Abstract
Phosphorylation of proteins on serine/threonine residues preceding proline is a key signaling mechanism. The conformation and function of a subset of these phosphorylated proteins is regulated by the prolyl isomerase Pin1 through isomerization of phosphorylated Ser/Thr-Pro bonds. Although young Pin1−/− mice have been previously shown to develop normally, we show here that they displayed a range of cell-proliferative abnormalities, including decreased body weight and testicular and retinal atrophies. Furthermore, in Pin1−/− adult females, the breast epithelial compartment failed to undergo the massive proliferative changes associated with pregnancy. Interestingly, many of these Pin1-deficient phenotypes such as retinal hypoplasia and mammary gland impairment are also the characteristic of cyclin D1-deficient mice. Cyclin D1 levels were significantly reduced in many tissues in Pin1-deficient mice, including retina and breast epithelial cells from pregnant mice. Moreover, Pin1 directly bound to cyclin D1 phosphorylated on Thr-286–Pro increased cyclin D1 in the nucleus and stabilized cyclin D1. These results indicate that Pin1 positively regulates cyclin D1 function at the transcriptional level, as demonstrated previously, and also through posttranslational stabilization, which together explain why Pin1 loss-of-function phenotypes in the mouse resemble cyclin D1-null phenotypes. Our results provide genetic evidence for an essential role of Pin1 in maintaining cell proliferation and regulating cyclin D1 function.
Phosphorylation of proteins on serine/threonine residues preceding proline (pSer/Thr-Pro) is a key regulatory mechanism for the control of various cellular processes, including cell division and transcription (for reviews see refs. 1–3). The pSer/Thr-Pro moiety in peptides and proteins exists in two distinct cis and trans conformations, whose conversion is catalyzed specifically by Pin1 (4, 5). Pin1 is a cis/trans peptidyl-prolyl isomerase that acts only on phosphorylated Ser/Thr-Pro bonds (6–8). In addition, Pin1 contains an N-terminal WW domain, which functions as a phosphorylated Ser/Thr-Pro binding module (9, 10). This phosphorylation-dependent interaction targets Pin1 to a defined subset of phosphorylated substrates facilitating conformational changes in phosphorylated proteins, thereby regulating their biological function (7, 11–20). Thus, Pin1-dependent prolyl isomerization is an essential and novel postphosphorylation regulatory mechanism.
Given its phosphorylated Ser/Thr-Pro substrate specificity, Pin1 has also been shown to be essential for maximal cell growth in different systems (4, 5). Interestingly, we have recently found that Pin1 is strikingly overexpressed in most human breast cancer tissues (21, 22). Pin1 levels are correlated with cyclin D1 mRNA and protein levels in human cancer tissues. Moreover, Pin1 can activate the cyclin D1 promoter in cell lines via binding phosphorylated c-Jun and β-catenin and increasing their transcriptional activity (21, 22). These results suggest that Pin1 may play an important role in regulation of cyclin D1 expression and also contribute to neoplastic transformation. Interestingly, disruption of cyclin D1 results in several prominent phenotypes, including retinal degeneration and mammary gland impairment (23, 24). However, disruption of the Pin1 gene in mice has been previously reported to develop normally (25). Therefore, the genetic connection between Pin1 and cyclin D1 remains to be established. Furthermore, although turnover and subcellular localization of cyclin D1 is regulated by phosphorylation on Thr-286–Pro motif by GSK-3β (26–28), it is unknown whether it is further regulated after phosphorylation.
Here, we found a range of cell-proliferative abnormalities, including decreased body weight and testicular and retinal atrophies. Interestingly, some of these phenotypes are also characteristic of cyclin D1-deficient mutant mice. In addition, we found that Pin1 directly bound to and stabilized cyclin D1 in nucleus, indicating that Pin1 regulates stability and subcellular localization of cyclin D1, in addition to the transcriptional regulation of the cyclin D1 gene we reported previously (21, 22). This study provides direct evidence that Pin1 plays a critical role in the regulation of cyclin D1 and suggests a novel mechanism for regulating cyclin D1 function.
Materials and Methods
Immunohistochemical Analysis.
