Abstract
The transition from the juvenile to the mature phase during vegetative development in plants is characterized by changes in leaf shape. We show that GENERAL TRANSCRIPTION FACTOR GROUP E6 (GTE6) regulates differences in leaf patterning between juvenile and mature leaves in Arabidopsis. GTE6 encodes a novel small bromodomain-containing protein unique to plants. Mutations in GTE6 disrupt the formation of elliptical leaf laminae in mature leaves, whereas overexpression of GTE6 resulted in elongated juvenile leaves. GTE6 positively regulates the expression of ASYMMETRIC LEAVES1 (AS1), which encodes a myb-domain protein that controls proximodistal patterning of leaves. Using chromatin immunoprecipitation (ChIP) assays, we show that GTE6 is associated with the promoter and the start of the transcribed region of AS1 and up-regulates expression of AS1 through acetylation of histones H3 and H4. Genetic studies demonstrated that AS1 is epistatic to GTE6, indicating that GTE6 regulates AS1 during leaf morphogenesis. Chromatin remodeling at AS1 is a key regulatory mechanism in leaf development, which ensures the continual production of mature leaves following juvenile-adult transition, thereby maintaining the identity of the mature vegetative phase.
Keywords: Arabidopsis, bromodomain protein, heteroblasty, histone acetylation, phase transition
Plant development follows a coordinated and progressive transition from the reproductively incompetent juvenile phase to the mature phase, which is competent for flowering (for review, see Kerstetter and Poethig 1998). The transition from juvenile to mature development involves various physiological and morphological changes, including alterations in leaf structure. Variation in leaf morphology between vegetative phases, also known as heteroblasty, is observed in many plant species, including the model plant Arabidopsis (Lawson and Poethig 1995; Telfer et al. 1997). In Arabidopsis, juvenile leaves are round with a sharp boundary between the lamina and petiole, whereas the lamina of mature leaves is larger and elliptical, with an obtuse angle between the lamina and petiole (Lawson and Poethig 1995; Theodoris et al. 2003). Heteroblasty is an important developmental process, whereby leaf morphology is modified for efficient light capture, because of differences in the light environment and the size of the shoot between juvenile and mature vegetative phases (Day et al. 1997; Kerstetter and Poethig 1998).
Recent studies have indicated that leaf morphogenesis is regulated by post-translational modifications of histones and chromatin remodeling (Wagner 2003). Histones are modified by acetylation, methylation, phosphorylation, ubiquitination, and ADP-ribosylation, which constitutes a code for various regulatory proteins to carry out specific functions, such as transcriptional activation or repression (Strahl and Allis 2000). In Arabidopsis, mutations in genes involved in epigenetic silencing, such as PKL, which encodes the plant homolog of the mammalian Mi2 (Eshed et al. 1999), and the polycomb-group genes CLF (Goodrich et al. 1997), EMF2 (Chen et al. 1997), and FIE (Katz et al. 2004), resulted in aberrant leaf structures. In addition, mutations in genes involved in heterochromatin formation, such as LHP1 (Gaudin et al. 2001), TFL2 (Larsson et al. 1998), and HISTONE H1 (Wierzbicki and Jerzmanowski 2004), also lead to changes in leaf shape and size. In transgenic Arabidopsis containing an antisense construct of the histone deacetylase gene ATHD1, which resulted in increased histone acetylation, leaf shape was dramatically altered, and juvenile leaves resembled mature leaves (Tian and Chen 2001). This suggests that the expression of genes involved in leaf-axis specification is regulated by acetylation of histones and chromatin remodeling (Tian and Chen 2001). Acetylated histone tails are recognized by the bromodomain, which is a conserved protein module of ∼110 amino acid residues that binds acetylated lysine residues (Dhalluin et al. 1999; Ladurner et al. 2003). The bromodomain module is present in various basal transcription factors and coactivators, and links acetylated histones and transcriptional activation (Dhalluin et al. 1999; Ladurner et al. 2003).
Plants possess an open growth system and are constantly forming new vegetative organs (Poethig 1990; Telfer et al. 1997). Leaf primordia that are initiated from the shoot apical meristem have to be specified to develop into a juvenile leaf or a mature leaf, depending on the vegetative phase of the plant. This regulatory process is nonexistent in animals, because maturation involves differentiation of organ primordia that are formed during embryogenesis, along with recruitment of new cells (Thisse and Zon 2002). Here, we show that GENERAL TRANSCRIPTION FACTOR GROUP E6 (GTE6), which encodes a bromodomain-containing protein, plays an essential role in controlling differences in the development of primordia produced during juvenile and mature phases. GTE6 regulates the development of mature leaf shape through acetylation of histones present on ASYMMETRIC LEAVES1 (AS1), a gene involved in leaf-axis specification in Arabidopsis (Byrne et al. 2000, 2001). Our data reveal the functions of a small bromodomain gene unique to plants and indicate a network of transcription factors that regulates leaf patterning during juvenile-adult transition.
Results
High GTE6 expression is correlated with mature leaf shape
GTE6 has eight exons and encodes a protein of 369 amino acid residues, with the bromodomain motif spanning residues 116-194 (Fig. 1A). To identify the transcription start site of GTE6, the 5′ end of the GTE6 transcript was examined by 5′ rapid amplification of cDNA ends (RACE), followed by PCR. The transcription start site was mapped to 73 bp upstream of the translation start site (Fig. 1A).
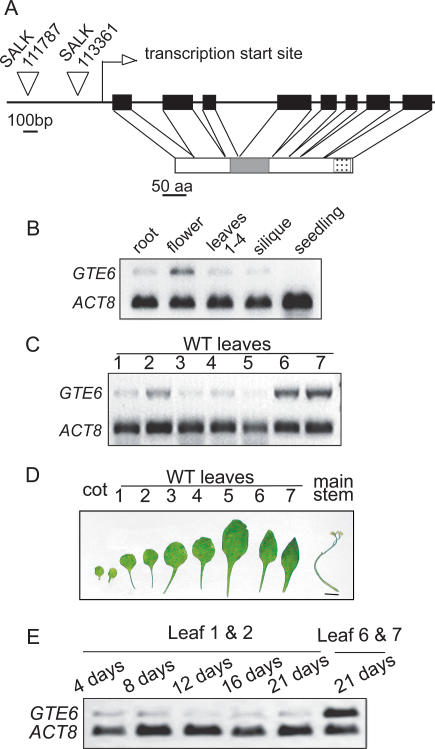
Figure 1.
