-NS both facilitates transpososome unfolding and helps to maintain the transpososome in this state through the course of the reaction; this state is favorable for intermolecular insertion events. Thus, H-NS acts as a modulator of Tn10 transposition by influencing the target interaction pathway of the transpososome.
H-NS and transpososome unfolding
We have provided two independent pieces of evidence supporting the contention that H-NS induces transpososome unfolding. First, it has been shown that when the initial transpososome unfolds, the nucleotides at the transposon-donor junction become hypersensitive to cleavage by chemical nucleases (Crellin and Chalmers 2001; Allingham and Haniford 2002). We have shown that when H-NS binds to the folded transpososome the same nucleotides become hypersensitive to OP-Cu (Fig. 4). Second, the extent of the band shift for folded and unfolded transpososomes was very similar when an equal amount of H-NS was added to these transpososomes. If H-NS were to bind and unfold the folded transpososome, then this transpososome would be expected to have a structure equivalent to that of the H-NS-bound unfolded transpososome, and consequently the two species would comigrate in the native gel. An alternative explanation for the observed comigration might be that the folded transpososome binds more H-NS compared with the unfolded transpososome. In this case the increased mass of the former, compared with the latter, could account for the comigration. However, H-NS is a relatively small protein and to have such a dramatic effect on mobility, substantially more H-NS would have to bind to the folded relative to the unfolded transpososome. We have seen no evidence of this in footprinting studies (data not shown). Moreover, since we know that the conformation of the OE substrate is a major determinant of transpososome mobility (Sakai et al. 2000), we favor the idea that comigration of H-NS-bound forms of folded and unfolded transpososomes reflects H-NS-directed transpososome unfolding.
We have also presented two pieces of evidence supporting the contention that H-NS helps to maintain the transpososome in an unfolded form. First, when H-NS was added to folded transpososome that had been treated with heparin, the entire transpososome population was supershifted even though the heparin treatment was sufficient to convert only about half of the folded transpososome population to an unfolded form (Fig. 5A). Furthermore, IHF had been ejected from the supershifted species, a clear indication that this transpososome is unfolded (Crellin and Chalmers 2001). The flanking donor DNA was only 15 bp in the substrate used, and consequently H-NS would not be able to act directly on the folded transpososome population. We therefore infer that H-NS shifts the folding-unfolding equilibrium (set by heparin treatment) towards the unfolded form, by binding to a site that is accessible only in unfolded transpososomes. Thus, transpososomes, which normally refold at the concentration of heparin used, would be locked by H-NS binding in the unfolded conformation.
Second, we demonstrated that strand transfer in a reaction with folded cleaved transpososome was stimulated upon adding H-NS, after adding limiting amounts of target DNA (Fig. 6A). As in the situation described above, the addition of limiting amounts of target DNA establishes a new folding-unfolding equilibrium, or biases the equilibrium to the unfolded form, by titrating IHF out of the transpososome (Junop and Haniford 1997). H-NS binding to the unfolded transpososome would prevent refolding, and this would increase the amount of intermolecular target capture and strand transfer events. Importantly, when the cleaved transpososome was unfolded prior to H-NS treatment, no stimulation of strand transfer was observed. This is consistent with our model. Heparin treatment would perturb the equilibrium towards unfolding, and no stimulation in strand transfer would be expected. In addition to supporting the idea that H-NS functions in stabilizing the cleaved transpososome in the unfolded form, the results here also indicate that H-NS does not significantly affect other steps in the strand transfer reaction.
A corollary to H-NS acting as a modulator of Tn10 transposition, by stabilizing the unfolded cleaved transpososome, is that production of the unfolded from folded transpososome should be a rate-limiting step in the in vitro transposition reaction. In support of this, heparin addition increases the level of STPs in an unstaged reaction without affecting transposon excision (Fig. 2A). Furthermore, it has been shown through heparin titration of reactions staged to form only folded uncleaved transpososome, folded transpososome in which flanking donor DNA is present at only one end, or fully cleaved folded transpososome that the fully cleaved transpososome is the most difficult transpososome to unfold. This is consistent with unfolding of the fully cleaved transpososome being a rate-limiting step (Liu et al. 2005).