For immunohistochemical analysis, both age-matched wild-type and knockout mice tissues were perfused and fixed by using Bouin's fixation solution. The immunostaining was carried out as described (13). Briefly, the fixed tissues were embedded in paraffin and sectioned at 6 μm. The dissected sections were deparaffinized in xylene, hydrated through an alcohol series from 100 to 50%. To inhibit endogenous peroxidase, sections were treated with H2O2. Antigen recapture was done by boiling slides in 1× antigen retrieval Citra (Biogenex Laboratories, San Ramon, CA). Primary antibody incubations were performed overnight at 4°C in a humidified chamber. Affinity-purified anti-Pin1 antibodies were as described (13), and polyclonal cyclin D1 antibodies were raised against a C-terminal peptide and purified by using the antigen peptide, as described (13). For the cyclin D1 control slides, cyclin D1 primary antibody was incubated with the excess antigenic peptide for 2 h before use. Immunohistochemical analysis and DAB staining were performed by using a Vectastain ABC kit (Vector Laboratories) as described (21, 22).
Mammary Gland Whole Mounts.
To examine the development of the mammary epithelium during pregnancy, the no. 4 mammary glands of nulliparous and 1-day postpartum wild-type and Pin1−/− mice were dissected, spread onto a glass slide, and fixed overnight in 6:3:1 methanol:chloroform:acetic acid buffer. The fixed glands were washed by using 70% alcohol with several changes, then defatted with acetone once or twice for 2 h each. The glands were stained overnight in 0.2% carmine red (Sigma) and 0.5% AlK(SO4)2, dehydrated in graded ethanol solutions, followed by clearing in toluene and methyl salicylate. Photos were taken by using a dissecting microscope (23, 29).
Glutathione S-Transferase (GST)-Pull Down Assay.
HeLa cells were transiently transfected either hemagglutinin (HA)-tagged cyclin D1-wild type or HA-tagged cyclin D1T286A mutant for 24 h. Cell lysates were incubated with 20 μl of agarose beads containing GST-Pin1 or GST for 2 h at 4°C as described (22). The precipitated proteins were washed three times in wash buffer containing 1% Triton X-100 and subjected to SDS/PAGE, as described (11).
Pulse–Chase Analysis.
Primary embryonic fibroblasts were prepared from 14.5-day embryos. MEF cells were grown in 60-mm dishes to 60% confluence in normal growth medium. Cells were transfected with HA-cyclin D1 and CDK4 by using Effectene (Qiagen, Chatsworth, CA). After 16 h of transfection, cells were washed twice with Hanks' balanced salt solution and pulse-labeled for 40 min in 1 ml of methionine- and glutamine-free MEM (GIBCO/BRL) supplemented with 4 mM glutamine/10% dialyzed FCS/100 μCi of [35S]methionine, as described (22, 26). Labeled cells were washed twice with Hanks' balanced salt solution and rinsed with normal growth medium. Cells were harvested at various time points and subjected to immunoprecipitation with the 12CA5 monoclonal antibody.
Expression and Localization of Cyclin D1 in MFE Cells.
Exponentially growing Pin1−/− or wild-type MEF cells were placed on glass plates and stained with anti-cyclin D1 polyclonal antibody (Santa Cruz Biotechnology). Total DNA was visualized with 4′,6-iamidino-2-phenylindole (DAPI) staining followed by the fluorescent microscopic analysis as described (22, 28). For localization experiment, Pin1−/− MEF cells transfected with either GFP or GFP-Pin1 were arrested in G0 by serum deprivation and contact inhibition, and then cells were fixed with 3.7% formaldehyde at 18 h after serum addition. Subcellular localization of cyclin D1 was determined by staining with cyclin D1 antibodies and DAPI as described above.
Preparation of Nuclear Extracts.
A nuclear fraction was prepared as described (22). Briefly, cells were washed with PBS and resuspended in hypotonic solution (10 mM Hepes, pH 7.8/10 mM KCl/2 mM MgCl2/1 mM DTT/0.1 mM EDTA supplemented with protease inhibitors). After 10 min at 4°C, Nonidet P-40 was added to 1%; the cells were centrifuged for 1 min, and the nuclear pellet was briefly washed with hypotonic buffer and resuspended in SDS sample buffer.
Results
Phenotype of Pin1−/− Mice.