GTE6 and its expression in Arabidopsis. (A) Schematic diagram of GTE6. Solid rectangles indicate exons. Arrow-heads mark the sites of T-DNA insertions. The open rectangle represents amino acid residues 1-369 of GTE6, showing the positions of the bromodomain (shaded box) and the nuclear targeting signal (dotted box). (B) Transcript abundance of GTE6, as analyzed by RT-PCR using ACTIN8 (ACT8) as an internal loading control, in various tissues of 21-d-old wild-type Columbia plants. Seedlings were 5 d old. (C) RT-PCR analyses of GTE6 transcripts in individual leaves of the basal rosette of 21-d-old wild-type Columbia plants. Leaves are numbered in order of appearance, with leaf 7 being the youngest leaf. (D) Individual leaves of the basal rosette of 21-d-old wild-type Columbia. Bar, 1 cm. (E) RT-PCR analyses of GTE6 transcripts in leaves at various times during leaf elongation and maturation. Leaves 1 and 2 were collected 4, 8, 12, 16, or 21 d after appearance.
In wild-type Arabidopsis, GTE6 transcripts are most abundant in flowers; low in roots, leaves, and siliques; and undetectable in 5-d-old seedlings (Fig. 1B). In the basal rosette leaves of 21-d-old wild-type plants, GTE6 transcripts are more abundant in leaves 6 and 7, which possess narrow elliptical laminae, than in leaves 1-4, which have round laminae (Fig. 1C,D), suggesting a possible correlation between GTE6 expression and the formation of elliptical leaf laminae in mature leaves.
Differences in GTE6 expression between the juvenile and mature leaves in 21-d-old plants are not likely to be due to differences in leaf age, because GTE6 transcript levels are similar in leaves 1 and 2 at 4-21 d after leaf initiation, suggesting that GTE6 expression does not change during various times of leaf elongation and maturation (Fig. 1E).
Mutations in GTE6 result in alteration of leaf shape
To investigate whether GTE6 may control the development of leaf shape, we analyzed mutants from the SALK collection (Alonso et al. 2003) that contained transferred DNA (T-DNA) insertions in the promoter region of the GTE6 gene (Fig. 1A). The effect of the mutations is most obvious in leaves 6 and 7, which have round laminae (Fig. 2A; width/length ratios in Supplementary Fig. S1), rather than the elliptical laminae of leaves 6 and 7 of the wild-type plants (Fig. 1D). The T-DNA insertions resulted in fewer GTE6 transcripts in leaf 7 (Fig. 2B), suggesting misregulation of GTE6 expression. The transcript abundance of GTE6 in leaf 7 of the insertion mutants was similar to leaf 4 of the wild-type and the insertion mutant plants, suggesting that the T-DNA insertions have disrupted the activation of GTE6 expression in the mature leaves (Fig. 2B). Thus, elevated expression of GTE6 in leaves 6 and 7 of wild-type plants is important for the development of the elliptical leaf lamina.
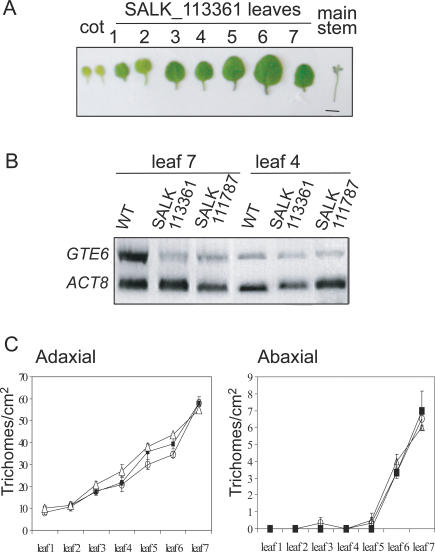
Figure 2.
T-DNA insertion mutants of GTE6.(A) Basal rosette leaves of 21-d-old insertion mutant SALK_113361. Bar, 1 cm. (B) RT-PCR analysis of GTE6 transcripts in leaves 7 and 4 of 21-d-old Columbia wild-type and T-DNA insertion mutants. (C) Trichome numbers on the adaxial (left) and abaxial (right) surfaces of wild-type Columbia (squares), SALK_13361 (triangles), and SALK_111787 (circles). The plants were germinated on MS agar, and trichomes were counted 4 wk later. The plants flowered after producing seven leaves. Error bars indicate standard error of the mean.
Phase transition during vegetative development is also characterized by changes in trichome distribution (Telfer et al. 1997). In Arabidopsis, juvenile leaves (leaves 1 to 4) have trichomes present only on the adaxial (upper) surface, whereas mature leaves (leaves 5 and above) possess trichomes on both adaxial and abaxial (lower) surfaces (Telfer et al. 1997). Trichome distribution on the rosette leaves of the insertion mutants was similar to that of the wild-type plants (Fig. 2C), indicating that GTE6 does not regulate phase transition per se but is involved in controlling development of mature leaf shapes.
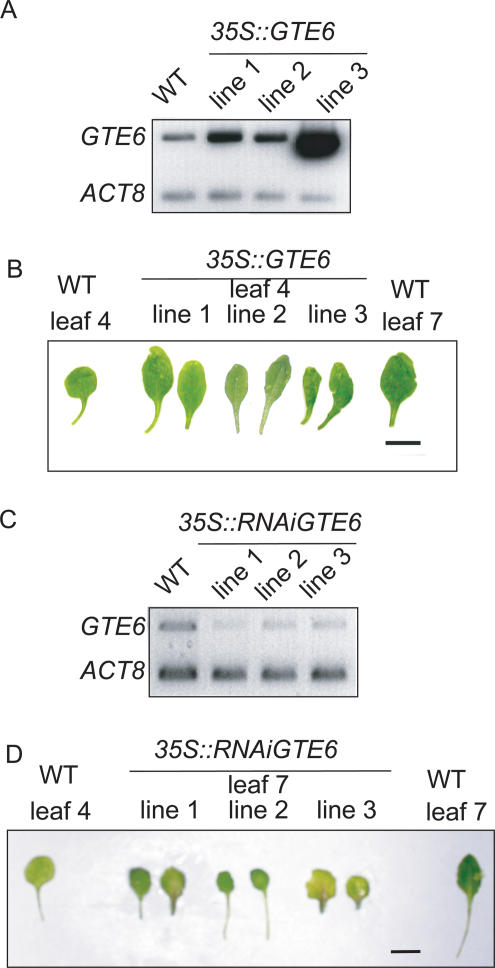
To investigate further the regulation of leaf development by GTE6, we transformed wild-type plants with a 35S::GTE6 construct containing the cauliflower mosaic virus (CaMV) 35S promoter fused to the GTE6 cDNA, resulting in more GTE6 transcripts (Fig. 3A). Elevated GTE6 expression resulted in juvenile leaves showing an elongated and elliptical shape (Fig. 3B; width/length ratios in Supplementary Fig. S1). We also analyzed mutants possessing decreased amounts of GTE6 transcripts, generated using a 35S::RNAiGTE6 construct containing the CaMV 35S promoter linked to two copies of a 524-bp region of the GTE6 cDNA in inverted orientations. Database analysis of this 524-bp sequence did not identify any other Arabidopsis genes containing similar nucleotide sequences of 15 bp or more, suggesting that the 35S::RNAiGTE6 construct was specific for GTE6. Decreased GTE6 transcript levels in the 35S::RNAiGTE6 plants (Fig. 3C) resulted in shorter and broader leaf laminae in mature leaves, as compared with the corresponding leaves in the wild-type plants (Fig. 3D; width/length ratios in Supplementary Fig. S1). Control plants transformed with empty vectors were morphologically similar to wild-type plants, indicating that the phenotype of the leaves of plants containing the 35S::GTE6 and 35S::RNAiGTE6 constructs was not the result of transformation procedures (data not shown). Altered amounts of GTE6 transcripts in the 35S::GTE6 and 35S::RNAiGTE6 plants did not alter the rate of leaf production, the flowering time, or the total number of leaves in the basal rosette, suggesting that the vegetative phase was not affected. These data indicate that GTE6 is involved in the development of the leaf lamina in mature leaves. No change in flower morphology, silique structure, or root length was observed in the mutants.