It is also notable that H-NS did not have a significant effect on excision, because previous work has provided evidence that full excision is coupled to transpososome unfolding (Crellin et al. 2004; Humayun et al. 2005). Our failure in the present study to observe an H-NS effect on transposon excision could either be an indication that transpososome unfolding is a limiting step only at the transition from fully cleaved transpososome to strand transfer, or that full excision and transpososome unfolding are not actually coupled.
A two-step model for H-NS-directed transpososome unfolding
The initial interaction between H-NS and the folded transpososome appears to take place via the flanking donor DNA. It remains to be shown how this interaction ultimately affects the conversion of full excision to STPs, because the flanking donor DNA is lost from the transpososome prior to the target interaction and strand transfer steps (Sakai and Kleckner 1997). A likely explanation for this apparent paradox, and supported by data presented here, is that H-NS binding to the donor DNA initiates unfolding, and then additional molecules of H-NS bind within the terminal inverted repeat, effectively locking in the unfolded conformation. This would explain why the flanking donor-binding site is necessary for transpososome unfolding and stimulation of the in vitro reaction, as well as the dependence of H-NS binding to the terminal inverted repeat on transient unfolding. Consistent with this model, it was recently reported that flanking donor DNA is a key determinant in transpososome unfolding (Liu et al. 2005).
It is unclear how H-NS binding to the flanking donor site induces transpososome unfolding. We do not currently know the trajectory of the donor DNA as it projects out of the transpososome. H-NS preferentially binds to bent DNA (Yamada et al. 1991) and can itself induce DNA bending (Spurio et al. 1997). Thus, it is possible that H-NS situated at the donor flank is actually in close proximity to either the IHF-binding site or the subterminal transposase-binding site of the opposite end (see Fig. 1B). At this point we can only speculate that transpososome unfolding induced by H-NS binding to the donor flank creates a structural change in the terminal inverted repeat, such as DNA bending, that permits additional H-NS binding.
Does H-NS promote Tn10 transposition in vivo by inducing transpososome unfolding?
The conformation of the fully cleaved transpososome determines the mode of strand transfer (Fig. 1B; Chalmers et al. 1998). It follows that the types of STPs observed provide information regarding the conformation of the fully cleaved transpososome. We have shown that expression of a mutant form of H-NS (H-NSR12C) drastically alters the types of STPs formed. Some issues related to the interpretation of this experiment are discussed below.
We have argued based on in vitro data that H-NS promotes intermolecular Tn10 transposition by inducing transpososome unfolding and helping to maintain the transpososome in the unfolded state. Yet, the unknotted inversion circle, a product arising from a folded fully cleaved transpososome, is the predominant intramolecular transposition product observed in vivo in an E. coli strain expressing wild-type H-NS (Fig. 7). We suggest that not all of the fully cleaved transpososome interacts with H-NS and consequently the transpososomes enter different strand transfer pathways. Transpososomes that bind H-NS would undergo intermolecular transposition events, which are not detected in the in vivo product analysis assay. In contrast, transpososomes that do not bind H-NS would be subject to IHF channeling and therefore would give rise to unknotted inversion circles. H-NS may be limiting because, while it is an abundant protein, there are also a large number of potential H-NS-binding sites in the chromosome, and its concentration varies with growth phase. Moreover, much of the H-NS may be present in higher-order oligomeric forms, from which it may not be able to bind to Tn10 transpososomes.
The H-NSR12C mutant was initially identified in a screen for H-NS mutants that were defective at repressing the proVWX operon in E. coli. While the R12C protein is a poor repressor, it retains both nonspecific and structure-specific DNA binding activities. It is also proficient in dimerization and in forming higher-order H-NS oligomers (Ueguchi et al. 1996). Since there are no obvious defects in DNA binding or in self-association, it was suggested that the loss of repressor function might be due to a defective interaction with RNA polymerase (Ueguchi et al. 1996). Notably, R12 is located on a surface-exposed segment of H-NS (Bloch et al. 2003), and thus is in a good position to interact with residues in other proteins.