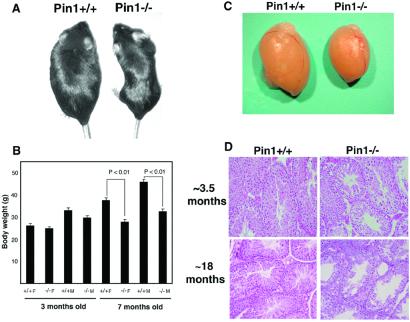
By careful analysis of Pin1−/− mice generated previously (25), we found that they remarkably displayed a range of abnormalities, including decreased body weight, testicular atrophy, retinal degeneration, and mammary gland proliferative impairment. From birth to ≈3 months, Pin1−/− mice were indistinguishable from their wild-type controls, but adult Pin1−/− mice appeared smaller than wild-type controls (Fig. 1 A and B). At the age of ≈7 months, the body weight of Pin1−/− mice was significantly smaller for both male and female animals, with the average weight being only 71% of that of Pin1+/+ mice (Fig. 1 A and B).
Figure 1.
Reduced body weight and testicular atrophy in Pin1−/− mice. (A and B) Reduced body weight. Representative adult wild-type mouse (Left) and Pin1−/− mouse (Right) are shown in A. A comparison of body weight of 10 wild-type and Pin1−/− mutant male and female mice at ≈3.5 and ≈7 months is shown in B. (C and D) Testicular atrophy, as indicated by representative testis from wild-type or Pin1−/− mouse at ≈3.5 months old (C) or by histopathological comparison (D). Testicular sections obtained from ≈3.5- and ≈16-month-old mice were stained with hematoxylin and eosin.
Effects on Seminiferous Tubules.
Although Pin1−/− female and male mice were fertile, the fact that the success rate of homozygous cross breeding was much lower or took much longer than that of wild-type or heterozygous mice led us to suspect that Pin1−/− mice might develop fertility problems. To determine whether Pin1 affects sexual maturation, we first examined the development of the ovary and testis. Ovarian tissues lacking Pin1 appeared to have normal morphology and histology. On the other hand, all autopsied Pin1−/− males had testicular atrophy. By 3–5 months of age, the average weight of six Pin1−/− testes was only 56% of that of wild-type controls (Fig. 1C, data not shown). Moreover, this striking weight difference of testis was not due to the smaller body weight because Pin1−/− males were not significantly smaller than the wild-type controls at this age (Fig. 1B). Histological examination revealed that the seminiferous-tubule degeneration could be detected at the age of ≈3.5 months old (Fig. 1D). By ≈15 months and even more pronounced by 18 months old, the seminiferous tubules in Pin1−/− mice degenerated, with basically no mature sperm in the lumen, whereas age-matched wild-type mice exhibited healthy seminiferous tubules (Fig. 1D). These observations suggest that Pin1 may play a critical maintenance role in adult spermatogenesis, and its absence of Pin1 may result in defects in spermatogonial cell division, meiosis, or sperm maturation.
Effects on Retinal Tissue.
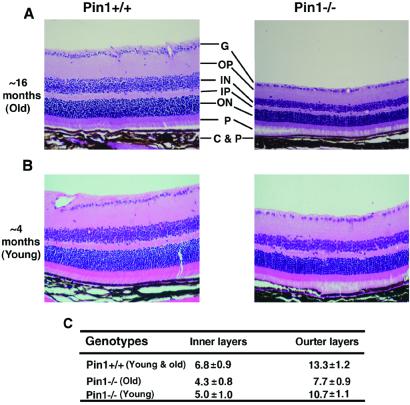
As part of our analysis of the phenotypes of Pin1−/− mice, we found that Pin1−/− mice exhibited retinal degeneration at ≈16 months of age. The thickness of retinal layers in Pin1−/− mice was strikingly reduced (Fig. 2A). To quantify the retinal degeneration, we determined the number of nuclear cell layers in six retinas. Wild-type mice contained an average of ≈7-cell diameter thickness in the inner and 13-cell diameter thickness in the outer nuclear layers, whereas Pin1−/− mice contained only ≈4-cell diameter thickness in the inner and 7-cell diameter thickness in the outer nuclear layers (Fig. 2C). Given the retinal degenerative phenotype in old Pin1−/− mice, we also examined the eyes at 1 day after delivery and at 4–6 months to examine whether younger Pin1−/− mice have a similar abnormality. Although the retinal layers in Pin1−/− mice were similar to those of Pin1+/+ mice at birth (data not shown), about 50% of Pin1−/− eyes displayed mild retinal degradation by 4–6 months of age (Fig. 2B). These results indicate that the Pin1−/− retina undergoes degeneration and that, like the testicular atrophy, this is also age-dependent (Fig. 1 C and D), suggesting that Pin1 may play an important role in survival or cell proliferation in the retina.
Figure 2.