Figure 3.
Altered expression of GTE6. (A) Transcript abundance of GTE6 in shoots of three independently transformed lines of 35S::GTE6 plants, analyzed by quantitative RT-PCR using ACT8 as an internal control. (B) Leaves of three individually transformed lines of 35S::GTE6 plants. Bar, 1 cm. (C) Transcript abundance of GTE6 in shoots of three independently transformed lines of 35S::RNAiGTE6 plants, transformed with an RNAi construct containing two copies of GTE6 cDNA sequences in reverse orientations, separated by a 1-kbp intron of the pea Lip1 gene. This construct should produce a double-stranded RNA upon transcription, which should target the RNA degradation machinery to GTE6 transcripts to induce silencing. (D) Leaves of three individually transformed lines of 35S::RNAiGTE6 plants. Bar, 1 cm.
GTE6 positively regulates the expression of AS1
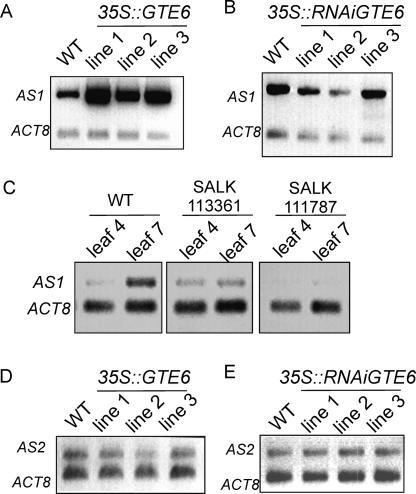
The proximodistal axis of the leaf is established in part by AS1 in Arabidopsis (Byrne et al. 2000, 2001). Loss-of-function mutations in AS1 led to shorter, broader, and occasionally lobed laminae, whereas overexpression resulted in elongated and elliptical juvenile leaves (Byrne et al. 2000; Theodoris et al. 2003). Because the phenotypes of the GTE6 mutants are similar to the AS1 mutants and GTE6 is likely to regulate leaf shape by regulating the expression of genes involved in leaf development, we investigated AS1 transcript levels in the GTE6 mutants. Elevated GTE6 expression in the 35S::GTE6 plants resulted in more AS1 transcripts (Fig. 4A), whereas decreased expression of GTE6 in the RNAi plants resulted in fewer AS1 transcripts (Fig. 4B). In wild-type plants, AS1 transcripts in leaf 7 were more abundant than in leaf 4 (Fig. 4C), consistent with the observation that GTE6 transcripts were more abundant in leaf 7 than leaf 4 (Fig. 1C). No difference in the amount of AS1 transcripts was observed between leaves 7 and 4 of the T-DNA insertion mutants, suggesting misregulation of AS1 expression in the insertion mutants (Fig. 4C). We also examined the expression of AS2, which genetically interacts with AS1 to regulate leaf patterning in Arabidopsis (Semiarti et al. 2001). The expression of AS2 was not altered in the GTE6 overexpressing mutants (Fig. 4D) or in GTE6 RNAi plants (Fig. 4E). The expression of other genes involved in leaf development, such as AN (Tsuge et al. 1996), CLF (Goodrich et al. 1997), DRL (Nelissen et al. 2003), ROT3 (G.-T. Kim et al. 1999), and TPC2 (Palatnik et al. 2003) was also investigated. However, no change in the transcript levels of these genes was observed in the GTE6 mutants, suggesting that GTE6 is not regulating the expression of these genes (Supplementary Fig. S2).
Figure 4.
Expression of AS1 and AS2 in GTE6 mutants. (A) Expression of AS1 in shoots of 21-d-old 35S::GTE6 plants, analyzed by RT-PCR. (B) RT-PCR analyses of AS1 in shoots of 21-d-old 35S::RNAiGTE6 plants. (C) RT-PCR analyses of AS1 transcripts in leaves 4 and 7 of wild-type Columbia and T-DNA insertion mutants. (D) Expression of AS2 in shoots of 21-d-old 35S::GTE6 plants. (E) RT-PCR analyses of AS2 in shoots of 21-d-old 35S::RNAiGTE6 plants.
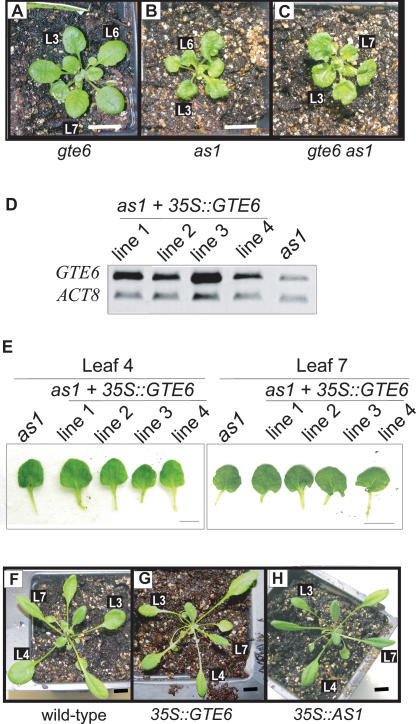
Although as1 and GTE6 T-DNA insertion mutants have similar altered mature leaf phenotypes, they are different in other aspects of leaf development; e.g., GTE6 T-DNA insertion mutants have flat leaf laminae whereas as1 has curly leaf blades (Fig. 5A,B). Double mutants of as1 and GTE6 T-DNA insertion mutant have curly leaves and are morphologically more similar to as1 than GTE6 mutants, suggesting that AS1 is epistatic to GTE6 (Fig. 5C).