In the hns12 strain, knotted inversion circles (and deletion circles) were as abundant as unknotted inversion circles. This dramatic difference in intramolecular STP distribution relative to the wild-type strain could be explained if the H-NSR12C protein were to bind a sub-population of transpososomes and only partially constrain the transposon ends. Intramolecular target interactions would be less restricted, and this would increase the probability of DNA supercoils present in the fully cleaved transpososome being trapped in the STP in the form of knots (Chalmers et al. 1998). The production of unknotted inversion circles in the hns12 strain could simply reflect the distribution of limiting H-NS between transpososomes. One expectation of this hypothesis is that the intermolecular transposition frequency would be reduced in the hns12 strain because instead of the vast majority of H-NS-interacting transpososomes going on to form intermolecular STPs, some fraction of these would be partitioned to the intramolecular pathway to give knotted inversion circles. In support of this we have found that in a papillation assay (Swingle et al. 2004), intermolecular transposition events were greatly reduced in the hns12 strain (data not shown).
In considering both the effect of the H-NSR12C mutant on the Tn10 STP distribution in vivo and its physical properties, the possibility arises that H-NS influences the structure of the transpososome via an interaction with either transposase or IHF. The transposition phenotype of H-NSR12C could then be linked to the inability of this protein to participate in such an interaction. As the R12C mutation disrupts H-NS repressor function, it is also possible that H-NS R12C influences the transpososome structure by affecting the expression of another protein that influences transpososome structure.
Interestingly, it has been shown in vitro that when HU is used in place of IHF in transposition reactions, the occurrence of topologically complex STPs increases without a concomitant increase in intermolecular events (Chalmers et al. 1998). Unlike IHF, HU binds DNA in a sequence-nonspecific manner (Lavoie et al. 1996) and consequently it is likely that, in a transpososome formed with HU, the transposon ends are not constrained in precisely the same way as in a transpososome formed with IHF. The HU-transpososome therefore provides a precedent for the transpososome being able to adopt a conformation in vivo that is intermediate to the IHF-folded and unfolded forms.
Impact of H-NS on other transposition systems
Five different transposition systems, including Tn10, IS903, Tn552, IS1, and bacteriophage Mu, are modulated by H-NS. H-NS is inhibitory in the Mu system as it enhances the activity of the Mu repressor, thereby helping to block the transcription of genes whose products are required for transposition (Falconi et al. 1991). In IS1 transposition, H-NS promotes transposition as it affects the amount of transposase (InsAB protein) produced, either by aiding in InsAB translation or by protecting InsAB from proteolysis (Rouquette et al. 2004). H-NS also promotes IS903 and Tn552 transposition, but it is not yet known how it acts in these systems (Swingle et al. 2004). It seems unlikely that H-NS would influence IS903 or Tn552 transposition via the same mechanism as that proposed here for Tn10 transposition because there is no evidence for IHF-mediated transpososome unfolding in these systems and they lack obvious IHF-binding sites close to the terminal inverted repeats (Derbyshire and Grindley 1992; Leschziner et al. 1998).
It is likely that H-NS can act on a number of transposition systems in different ways because it is capable of interacting with other proteins and/or it has DNA structure-specific binding activity (Dorman 2004). It is not yet clear whether H-NS influences Mu and IS1 transposition through direct interactions with the Mu repressor protein and InsAB, respectively. With regard to structure-specific DNA binding, many transpososomes characterized to date are thought to possess bent DNA and thus could preferentially bind H-NS.
Could H-NS play a role in linking transposition to environmental stimuli?
Tn10/IS10 transposition is subject to several levels of negative regulation, which act primarily to inhibit transposase synthesis (for review, see Kleckner 1989). In the rare instances that the transposase protein is made, and is able to initiate a Tn10/IS10 transposition event, it would be beneficial to the transposon if intermolecular transposition events were favored. We suggest that by adapting to use a relatively abundant protein like H-NS to promote intermolecular transposition, the probability of Tn10/IS10 being maintained in bacterial hosts would be increased. Other transposons may use H-NS to promote transposition for the same reason, even though H-NS may act at different steps.
While H-NS is abundantly expressed in bacteria it is notable that its function can be modified in several different ways. For example, reduced transcriptional repressor function has been reported for H-NS at high temperature (Falconi et al. 1998) and increased osmolarity (Levinthal and Pownder 1996). Its interaction with several different proteins also modifies its function, sometimes in combination with altered environmental conditions (Zhang et al. 1996; Free et al. 1998; Nieto et al. 2000; Deighan et al. 2003; Paytubi et al. 2004). In addition, the concentration of H-NS is growth rate-regulated (Ali Azam et al. 1999). This raises the possibility that the transposition frequency of some bacterial transposons, such as Tn10/IS10, could be modulated by environmental conditions through H-NS.