Retinal atrophy in Pin1−/− mice. (A and B) Histopathological examination of retinas. Sections through age-matched wild-type (Left) and Pin1−/− (Right) retinas at ≈16-month-old (A) or ≈4-month-old (B) mice were stained with hematoxylin and eosin. The layers indicated are as follows: G, surface ganglion cell layers; OP, outer plexiform layer; ON, outer nuclear layer; IN, inner nuclear layer; P, photoreceptor cell layer; C&P, choroid and pigment cells. (C) Comparison of the number of the nuclear layers. The numbers of the inner and outer nuclear layers were counted from six age-matched wild-type (Left) and Pin1−/− (Right) retinas, with the average and standard deviations being presented. Because there is no difference between young and old animals, the results are combined.
Effects on Mammary Gland.
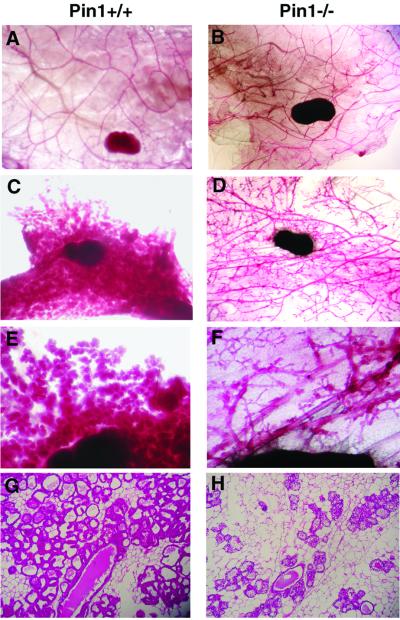
One of the most striking ailments found in cyclin D1 mutant mice was that the mammary gland fails to undergo full lobuloalveolar development during pregnancy. These observations prompted us to examine whether Pin1-deficient mice also display impaired mammary gland associated with pregnancy. As shown in Fig. 3 A and B, the adult epithelial ducts from both Pin1+/+ and Pin1−/− female mice developed normally and formed side branches before pregnancy. As seen in normal mice (23, 24), the Pin1+/+ females underwent the normal massive expansion during pregnancy; the mammary epithelial ducts significantly extended their side branches and built up numerous alveolar structures, which replaced the mammary fat pad and formed lobules (Fig. 3 C and E). In sharp contrast to this normal pregnancy-induced response, in Pin1−/− female mice a severe reduction in mammary epithelial duct development was observed during pregnancy, and the mammary gland failed to undergo the usual massive expansion (Fig. 3 D and F). Consistent with these whole mount results, histological sections revealed that the pregnant wild-type mice displayed a massive proliferation of full-developed mammary epithelial cells (Fig. 3G). In contrast, the mammary gland of Pin1−/− pregnant mice showed a severe impairment in the development and proliferation of mammary epithelial cells (Fig. 3H).
Figure 3.
Impaired mammary epithelial expansion during pregnancy in Pin1−/− mice. The whole mount (A–F) and histological (G and H) appearance of mammary glands derived from 3–4-month-old wild-type (A, C, E, and G) or Pin1−/− (B, D, F, and H) mice of ≈4 months of age. The whole mounts of inguinal mammary glands were prepared, and the epithelial component was stained with carmine red. Histological sections were stained with hematoxylin and eosin. (A) Nulliparous wild-type mouse. (B) Nulliparous mutant mouse. (C, E, and G) Wild-type mouse, 1 day after delivery. (D, F, and H) Mutant mouse, 1 day after delivery.
Effects on Cyclin D1 Levels in Various Tissues.