Figure 5.
Genetic interactions between AS1 and GTE6. (A) GTE6 T-DNA insertion mutant SALK_113361. (B) as1 mutant. (C) Double mutant of as1 and GTE6 T-DNA insertion mutant (SALK_113361). (D) RT-PCR analysis of transcript abundance of GTE6 in four lines of as1 transformed with a 35S::GTE6 construct. (E) Leaves of the four lines of as1 overexpressing GTE6. Bar, 1 cm. (F) Wild-type Columbia. (G) Columbia containing 35S::GTE6. (H) Columbia containing 35S::AS1. Bar, 1 cm. (L) Leaf.
When GTE6 was overexpressed in as1, no change in leaf shape was observed in any of the overexpressing plants, further suggesting that GTE6 regulates leaf development through AS1 (Fig. 5D,E). Wild-type plants overexpressing AS1 have a phenotype similar to 35S::GTE6 plants; i.e., elliptical leaves 3 and 4, implicating both genes in control of mature leaf shape (Fig. 5F,G). These results provide genetic evidence that AS1 acts downstream of GTE6.
GTE6 associates with the promoter and intron of AS1
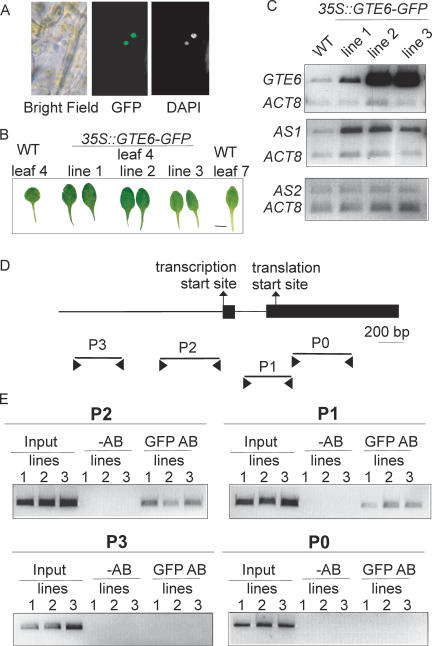
The location of the GTE6 protein in the cell was examined using Arabidopsis plants stably transformed with a 35S::GTE6-GFP construct, which expresses a GTE6-green fluorescent protein (GFP) fusion protein. The GTE6-GFP fusion protein is localized to the nucleus (Fig. 6A), which is consistent with its predicted role as a transcription factor. A nuclear-targeting signal is predicted in the C-terminal region of GTE6 (Fig. 1A).
Figure 6.
Association of GFP-tagged GTE6 with the AS1 gene in 35S::GTE6-GFP plants. (A) Confocal images of hypocotyl cells in a seedling of a 35S::GTE6-GFP plant (line 2), showing GFP fluorescence and DAPI staining of DNA. The GTE6-GFP fusion in the other lines of 35S::GTE6-GFP plants were similarly localized to the nucleus. (B) Leaf 4 of two plants from each of the three independently transformed lines of 35S::GTE6-GFP plants. Bar, 1 cm. (C) Analysis of GTE6, AS1, and AS2 transcripts in shoots of 21-d-old plants expressing the GTE6-GFP fusion by quantitative RT-PCR. ACT8 was used as an internal loading control. (D) Location of primers designed to amplify the upstream region (P3), the promoter (P2), the 3′ end of the intron and 5′ end of exon 2 (P1), and exon 2 (P0) of AS1 for ChIP assays. Exons are indicated by solid rectangles. (E) ChIP was performed using three independently transformed lines of 21-d-old 35S::GTE6-GFP plants. (Input) PCR on DNA present in the chromatin extract used for immunoprecipitation; (-AB) ChIP with no antibody; (GFP AB) ChIP with antibodies to GFP. Similar results were obtained in triplicates of ChIP reactions performed on each of the three lines.
To investigate whether the expression of AS1 is directly regulated by GTE6, we examined the association of GTE6 with various regions of AS1 with chromatin immunoprecipitation (ChIP) experiments, using Arabidopsis plants expressing a GTE6-GFP fusion protein. Linking GFP to the C terminus of GTE6 did not abolish the function of the protein, as shown by the phenotypes of wild-type plants transformed with 35S::GTE6-GFP, which were similar to wild-type plants transformed with 35S::GTE6; i.e., elongated and elliptical laminae in leaves 1-4. Figure 6B shows leaf 4 of three independently transformed lines of 35S::GTE6-GFP plants. Furthermore, AS1 transcripts were increased in the 35S::GTE6-GFP plants (Fig. 6C), as well as in the 35S::GTE6 plants (Fig. 4A), indicating that both GTE6-GFP and GTE6 up-regulated the expression of AS1. AS2 transcripts remained unchanged in the 35S::GTE6-GFP plants (Fig. 6C), as in the 35S::GTE6 plants (Fig. 4D).
ChIP was performed using antibodies to GFP, as described previously (Chua et al. 2001; Gendrel et al. 2001). Coprecipitated DNA was analyzed by PCR using primer pairs that amplify various regions of AS1 (Fig. 6D). In three independently transformed lines of 35S::GTE6-GFP plants, the GFP antibodies coprecipitated the promoter (region P2), and the 3′ end of the intron and the 5′ end of exon 2 (region P1) of AS1 (Fig. 6E). The GFP antibodies did not coprecipitate regions P3 and P0 of AS1; these sequences were undetected even after forty rounds of PCR. GTE6 thus regulates expression by associating with a 1-kbp region containing the promoter and the start of the transcribed region of AS1, which are likely to be important regulatory regions for transcriptional control (Fig. 6E).
GTE6 activates expression by mediating acetylation of histones
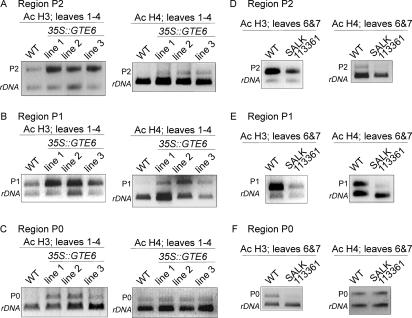
We next examined histone acetylation states at AS1 to investigate whether GTE6 may regulate expression through acetylation of histones. ChIP was performed using leaves 1-4 of wild-type Columbia and 35S::GTE6 plants. Histones H3 associated with the promoter and the transcribed region of AS1 (regions P2, P1, and P0) were more acetylated in leaves 1-4 of the 35S::GTE6 plants, compared with the wild-type plants (Fig. 7A-C). Histone H4 acetylation was also increased at regions P2 and P1 of AS1 in the 35S::GTE6 plants (Fig. 7A,B). Histone H4 acetylation at region P0 of exon 2 was unaltered (Fig. 7C). Elevated GTE6 expression in the 35S::GTE6 plants thus resulted in increased histone H3 and H4 acetylation at the promoter and the start of the transcribed region of AS1, regions where the GTE6-GFP fusion protein binds (Fig. 6E). No change in histone H3 and H4 acetylation at the upstream region P3 (-1855 to -1367, with +1 as translation start site) was observed (data not shown).