Materials and methods
Proteins and OE substrates
Tn10 transposase, IHF, and H-NS were purified as described by Chalmers and Kleckner (1994), Surette and Chaconas (1989), and Cusick and Belfort (1998), respectively. Protein concentrations were determined using the BCA assay. For in vivo analysis of transposition products, Tn10 transposase was synthesized from pDH10 (Haniford et al. 1989).
OE substrates with 49 and 0 bp of donor DNA were generated by SalI-BglII digestion of pNK1935 and PvuII digestion of pWY1005, respectively (details of plasmid constructs are available upon request). OE substrates with 29 and 39 bp of donor DNA were isolated by BstBI and EcoRV digestion of pWY1005, respectively. OE substrate with 15 bp of flanking donor DNA was generated by annealing oligonucleotides 78-NTS and 80-TS, as described by Allingham et al. (2001). Target DNA containing the HisG1 hotspot for Tn10 insertion (Halling and Kleckner 1982) was generated by annealing oligonucleotides HisG1-40mer-T and HisG1-40mer-B, as described by Pribil and Haniford (2000).The mini-Tn10 element used for the in vivo analysis was present on pDH50 (Haniford and Kleckner 1994).
Time-course experiments
32P-labeled OE DNA (6 nM), IHF (18 nM), H-NS (30 nM), transposase (20 nM), target DNA (250 nM), and MgCl2 (5 mM) were mixed in the order listed in a Tris-HCl-buffered KCl solution at 25°C (Sakai et al. 1995). When heparin sulfate was used in place of H-NS, heparin was present at 170 nM. At the indicated times aliquots were removed and treated with phenol, and the DNA was recovered by ethanol precipitation. DNA pellets were resuspended in denaturing loading dye and applied to an 8% sequencing gel. Gels were analyzed by phosphorimaging, and quantification of product levels was carried out using ImageQuant software.
Electrophoretic mobility shift assays
32P-labeled OE DNA (6 nM) was mixed in a Tris-HCl buffered KCl solution (Sakai et al. 1995) with IHF (18 nM) and incubated for 30 min at 25°C. Transposase (20 nM) was then added and after an additional 2 h of incubation, either H-NS (7-60 nM) or heparin (170-330 nM) followed by H-NS was added. Following a further 2 h of incubation, samples were mixed with nondenaturing loading dye and applied to a 5% native polyacrylamide gel as described by Sakai et al. (1995). In the experiments shown in Figure 3, CaCl2 (5 mM) was added in addition to heparin to induce transpososome unfolding, whereas in the experiment shown in Figure 5, no CaCl2 was added. All incubations were at 25°C.
OP-Cu footprinting
Transpososome assembly reactions were carried out as described for the electrophoretic mobility shift assays, except that in Figure 4, H-NS-shifted IHF complex was generated by adding H-NS to 150 nM H-NS instead of 30 nM H-NS. Assembly reactions were subjected to electrophoresis on a 5% native polyacrylamide gel, and “in-gel” footprinting was performed as previously described (Allingham et al. 2001). Briefly, this involved OP-Cu treatment of the entire gel, followed by isolating species of interest from the wet gel. The purified DNA was then subjected to electrophoresis through an 8% sequencing gel.
Strand transfer assay with precleaved OE substrate
Folded, fully cleaved transpososome was assembled essentially as described for the folded, uncleaved transpososome in the “gel mobility shift” section. Target DNA (HisG1-containing fragment) (5-1000 nM) was added along with MgCl2 (5 mM), and where indicated, H-NS (30 nM). Alternatively, heparin (330 nM), target DNA (2500 nM), and MgCl2 were added prior to H-NS (30-60 nM) addition. Two hours after all of the components were added, reactions were phenol-extracted, and the purified DNA was analyzed on an 8% sequencing gel.