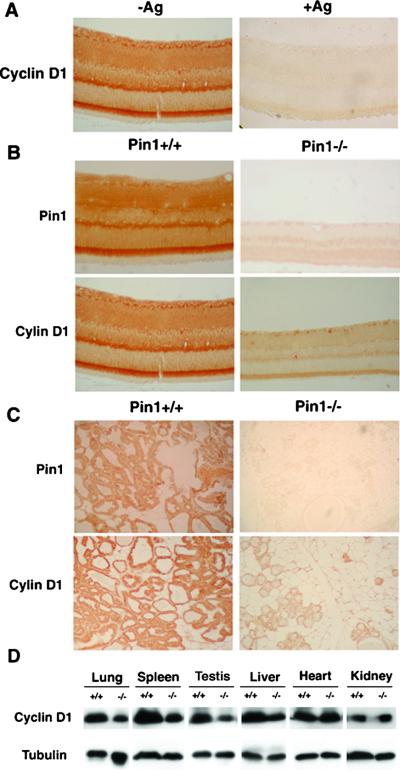
To detect the cyclin D1 protein in retina, we used immunostaining with affinity-purified anti-cyclin D1 antibodies. The cyclin D1 antibodies strongly stained the wild-type retina of 4–16 months of age (Fig. 4A; data not shown), which is consistent with high levels of cyclin D1 mRNA in this tissue as shown by in situ hybridization (23). In contrast, the antigenic peptide-blocked antibodies failed to detect any specific signal on retinal sections (Fig. 4A). In addition, the strong Pin1-staining signals obtained with affinity-purified anti-Pin1 antibodies (13) on wild-type tissues were in contrast to the lack of staining of Pin1−/− tissues (Fig. 4 B and C), confirming that Pin1−/− mice do not express Pin1 protein. Similar strong staining patterns were observed for both Pin1 and cyclin D1 in the retina (Fig. 4 B and C). More importantly, the cyclin D1 protein level in Pin1-deficient mice was strikingly lower than that in the age-matched wild-type mice both in the retina and mammary glands in pregnant mice (Fig. 4 B and C). To examine the effects on cyclin D1 levels in other tissues, several selected tissues from wild-type and Pin1−/− mice were subjected to immunoblotting analysis with anti-cyclin D1 antibodies. Although cyclin D1 levels appeared not to be affected in heart and kidney, cyclin D1 was significantly reduced in testis, spleen, liver, and lung in Pin1-deficient mice (Fig. 4D). These results indicate that mice lacking Pin1 display a significant reduction in cyclin D1 protein level in many tissues, including retina, spleen, and testis, and in the mammary gland in pregnant females, and suggest that Pin1 affects cyclin D1 levels in tissues that contain actively dividing cells.
Figure 4.
Expression of Pin1 and cyclin D1 in various mouse tissues. (A) Specificity of immunostaining with anti-cyclin D1 antibodies. Anti-cyclin D1 antibodies that raised against the C-terminal peptide of cyclin D1 were affinity purified by using the cyclin D1 peptide. A retinal paraffin section was stained with the affinity-purified anti-cyclin D1 antibodies in the absence (Left) or presence (Right) of the cyclin D1 peptide (Ag) that was used as the antigen. (B and C) Sections of retina (B) and mammary gland in pregnant females (C) derived from Pin1+/+ and Pin1−/− mice were stained with affinity-purified anti-Pin1 or anti-cyclin D1 antibodies. (D) Immunoblotting analysis of selected tissues with anti-cyclin D1 antibodies. Several selected tissues from Pin1+/+ and Pin1−/− mice were lysed in SDS-sample buffer and subjected to immunoblotting analysis with anti-cyclin D1 antibodies. The same membranes were also probed with anti-tubulin antibodies as a loading control.
Pin1 Regulates Cyclin D1 Turnover and Subcellular Localization in Addition to Its Transcription.
The above results indicate that Pin1 loss-of-function in the mouse resembles many of the cyclin D1-null mouse phenotypes. Our previous studies have shown that Pin1 enhances the transcription of cyclin D1 through c-Jun/AP-1 and β-catenin/TCF in cancer cells. However, mouse models in which AP-1 or β-catenin/APC function is perturbed do not display strong cyclin D1-related phenotypes (30–32). This led us to speculate that an additional molecular mechanism may contribute to the drastic phenotypes observed in the Pin1-deficient mice.