Figure 7.
Histone acetylation at AS1 in GTE6 mutants. ChIP assays were performed using antibodies to acetylated N-terminal tails of histones H3 (Ac H3), or H4 (Ac H4). (A-C) Histone acetylation at P2, P1, and P0 of AS1 was investigated in leaves 1-4 of low-expressing (line 2), medium-expressing (line 1), and high-expressing (line 3) lines of 21-d-old 35S::GTE6 plants. The positions of primers used to amplify regions P2, P1, and P0 of AS1 are indicated in Figure 6D. 18S rDNA was used as an internal control. Similar results were obtained in triplicates of ChIP reactions performed on each of the three lines. (D-F) Effect of misregulation of GTE6 expression on histone acetylation status of regions P2, P1, and P0 of AS1 in the insertion mutant SALK_113361. ChIP assays were performed using leaves 6 and 7 of 21-d-old plants. Similar results were obtained in three independent replicates of ChIP.
Histone acetylation at AS1 was examined in leaves 6 and 7 of a T-DNA insertion mutant to determine whether decreased GTE6 expression resulted in decreased histone acetylation. Leaves 6 and 7 were investigated because the shapes of these leaves were most different between the insertion mutant and the wild-type plants (Figs. 1D, 2A). The promoter and the transcribed regions of AS1 (regions P2, P1, and P0) in leaves 6 and 7 of wild-type plants were associated with high histone H3 and H4 acetylation, compared with leaves 1-4 of wild-type (Fig. 7A-F), again correlating increased histone acetylation with mature leaf shape. In leaves 6 and 7 of the insertion mutant, histone H3 acetylation at the promoter and transcribed region of AS1 (regions P2, P1, and P0) was lower than in the wild-type plants (Fig. 7D-F). Histone H4 acetylation at regions P2 and P1 of AS1 was also decreased in the insertion mutant (Fig. 7D,E). Thus, lower GTE6 expression in leaves 6 and 7 of the T-DNA insertion mutant is associated with decreased acetylation of histones H3 and H4 at AS1, as compared with wild-type plants. No difference in histone H4 acetylation at region P0 of exon 2 was observed between the insertion mutant and the wild-type plants (Fig. 7F).
GTE6 encodes a nuclear protein unique to plants
Following the complete sequencing of the Arabidopsis genome, GTE6 was annotated as a BET (bromodomain-Extraterminal) gene, because it contains a bromodomain and an Extraterminal (ET) domain, and was classified as a member of the GTE family of transcription factors (Pandey et al. 2002). The function of the ET domain is unknown but has been proposed to be involved in protein-protein interactions (Lygerou et al. 1994; Jeanmougin et al. 1997). The BET class of proteins includes the yeast BDF1 (Lygerou et al. 1994), the Drosophila FSH (Chua and Roeder 1995), and the vertebrate Brd2/Ring3/fsrg2, Brd4/HUNKI/MCAP, and Brd5/BRDT (Houzelstein et al. 2002). However, GTE6 was not among the 11 putative BET proteins proposed for Arabidopsis (Florence and Faller 2001). The C terminus of GTE6 contains only 9 of the 30 residues conserved in the ET domain and lacks the serine-rich motif, known as the SEED motif, that is characteristic of BET proteins (Supplementary Fig. S3; Florence and Faller 2001). The sequences flanking the bromodomain of GTE6 also do not possess similarity to animal and yeast BET proteins. Furthermore, animal and yeast BET proteins contain 2 bromodomains, whereas GTE6 contains a single bromodomain. Thus, GTE6 is not likely to be a plant homolog of the animal BET proteins. Database searches indicate that genes that possess high sequence similarities with GTE6 are present in pea, maize, and rice but not in unicellular organisms and animals, suggesting that this class of genes is unique to plants.
Discussion
GTE6 up-regulates expression of AS1 during leaf development
Our findings indicate that AS1 is a target gene of GTE6, and its expression is regulated by GTE6 through acetylation of histones. In wild-type Arabidopsis, high GTE6 expression in mature leaves, as compared with juvenile leaves, results in increased histone H3 and H4 acetylation at the promoter and transcribed region of AS1, leading to activation of AS1 expression. This leads to lamina elongation along the proximodistal axis and contributes to the elliptical shape in mature leaves.
GTE6 associates with the promoter and the first exon and intron of AS1 in a 1-kbp region, and increases the acetylation states of histones H3 and H4 present on this region. Increased histone H3 acetylation was also observed at exon 2, which is not associated with GTE6, in overexpressing mutants. GTE6 may increase histone acetylation at AS1 by binding to acetylated histones and protecting them from the enzymatic actions of histone deacetylases. This has been observed in yeast, where the BET protein Bdf1p shields acetylated histone H4 from deacetylation by Sir2p, which is a histone deacetylase (Ladurner et al. 2003). Alternatively, GTE6 may function as an adaptor protein that associates with histone acetyltransferases to increase acetylation levels (Hassan et al. 2001). An investigation of the proteins that interact with GTE6 in vivo may identify the mechanism of GTE6-mediated histone acetylation.
The mature leaves of as1 and gte6 homozygous plants display round laminae, suggesting that both genes are involved in the specification of adult leaf phenotype. The observation that the as1 gte6 double mutant is more similar to as1 than gte6 indicates that as1 is epistatic to gte6 in the development of mature leaves. We showed that AS1 is indeed a downstream target gene of GTE6 using ChIP experiments. Although the mature leaves are similar in both the as1 and gte6 mutants, there are slight phenotypic differences; as1 has curly leaf laminae, and gte6 has flat leaves (Fig. 5A,B). This is likely to be due to differences in AS1 expression; as1 is a null mutant and does not contain any functional AS1 protein (Byrne et al. 2000), whereas gte6 expresses AS1, albeit at a lower level than wild-type plants (Fig. 4C). Thus, the phenotypes of as1, such as broader and curly leaf lamina and shorter petioles, are stronger than those of gte6.