In vivo analysis of transposition products
Isogenic strains of E. coli (DH10B) were transformed with compatible plasmids encoding ptac-Tn10 transposase (pDH10) and a mini-KanR-Tn10 (pDH50). Transformants were grown to an OD600 of 0.4 in LB media supplemented with ampicillin (100 μg/mL) and chloramphenicol (20 μg/mL). Transposase synthesis was then induced by addition of IPTG (1 mM) for 1 h, before cells were harvested and DNA was purified by a minicleared lysate protocol (Clewell and Helinski 1970). DNA was detected by Southern blot analysis as previously described using a KanR fragment probe (Haniford and Kleckner 1994). Four different “H-NS mutant strains” were analyzed: hns12 (R12C), hns52 (G113D), hns60 (1-93), and hns205 (1-91). Only hns12 gave a product distribution that looked significantly different from the strain encoding wild-type H-NS.
Acknowledgments
We thank D. Edgell for critical reading of the manuscript and K. Kobryn for helpful suggestions throughout the course of this work. We also thank M. Belfort for providing an H-NS overexpression plasmid. This research was supported by a grant to K.M.D from the National Institutes of Health (GM50699) and by a grant to D.B.H from the Canadian Institutes of Health Research (MOP11281).
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1338905.
References
- Ali Azam T., Iwata, A., Nishimura, A., Ueda, S., and Ishihama, A. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181: 6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allingham J.S. and Haniford, D.B. 2002. Mechanisms of metal ion action in Tn10 transposition. J. Mol. Biol. 319: 53-65. [DOI] [PubMed] [Google Scholar]
- Allingham J.S., Wardle, S.J., and Haniford, D.B. 2001. Determinants for hairpin formation in Tn10 transposition. EMBO J. 20: 2931-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T. and Ingmer, H. 1997. H-NS: A modulator of environmentally regulated gene expression. Mol. Microbiol. 24: 7-17. [DOI] [PubMed] [Google Scholar]
- Benjamin H.W. and Kleckner, N. 1989. Intramolecular transposition by Tn10. Cell 59: 373-383. [DOI] [PubMed] [Google Scholar]
- Bloch V., Yang, Y., Margeat, E., Chavanieu, A., Auge, M.T., Robert, B., Arold, S., Rimsky, S., and Kochoyan, M. 2003. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat. Struct. Biol. 10: 212-218. [DOI] [PubMed] [Google Scholar]
- Chalmers R.M. and Kleckner, N. 1994. Tn10/IS10 transposase purification, activation, and in vitro reaction. J. Biol. Chem. 269: 8029-8035. [PubMed] [Google Scholar]
- Chalmers R., Guhathakurta, A., Benjamin, H., and Kleckner, N. 1998. IHF modulation of Tn10 transposition: Sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell 93: 897-908. [DOI] [PubMed] [Google Scholar]
- Clewell D.B. and Helinski, D.R. 1970. Properties of a super-coiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry 9: 4428-4440. [DOI] [PubMed] [Google Scholar]
- Craigie R., Arndt-Jovin, D.J., and Mizuuchi, K. 1985. A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: Protein and DNA substrate requirements. Proc. Natl. Acad. Sci. 82: 7570-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crellin P. and Chalmers, R. 2001. Protein-DNA contacts and conformational changes in the Tn10 transpososome during assembly and activation for cleavage. EMBO J. 20: 3882-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crellin P., Sewitz, S., and Chalmers, R. 2004. DNA looping and catalysis; the IHF-folded arm of Tn10 promotes conformational changes and hairpin resolution. Mol. Cell 13: 537-547. [DOI] [PubMed] [Google Scholar]
- Cusick M.E. and Belfort, M. 1998. Domain structure and RNA annealing activity of the Escherichia coli regulatory protein StpA. Mol. Microbiol. 28: 847-857. [DOI] [PubMed] [Google Scholar]
- Deighan P., Beloin, C., and Dorman, C.J. 2003. Three-way interactions among the Sfh, StpA and H-NS nucleoid-structuring proteins of Shigella flexneri 2a strain 2457T. Mol. Microbiol. 48: 1401-1416. [DOI] [PubMed] [Google Scholar]
- Derbyshire K.M. and Grindley, N.D. 1992. Binding of the IS903 transposase to its inverted repeat in vitro. EMBO J. 11: 3449-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman C.J. 2004. H-NS: A universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2: 391-400. [DOI] [PubMed] [Google Scholar]