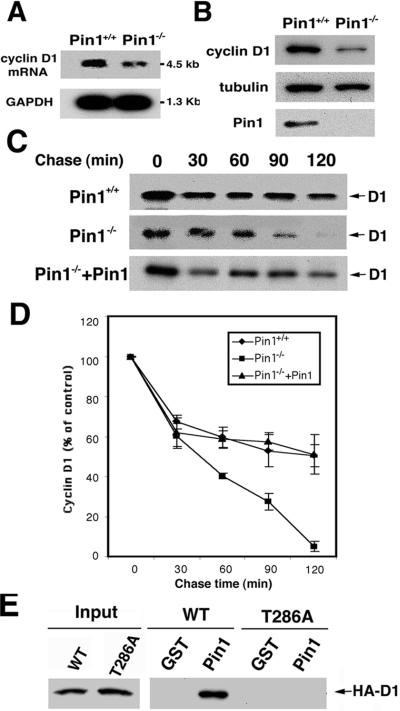
To test this hypothesis, we first examined cyclin D1 mRNA and protein levels in Pin1−/− embryonic fibroblasts (MEFs). The cyclin D1 mRNA level was lower in Pin1−/− MEFs than that in wild-type MEFs (Fig. 5A), consistent with Pin1 being involved in regulation of cyclin D1 transcription (21, 22). However, the cyclin D1 protein level was decreased to a significantly greater extent in Pin1−/− MEFs (Figs. 5B and 6A), suggesting that Pin1 might also affect the stability of cyclin D1 protein. To test this idea, we investigated the stability of exogenously expressed cyclin D1 by using pulse–chase experiments. Although the stability of cyclin D1 in wild-type MEFs was almost the same as those in other cells reported earlier (26, 27), its stability in Pin1−/− MEFs was significantly decreased (Fig. 5 C and D). Moreover, reexpression of Pin1 in Pin1−/− MEFs restored the stability of cyclin D1 (Fig. 5 C and D). These results indicate that Pin1 increases the stability of cyclin D1 protein.
Figure 5.
Pin1 binds cyclin D1 phosphorylated on Thr-286 and stabilizes cyclin D1 protein. (A) Total RNA was isolated from wild-type (Pin1+/+) or Pin1−/− MEFs and then subjected to Northern blot analysis, with glyceraldehyde-3-phosphate dehydrogenase as a loading control. (B) Reduced cyclin D1 protein in Pin1−/− MEFs. The same MEFs as described in A were subjected to immunoblot analysis with anti-cyclin D1, anti-tubulin, and anti-Pin1 antibodies. (C and D) Pin1 stabilizes cyclin D1 protein. Pin1+/+, Pin1−/− MEFs, or Pin1−/− MEFs engineered to express Pin1 were transfected with HA-tagged cyclin D1 and CDK4. After 16 h, cells were metabolically labeled with [35S]Met for 40 min. Cells were washed with complete medium containing excess unlabeled Met and collected at indicated times. Cells were lysed and immunoprecipitated with 12CA5 antibody, followed by autoradiography (C). The radioactivity of immunoprecipitated cyclin D1 was quantified with a Phosphoimager and normalized to the 0-h point. Results shown are means ± SD for three independent experiments (D). (E) Pin1 binds cyclin D1 phosphorylated on Thr-286. Cells were transfected with either wild-type HA-tagged cyclin D1 or HA-tagged cyclin D1 mutant (T286A) for 24 h. Cell extracts were incubated with glutathione agarose beads containing GST or GST-Pin1. After washing, binding proteins were subjected to immunoblotting analysis with 12CA5 mouse monoclonal antibody to HA peptide.
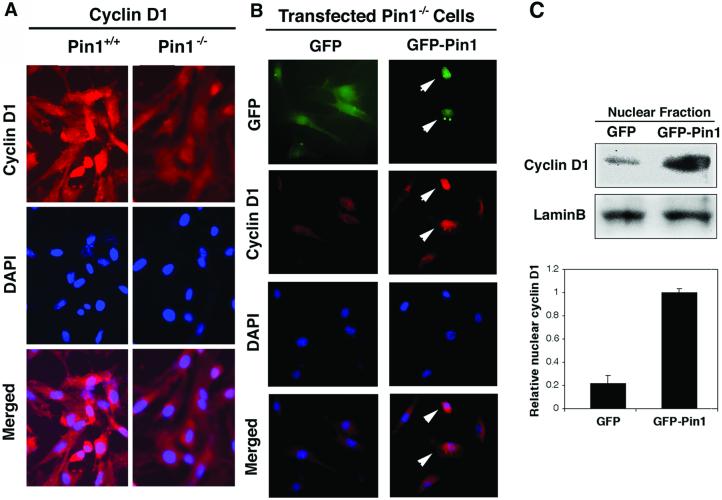
Figure 6.
Pin1 increases cyclin D1 protein level and increases its nuclear localization. (A) Reduced cyclin D1 in Pin1−/− MEFs. Growing wild-type or Pin1−/− MEFs were fixed and stained with anti-cyclin D1 antibodies and DAPI. (B) Pin1 increases the nuclear localization of cyclin D1. Pin1−/− MEF cells transfected with either GFP or GFP-Pin1 were arrested in G0 by serum deprivation and contact inhibition, and then cells were harvested at 18 h after serum addition. Subcellular localization of cyclin D1 was determined by immunostaining with cyclin D1 antibodies and DAPI. Arrows point to GFP-Pin1-expressing cells. (C) The nuclear fraction was isolated from cells transfected as described in B by using hypotonic buffer and then subjected to immunoblotting analysis with anti-cyclin D1 and anti-lamin B antibodies. Relative amounts of nuclear cyclin D1 was semiquantified and normalized with lamin.