Overexpression of GTE6 in Arabidopsis resulted in elongated juvenile leaves, whereas RNAi depletion resulted in round mature leaves. However, overexpression of GTE6 in an as1 background did not alter leaf shape, further supporting the notion that GTE6 and AS1 are in the same genetic pathway, and that GTE6 requires a functional AS1 gene to regulate leaf morphogenesis. AS1 is a myb-domain protein and functions as a transcription factor (Byrne et al. 2000). AS1 and its homologs ROUGH SHEATH2 in maize and PHANTASTICA in Antirrhinum, play central roles in leaf patterning (Byrne et al. 2000, 2002). AS1 may modulate leaf shape by affecting the direction of cell division and/or elongation. In wild-type leaves, epidermal cell files run almost parallel to the petiole, indicating that cells elongate in one general direction to give an elliptical leaf lamina (Meijer and Murray 2001). However, in as1 mutants, files of elongated cells are arranged perpendicular to the petiole, resulting in round leaf laminae (Byrne et al. 2000). AS1 forms a complex with AS2 to repress the transcription of the KNOX genes KNAT1 and KNAT2, which encode homeobox transcription factors in the apical meristem (Byrne et al. 2002; Theodoris et al. 2003). Consistently, the leaf phenotypes of gte6 mutants are similar to as2 mutants (data not shown), suggesting that the downstream target genes of GTE6 and AS2 are common. However, because GTE6 does not regulate the expression of AS2 (Fig. 4D,E), the genes downstream of AS2 are likely to be controlled by GTE6 through AS1.
GTE6 specifies mature leaf development
Leaf morphogenesis is regulated by various chromatin remodeling genes. For example, mutations in PKL (Eshed et al. 1999), which is similar to the mammalian repressor Mi2, and CLF (Goodrich et al. 1997), EMF2 (Chen et al. 1997), and FIE (Katz et al. 2004), which encode polycomb-group proteins, result in aberrant leaf structures. In addition, abnormalities in leaf development were observed in mutants of LHP1 (Gaudin et al. 2001), TFL2 (Larsson et al. 1998), and HISTONE H1 (Wierzbicki and Jerzmanowski 2004), which encode proteins involved in heterochromatin formation. These genes repress transcription through histone deacetylation and DNA methylation and regulate leaf morphogenesis by silencing flowering and embryonic genes in leaves (Goodrich et al. 1997; Eshed et al. 1999). Our data show that acetylation of histones regulates a very different aspect of leaf morphogenesis; i.e., specification of differences between juvenile and mature leaf patterning. Interestingly, antisense inhibition of the histone deacetylase gene ATHD1 in Arabidopsis resulted in increased amounts of acetylated histones H3 and H4, and also elongated and serrated juvenile leaves that resembled mature leaves, further correlating mature leaf development with acetylation of histones (Tian and Chen 2001). Chromatin remodeling thus plays a central role in regulation of leaf development, where mechanisms of gene silencing and heterochromatin formation regulate tissue-specific gene expression, and histone acetylation controls leaf patterning.
Plants possess an open growth system and are constantly forming new leaf primordia at the meristems (Poethig 1990; Byrne et al. 2000). During transition into the mature vegetative phase, mature leaves are not derived from juvenile leaves but develop from newly formed leaf primordia. A mature plant thus possesses both juvenile and mature vegetative organs, with the older juvenile leaves present at the base of the plant and the younger mature leaves at the apices (Poethig 1990; Telfer et al. 1997). Both the juvenile and the mature vegetative developmental phases are maintained in the plant, because shoots initiated from the juvenile parts possess juvenile traits, whereas shoots arising from the mature parts produce mature leaves (Telfer et al. 1997). Moreover, it is extremely difficult to convert the mature phase back to the juvenile phase (Poethig 1990). It has long been proposed that the mechanism that maintains developmental phases in plants is epigenetic (Poethig 1990), and phase transition is associated with changes in genomic DNA methylation (Fraga et al. 2002; Baurens et al. 2004), although the regulatory process involved has not been elucidated. Our data indicate that chromatin remodeling at AS1 is necessary for the adult leaf phenotype. This epigenetic control of mature leaf shape by acetylation of histones would provide plants with the mechanism to continue producing mature leaves and not to revert into initiating juvenile leaves, following transition into the mature vegetative phase.
Heteroblasty reflects plasticity in plant development, where leaf morphology is modified during the mature vegetative phase for efficient light capture for photosynthesis (Day et al. 1997). In Arabidopsis, the elliptical mature leaf possesses almost twice as much lamina as a round juvenile leaf of the same width. Thus, the adult leaf shape minimizes overlap between leaves while increasing lamina area available for light capture. Heteroblasty is associated with aging, which occurs after the juvenile vegetative phase (Kerstetter and Poethig 1998). Interestingly, treatment with histone deacetylase inhibitors increases life span and retards aging in Drosophila (Chang and Min 2002; Kang et al. 2002). In addition, overexpression of the histone deacetylase gene Sir2 in yeast and Caenorhabditis elegans (Imai et al. 2000; Tissenbaum and Guarente 2001), and deletion of the histone deacetylase gene RPD3 in yeast (S. Kim et al. 1999), delay the aging process, suggesting that genes associated with maturation and aging are regulated by deacetylation of histones. We show that elongated elliptical leaf lamina, an adult trait in Arabidopsis, is controlled by acetylation of histones. It is interesting that regulation of gene expression during maturation appears to involve changes in histone acetylation levels in both animals and plants.
In summary, we propose that GTE6 plays a fundamental role in regulating leaf patterning during juvenile-to-adult transition in plants, and the mechanism involves chromatin remodeling at AS1 through acetylation of histones. AS1 in turn represses the transcription of KNAT1 and KNAT2, which encode homeobox proteins. Regulation of leaf shape associated with heteroblasty is thus a highly regulated process, involving a network of transcription factors. GTE6 and its plant homologs are likely to be plant-specific proteins adapted to maintain adult leaf shapes during the mature vegetative phase through acetylation of histones.
Materials and methods
Plants and growth conditions
Seeds of mutant and wild-type Columbia were surface-sterilized and germinated on 1/2 MS agar (0.24% MS salts, 0.8% agar at pH 5.9). The seeds were treated in the dark for 4 d at 4°C and transferred to 25°C under a 24-h cycle of 16 h PAR (150 μmol photons m-2/s) and 8 h dark. Unless otherwise stated, all experiments were performed using 21-d-old plants grown on 1/2 MS agar in tissue culture plates. The T-DNA insertion mutants SALK_111787 and SALK_113361 were obtained from the SALK Institute (Alonso et al. 2003). Seeds of the as1 mutant were obtained from the Arabidopsis Biological Resource Centre. Seeds of Columbia containing 35S::AS1 were provided by Michael Freeling (University of California, Berkeley, CA).