- Falconi M., McGovern, V., Gualerzi, C., Hillyard, D., and Higgins, N.P. 1991. Mutations altering chromosomal protein H-NS induce mini-Mu transposition. New Biol. 3: 615-625. [PubMed] [Google Scholar]
- Falconi M., Colonna, B., Prosseda, G., Micheli, G., and Gualerzi, C.O. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 17: 7033-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free A., Williams, R.M., and Dorman, C.J. 1998. The StpA protein functions as a molecular adapter to mediate repression of the bgl operon by truncated H-NS in Escherichia coli. J. Bacteriol. 180: 994-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling S.M. and Kleckner, N. 1982. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell 28: 155-163. [DOI] [PubMed] [Google Scholar]
- Haniford D. 2002. Transposon Tn10. In Mobile DNA (eds. N.L. Craig et al.), Vol. II, pp. 457-483. ASM Press, Washington, D.C. [Google Scholar]
- Haniford D. and Kleckner, N. 1994. Tn 10 transposition in vivo: Temporal separation of cleavages at the two transposon ends and roles of terminal basepairs subsequent to interaction of ends. EMBO J. 13: 3401-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniford D.B., Chelouche, A.R., and Kleckner, N. 1989. A specific class of IS10 transposase mutants are blocked for target site interactions and promote formation of an excised transposon fragment. Cell 59: 385-394. [DOI] [PubMed] [Google Scholar]
- Haniford D.B., Benjamin, H.W., and Kleckner, N. 1991. Kinetic and structural analysis of a cleaved donor intermediate and a strand transfer intermediate in Tn10 transposition. Cell 64: 171-179. [DOI] [PubMed] [Google Scholar]
- Hommais F., Krin, E., Laurent-Winter, C., Soutourina, O., Malpertuy, A., Le Caer, J.P., Danchin, A., and Bertin, P. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40: 20-36. [DOI] [PubMed] [Google Scholar]
- Humayun S., Wardle, S.J., Shilton, B.H., Pribil, P.A., Liburd, J., and Haniford, D.B. 2005. Tn10 transposase mutants with altered transpososome unfolding properties are defective in hairpin formation. J. Mol. Biol. 346: 703-716. [DOI] [PubMed] [Google Scholar]
- Junop M.S. and Haniford, D.B. 1997. Factors responsible for target site selection in Tn10 transposition: A role for the DDE motif in target DNA capture. EMBO J. 16: 2646-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A.K., Guhathakurta, A., Kleckner, N., and Haniford, D.B. 1998. Tn10 transposition via a DNA hairpin intermediate. Cell 95: 125-134. [DOI] [PubMed] [Google Scholar]
- Kleckner N. 1989. Transposon Tn10. In Mobile DNA. (eds. D.E. Berg and M.M. Howe), Vol. I, pp. 227-268. ASM Press, Washington, D.C. [Google Scholar]
- Lavoie B.D., Shaw, G.S., Millner, A., and Chaconas, G. 1996. Anatomy of a flexer-DNA complex inside a higher-order transposition intermediate. Cell 85: 761-771. [DOI] [PubMed] [Google Scholar]
- Leschziner A.E., Griffin, T.J.t., and Grindley, N.D. 1998. Tn552 transposase catalyzes concerted strand transfer in vitro. Proc. Natl. Acad. Sci. 95: 7345-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinthal M. and Pownder, T. 1996. hns, rpoS and lrp mutations affect stationary-phase survival at high osmolarity. Res. Microbiol. 147: 333-342. [DOI] [PubMed] [Google Scholar]
- Liu D., Crellin, P., and Chalmers, R. 2005. Cyclic changes in the affinity of protein-DNA interactions drive the progression and regulate the outcome of the Tn10 transposition reaction. Nucleic Acids Res. 33: 1982-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisato D. and Kleckner, N. 1987. Tn10 transposition and circle formation in vitro. Cell 51: 101-111. [DOI] [PubMed] [Google Scholar]
- Nagy Z. and Chandler, M. 2004. Regulation of transposition in bacteria. Res. Microbiol. 155: 387-398. [DOI] [PubMed] [Google Scholar]
- Nieto J.M., Madrid, C., Prenafeta, A., Miquelay, E., Balsalobre, C., Carrascal, M., and Juarez, A. 2000. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol. Gen. Genet. 263: 349-358. [DOI] [PubMed] [Google Scholar]
- Paytubi S., Madrid, C., Forns, N., Nieto, J.M., Balsalobre, C., Uhlin, B.E., and Juarez, A. 2004. YdgT, the Hha paralogue in Escherichia coli, forms heteromeric complexes with H-NS and StpA. Mol. Microbiol. 54: 251-263. [DOI] [PubMed] [Google Scholar]