Because previous evidence indicates that the phosphorylation on Thr-286 by GSK-3β is a critical factor for cyclin D1 localization and turnover (26, 27), we asked whether Pin1 binds cyclin D1 phosphorylated on Thr-286 and whether Pin1 affects its subcellular localization. GST-pull down analyses revealed that Pin1 directly bound to cyclin D1 but not to its T286A mutant (Fig. 5E), indicating that Pin1 binds to cyclin D1 likely via phosphorylated Thr-286, which is succeeded by a proline and resided in a consensus Pin1-binding sequence (7, 9). Furthermore, in exponentially growing Pin1−/− MEFs, the level of endogenous cyclin D1 was not only significantly reduced compared with that in wild-type MEFs, but cyclin D1 was also primarily localized in cytoplasm of Pin1−/− MEFs, whereas it was largely nuclear in wild-type MEFs (Fig. 6A). In addition, the cyclin D1T286A mutant remained in the nucleus even in Pin1−/− MEFs (data not shown) (28). These results suggest that Pin1 might affect localization of cyclin D1. To confirm this, we looked at the cyclin D1 localization during S phase, when cyclin D1 is known to be exported to cytoplasm (26–28). Pin1−/− MEFs transfected with either GFP or GFP-Pin1 were arrested in G0 by serum starvation and allowed to enter the cell cycle by the addition of serum. At 18 h after the serum addition, when cells were in S phase, as monitored by BrdU incorporation (data not shown), cells were fixed and subjected to immunostaining with cyclin D1 antibodies (Fig. 6 A and B), or the nuclear fraction was isolated and subjected to immunoblotting analysis with cyclin D1 antibodies (Fig. 6C). As shown (26, 27), cyclin D1 was primarily localized in the cytoplasm in nontransfected or GFP-transfected Pin1−/− MEFs (Fig. 6 A and B). In contrast, cyclin D1 was mainly localized in the nucleus in over 90% GFP-Pin1-transfected cells (Fig. 6B, data not shown). Furthermore, levels of cyclin D1 in the nucleus were significantly higher in GFP-Pin1-transfected cells than control GFP-transfected Pin1−/− MEFs (Fig. 6C). Together, these results indicate that Pin1 regulates the turnover and subcellular localization of cyclin D1.
Discussion
We report here that Pin1−/− mice display many severe phenotypes, including decreased body weight, retinal degeneration, mammary gland retardation, and testicular atrophy. Most of these phenotypes are remarkably similar to those of cyclin D1-deficient mouse phenotypes. Of several phenotypes observed in Pin1−/− mice, the alterations in retina and mammary gland seemed to be most drastic. We found that Pin1−/− mice show dramatic impairments in cell survival or proliferation in the retina, especially at old age. Moreover, it is very clear and striking that in pregnant Pin1−/− female, mammary epithelia cells fail to undergo massive proliferation in the development of alveolar structures and ductal side branching. Our study demonstrated that Pin1 is highly expressed in retina and mammary gland compared with other tissues (data not shown), and the depletion of Pin1 causes a dramatic retinal atrophy and mammary gland impairment. Furthermore, disruption of Pin1 affects the level of cyclin D1 in the tissues that contain actively proliferative cells, such as spleen, retina, and mammary gland in pregnant females. Moreover, cyclin D1−/− mice also display a very similar phenotype in the retina and mammary gland. These results suggest that Pin1 could play an essential role in maintaining survival or proliferation of cells through regulating cyclin D1 expression. One notable difference between Pin1- and cyclin D1-deficient mice is that cyclin D1 mutant mice display a dramatic reduction in retinal layers at an early stage development because of proliferative failure (23, 24). However, in Pin1−/− mice, the thickness of the retina decreases slowly with age and becomes pronounced by over 1 year of age. These results indicate that the retinal hypoplasia in Pin1−/− mice is not because of proliferative failure during the embryonic development but rather likely because of a failure of cells to maintain cell survival and/or proliferation after birth because there is no obvious difference in the retina between Pin1−/− and wild-type mice at birth. This might be due to the fact that cyclin D1 level in Pin1−/− mice is lower, but not completely absent as in the case of cyclin D1 knockout mice. Thus, this level of cyclin D1 may become a limiting factor with aging and thereby affect survival or cell proliferation.