DNA constructs and plant transformation
The transcription start site of GTE6 was mapped by 5′ RACE using the FirstChoice RLM-RACE kit (Ambion, Inc.), using the method of Shaefer (1995). The primers used for the RACE reactions were GTE6raceR4 and GTE6raceR6 (Supplementary Table S1). The PCR products were sequenced to identify the transcription start site. The 35S::GTE6 construct was generated by inserting the cDNA of GTE6 into the BamHI site of pROK2 (Baulcombe et al. 1986). The 35S::RNAiGTE6 construct was generated using the intermediate vector pJH21, which contains intron 4 of the pea Lip1 gene (Sullivan and Gray 2000) inserted into the SalI-BamHI sites of pBCSK + phagemid (Stratagene). A 524-bp region of GTE6 (+587 to +1100 of GTE6 cDNA, with +1 as the translation start site) was inserted into the BglII-SalI sites of pJH21, and the same sequence, but in the reverse orientation, was inserted into the BamHI-SacI sites. The 524-bp region of GTE6 was obtained by PCR amplification. The DNA fragment containing the two copies of GTE6 separated by the intron was released by digestion with BglII and SacI, and inserted into pROK2. The 35S::GTE6-GFP construct was generated by inserting the cDNA of GTE6 into an intermediate vector pUCGFP (Haseloff et al. 1997), which contained the CaMV 35S promoter sequence, GFP, and the terminator of nos in pUC19. The 35S::GTE6-GFP sequence was excised by digestion with HindIII and EcoRI, and inserted into pROK2. Wild-type Col or as1-1 mutant was transformed with 35S::GTE6, 35S::RNAiGTE6, or 35S::GTE6-GFP with Agrobacterium strain C58C1 using the floral dip method (Clough and Bent 1998).
Generation of double mutant
The double mutant of as1-1 and GTE6 T-DNA insertion mutant (SALK_113361) was generated by pollinating as1 flowers with pollen from SALK_113361. Seeds collected from F1 plants were germinated, and the segregating F2 population was screened for double mutants by PCR using the primers LBB1, GTE6p-F2, GTE6p-F3, GTE6p-R2, AS1-f, and AS1-r (Supplementary Table S1).
Confocal microscopy
Epidermal peels from Arabidopsis leaves were stained with DAPI (10 mg/mL; Roche) for 15 min, washed with water, placed on glass slides, and examined by confocal laser scanning microscopy (Leica Microsystems) for GFP and DAPI-DNA fluorescence.
RNA analysis
RNA was purified using Tripure reagent (Roche), and reverse transcription was performed as previously described (Chua et al. 2004). Semi-quantitative duplex PCR was performed using primers specific for ACT8, AS1, GTE6, AN, CLF, ROT3, AS2, DRL, or TCP2 (Supplementary Table S1). PCR reactions were first performed with various dilutions of the template DNA to ensure that the PCR conditions were within the quantitative range of the amplification reaction. Typically, 28 cycles were used.
ChIP
ChIP experiments were performed as described previously (Chua et al. 2001; Gendrel et al. 2001). Approximately 0.1-0.8 g leaves (leaves 1-4 or 6 and 7, as described in the figure legends) of 21-d-old Arabidopsis plants grown on 1/2 MS agar in tissue culture plates were fixed with 1% formaldehyde (Sigma) under vacuum for 15 min. The plant material was ground in liquid nitrogen, and chromatin was extracted. The chromatin was sheared into fragments ranging from 400 bp to 1 kbp by sonication. The sonicated chromatin was immunoprecipitated with 10 μL of anti-acetylated histone H4 (Upstate Biotechnology), 10 μL of anti-acetylated histone H3 (Upstate Biotechnology), or 5 μL of anti-GFP (Abcam). DNA was amplified using primers specific for 18S rDNA, and various regions of AS1 (Supplementary Table S1). PCR reactions were first performed with various dilutions of the template DNA to ensure that the PCR conditions were within the quantitative range of the amplification reaction. Coprecipitated DNA was dissolved in 20 μL of TE, and typically 1 μL was used for PCR analyses. For total input samples, DNA was extracted from an aliquot of sonicated chromatin, dissolved in 80 μL of TE, and 1 μL was used for PCR. Twenty-eight cycles were used to amplify regions P1, P2, P3, or P0 of AS1, and 20 cycles were used to amplify the 18S rDNA sequence. Regions of AS1 were first amplified using AS1 primers for 8 cycles, followed by the addition of 18S rDNA primers, and the PCR was performed for another 20 cycles.
Acknowledgments
We thank J. Hibberd and J. Sullivan for providing vectors; G. Theodoris, M. Tsiantis, and M. Freeling for 35S::AS1 seeds; S. Natesan for help with confocal microscopy; J. Coates for helpful comments on the manuscript; C. Newell, T. Dyer, and T.W. Yeo for valuable discussions. This work was supported by research grants from the Biotechnology and Biological Sciences Research Council (UK).
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.352005.