- Plasterk R.H. 2002. RNA silencing: The genome's immune system. Science 296: 1263-1265. [DOI] [PubMed] [Google Scholar]
- Pribil P.A. and Haniford, D.B. 2000. Substrate recognition and induced DNA deformation by transposase at the target-capture stage of Tn10 transposition. J. Mol. Biol. 303: 145-159. [DOI] [PubMed] [Google Scholar]
- Rimsky S., Zuber, F., Buckle, M., and Buc, H. 2001. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 42: 1311-1323. [DOI] [PubMed] [Google Scholar]
- Rouquette C., Serre, M.C., and Lane, D. 2004. Protective role for H-NS protein in IS1 transposition. J. Bacteriol. 186: 2091-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai J. and Kleckner, N. 1997. The Tn10 synaptic complex can capture a target DNA only after transposon excision. Cell 89: 205-214. [DOI] [PubMed] [Google Scholar]
- Sakai J., Chalmers, R.M., and Kleckner, N. 1995. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 14: 4374-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai J.S., Kleckner, N., Yang, X., and Guhathakurta, A. 2000. Tn10 transpososome assembly involves a folded intermediate that must be unfolded for target capture and strand transfer. EMBO J. 19: 776-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewitz S., Crellin, P., and Chalmers, R. 2003. The positive and negative regulation of Tn10 transposition by IHF is mediated by structurally asymmetric transposon arms. Nucleic Acids Res. 31: 5868-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiga Y., Sekine, Y., Kano, Y., and Ohtsubo, E. 2001. Involvement of H-NS in transpositional recombination mediated by IS1. J. Bacteriol. 183: 2476-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker N.B. and Salyers, A.A. 1988. Tetracycline-dependent appearance of plasmidlike forms in Bacteroides uniformis 0061 mediated by conjugal Bacteroides tetracycline resistance elements. J. Bacteriol. 170: 1651-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman D.S., Kuwabara, M.D., Chen, C.H., and Bruice, T.W. 1991. Nuclease activity of 1,10-phenanthroline-copper in study of protein-DNA interactions. Methods Enzymol. 208: 414-433. [DOI] [PubMed] [Google Scholar]
- Signon L. and Kleckner, N. 1995. Negative and positive regulation of Tn10/IS10-promoted recombination by IHF: Two distinguishable processes inhibit transposition off of multicopy plasmid replicons and activate chromosomal events that favor evolution of new transposons. Genes & Dev. 9: 1123-1136. [DOI] [PubMed] [Google Scholar]
- Spassky A., Rimsky, S., Garreau, H., and Buc, H. 1984. H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 12: 5321-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurio R., Falconi, M., Brandi, A., Pon, C.L., and Gualerzi, C.O. 1997. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 16: 1795-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette M.G. and Chaconas, G. 1989. A protein factor which reduces the negative supercoiling requirement in the Mu DNA strand transfer reaction is Escherichia coli integration host factor. J. Biol. Chem. 264: 3028-3034. [PubMed] [Google Scholar]
- Swingle B., O'Carroll, M., Haniford, D., and Derbyshire, K.M. 2004. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. Mol. Microbiol. 52: 1055-1067. [DOI] [PubMed] [Google Scholar]
- Ueguchi C., Suzuki, T., Yoshida, T., Tanaka, K., and Mizuno, T. 1996. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 263: 149-162. [DOI] [PubMed] [Google Scholar]
- Wang Y., Rotman, E.R., Shoemaker, N.B., and Salyers, A.A. 2005. Translational control of tetracycline resistance and conjugation in the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 187: 2673-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H., Yoshida, T., Tanaka, K., Sasakawa, C., and Mizuno, T. 1991. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol. Gen. Genet. 230: 332-336. [DOI] [PubMed] [Google Scholar]
- Yoder J.A., Walsh, C.P., and Bestor, T.H. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13: 335-340. [DOI] [PubMed] [Google Scholar]
- Zhang A., Rimsky, S., Reaban, M.E., Buc, H., and Belfort, M. 1996. Escherichia coli protein analogs StpA and H-NS: Regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 15: 1340-1349. [PMC free article] [PubMed] [Google Scholar]