References
- Alonso J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653-657. [DOI] [PubMed] [Google Scholar]
- Baulcombe D.C., Saunders, G.R., Bevan, M.W., Mayo, M.A., and Harrison, B.D. 1986. Expression of biologically active viral satellite RNA from the nuclear genome of transformed plants. Nature 321: 446-449. [Google Scholar]
- Baurens F.-C., Nicolleau, J., Legavre, T., Verdeil, J.-L., and Monteuuis, O. 2004. Genomic DNA methylation of juvenile and mature Acacia mangium micropropagated in vitro with reference to leaf morphology as a phase change marker. Tree Physiol. 24: 401-407. [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. 2000. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967-971. [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Timmermans, M., Kidner, C., and Martienssen, R. 2001. Development of leaf shape. Curr. Opin. Plant Biol. 4: 38-43. [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Simorowski, J., and Martienseen, R.A. 2002. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 29: 1957-1965. [DOI] [PubMed] [Google Scholar]
- Chang K.T. and Min, K.-T. 2002. Regulation of lifespan by histone deacetylase. Ageing Res. Rev. 1: 313-326. [DOI] [PubMed] [Google Scholar]
- Chen L., Cheng, J.-C., Castle, L., and Sung, Z.R. 1997. EMF genes regulate Arabidopsis inflorescence development. Plant Cell 9: 2011-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua P. and Roeder, S.G. 1995. Bdf1, a yeast chromosomal protein required for sporulation. Mol. Cell. Biol. 15: 3685-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua Y.L., Brown, A.P.C., and Gray, J.C. 2001. Targeted histone acetylation and altered nuclease accessibility over short regions of the pea plastocyanin gene. Plant Cell 13: 599-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua Y.L., Mott, E., Brown, A.P., and Gray, J.C. 2004. Micro-array analysis of chromatin-immunoprecipitated DNA identifies specific regions of tobacco genes associated with acetylated histones. Plant J. 37: 789-800. [DOI] [PubMed] [Google Scholar]
- Clough S.J. and Bent, A.F. 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735-743. [DOI] [PubMed] [Google Scholar]
- Day J.S., Gould, K., and Jameson, P.E. 1997. Vegetative architecture of Elaeocarpus hookerianus. Transition from juvenile to adult. Ann. Bot. 79: 617-624. [Google Scholar]
- Dhalluin C., Carlson, J.E., Zeng, L., He, C., Aggarwal, A.K., and Zhou, M.-M. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491-496. [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum, S.F., and Bowman, J.L. 1999. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99: 199-209. [DOI] [PubMed] [Google Scholar]
- Florence B. and Faller, D.V. 2001. You bet-cha: A novel family of transcriptional regulators. Front. Biosci. 1: D1008-D1018. [DOI] [PubMed] [Google Scholar]
- Fraga M.F., Canal, M.J., and Rodriguez, R. 2002. Phase-change related epigenetic and physiological changes in Pinus radiata D. Don. Planta 215: 672-678. [DOI] [PubMed] [Google Scholar]
- Gaudin V., Libault, M., Pouteau, S., Juul, T., Zhao, G., Lefebvre, D., and Grandjean, O. 2001. Mutations in LIKE HETERO-CHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128: 4847-4858. [DOI] [PubMed] [Google Scholar]
- Gendrel A.-V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. 2001. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871-1873. [DOI] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. 1997. A polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44-51. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Siemering, K.R., Prasher, D.C., and Hodge, S. 1997. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic arabidopsis plants brightly. Proc. Natl. Acad. Sci. 94: 2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.H., Neely, K.E., and Workman, J.L. 2001. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell 104: 817-827. [DOI] [PubMed] [Google Scholar]
- Houzelstein D., Bullock, S.L., Lynch, D.E., Grigorieva, E.F., Wilson, V.A., and Beddington, R.S.P. 2002. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 22: 3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S.-I., Armstrong, C.M., Kaeberlein, M., and Guarente, L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795-780. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F., Wurtz, J.-M., Le Douarin, B., Chambon, P., and Losson, R. 1997. The bromodomain revisited. Trends Biochem. Sci. 22: 151-153. [DOI] [PubMed] [Google Scholar]
- Kang H.-L., Benzer, S., and Min, K.-T. 2002. Life extension in Drosophila by feeding a drug. Proc. Natl. Acad. Sci. 99: 838-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A., Oliva, M., Mosquna, A., Hakim, O., and Ohad, N. 2004. FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37: 707-719. [DOI] [PubMed] [Google Scholar]
- Kerstetter R.A. and Poethig, R.S. 1998. The specification of leaf identity during shoot development. Annu. Rev. Cell Dev. Biol. 14: 373-398. [DOI] [PubMed] [Google Scholar]
- Kim G.-T., Tsukaya, H., Saito, Y., and Uchimiya, H. 1999. Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proc. Natl. Acad. Sci. 96: 9433-9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Benguria, A., Lai, C.-Y., and Jazwinski, S.M. 1999. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol. Biol. Cell 10: 3125-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladurner A.G., Inouye, C., Jain, R., and Tjian, R. 2003. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell 11: 365-376. [DOI] [PubMed] [Google Scholar]
- Larsson A.S., Landberg, K., and Meeks-Wagner, D.R. 1998. The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics 149: 597-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson E.J.R. and Poethig, R.S. 1995. Shoot development in plants: Time for a change. Trends Genet. 11: 263-265. [DOI] [PubMed] [Google Scholar]
- Lygerou Z., Conesa, C., Lesage, P., Swanson, R.N., Ruet, A., Carlson, M., Sentenac, A., and Seraphin, B. 1994. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNA. Nucleic Acids Res. 22: 5332-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer M. and Murray, J.A.H. 2001. Cell cycle controls and the development of plant form. Curr. Opin. Plant Biol. 4: 44-49. [DOI] [PubMed] [Google Scholar]
- Nelissen H., Clarke, J.H., de Block, M., de Block, S., Vanderhaeghen, R., Zielinski, R.E., Dyer, T., Lust, S., Inze, D., and Lijsebettens, M.V. 2003. DRL, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell 15: 639-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257-263. [DOI] [PubMed] [Google Scholar]
- Pandey R., Muller, A., Napoli, C.A., Selinger, D.A., Pikaard, C.S., Richards, E.J., Bender, J., Mount, D.W., and Jorgensen, R.A. 2002. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30: 5036-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. 1990. Phase change and the regulation of shoot morphogenesis in plants. Science 250: 923-930. [DOI] [PubMed] [Google Scholar]
- Semiarti E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. 2001. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771-1783. [DOI] [PubMed] [Google Scholar]
- Shaefer B.C. 1995. Revolutions in rapid amplification of cDNA ends: New strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal. Biochem. 227: 255-273. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis, C.D. 2000. The language of covalent histone modifications. Nature 403: 41-45. [DOI] [PubMed] [Google Scholar]
- Sullivan J.A. and Gray, J.C. 2000. The pea light-independent photomorphogenesis1 mutant results from partial duplication of COP1 generating an internal promoter and producing two distinct transcripts. Plant Cell 12: 1927-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A., Bollman, K.M., and Poethig, R.S. 1997. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645-654. [DOI] [PubMed] [Google Scholar]
- Theodoris G., Inada, N., and Freeling, M. 2003. Conservation and molecular dissection of ROUGH SHEATH2 and ASYMMETRIC LEAVES1 function in leaf development. Proc. Natl. Acad. Sci. 100: 6837-6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C. and Zon, L.I. 2002. Organogenesis—Heart and blood formation from the zebrafish point of view. Science 295: 457-462. [DOI] [PubMed] [Google Scholar]
- Tian L. and Chen, J. 2001. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. 98: 200-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum H.A. and Guarente, L. 2001. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410: 227-230. [DOI] [PubMed] [Google Scholar]
- Tsuge T., Tsukaya, H., and Uchimiya, H. 1996. Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122: 1589-1600. [DOI] [PubMed] [Google Scholar]
- Wagner D. 2003. Chromatin regulation of plant development. Curr. Opin. Plant Biol. 6: 20-28. [DOI] [PubMed] [Google Scholar]
- Wierzbicki A.T. and Jerzmanowski, A. 2004. Suppression of histone H1 genes in Arabidopsis results in heritable developmental defects and stochastic changes in DNA methylation. Genetics 169: 997-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]