Our results strongly suggest that Pin1 regulates cyclin D1 not only through transcriptional regulation but also via a posttranslational mechanism. Previous studies by using human cancer tissues and cancer cell lines have demonstrated that Pin1 enhances cyclin D1 promoter activity through AP-1 and TCF sites as a result of activation and/or stabilization of phosphorylated c-Jun and β-catenin, respectively (21, 22). Here, we demonstrate that Pin1 also regulates the stability and subcellular localization of cyclin D1 itself. Phosphorylation of cyclin D1 on the Thr-286 site regulates turnover and localization of cyclin D1 by enhancing its nuclear export, which leads to degradation of cyclin D1 in the cytoplasm (26, 27). We have shown that Pin1 can bind to the phosphorylated Thr-286–Pro motif in cyclin D1 and stabilize cyclin D1, presumably by preventing nuclear export of cyclin D1 and proteolysis in cytoplasm. Interestingly, Pin1 regulates the turnover and localization of β-catenin in a similar manner (22). Thus, Pin1 positively regulates cyclin D1 function at both transcriptional and posttranslational levels (Fig. 7), and this may explain why Pin1 loss-of-function in the mouse resembles the cyclin D1-null phenotypes. Because cyclin D1 plays a critical role in oncogenesis (23, 24, 33–38), our current results further support a role of Pin1 in breast cancer (21, 22).
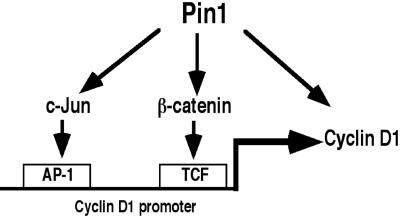
Figure 7.
A model for regulation of cyclin D1 function by Pin1. In addition to that Pin1 enhances cyclin D1 promoter activity through AP-1 and TCF sites as a result of activation and/or stabilization of phosphorylated c-Jun and β-catenin, respectively, our current results demonstrate that Pin1 directly regulates the stability and subcellular localization of cyclin D1 itself.
Pin1 has been demonstrated to have many functions; however, we have demonstrated here that Pin1−/− mice display rather restricted phenotypes that are related to those observed in cyclin D1−/− mice. The lack of more severe phenotypes can be explained by the idea that there are other Pin1-like genes that can compensate for the functions of Pin1. This idea is supported by our recent isolation of a second Pin1-like gene from Drosophila (A.R. and K.P.L., unpublished data). This finding strongly suggests the possibility of other existing Pin1-like genes in the higher species such as mouse and human. The strikingly testicular atrophy, which is not found in cyclin D1-null mice but seen in cyclin D2 knockout mice (39) or the cyclin D-dependent kinase inhibitors (40, 41), suggests that Pin1 may target other proteins, perhaps other D-type cyclins. Further studies are needed to illustrate the role of Pin1 in spermatogenesis.
In summary, we report that mice lacking Pin1 do display a range of severe cell-proliferative abnormalities, many of which resemble those in cyclin D1-deficient mice. Furthermore, disruption of Pin1 also causes a striking reduction in cyclin D1 level in many tissues, including the retina and mammary epithelial cells of pregnant females, the two most affected tissues. Using Pin1−/− MEF cells, we demonstrate that Pin1 binds and stabilizes cyclin D1 and increases its nuclear localization in addition to affecting cyclin D1 transcription. These results provide insight for an essential role of Pin1 in maintaining cell proliferation and regulating cyclin D1.
Acknowledgments
We are grateful to C. Sherr for constructive discussions, to W. Jiang for providing anti-cyclin D1 antibodies, to I. Kosugi for technical instruction, and to X. Zhou, O. Kops, S. Kishi, M. Nakamura, and G. Wulf in the Lu laboratory for their important contributions. Y.C.L., A.R., H.-K. H., and P.J.L. are fellows of the National Sciences and Engineering Research Council of Canada, the Japan Society for the Promotion of Science, the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation, and the Leukemia and Lymphoma Society, respectively. T.H. is a Frank and Else Schilling American Cancer Society Research Professor. K.P.L. is a Pew Scholar and a Leukemia and Lymphoma Society Scholar. This study was supported by National Institutes of Health Grants CA80100 (to T.H.) and GM56230, AG17870, and GM58556 (to K.P.L.).
Abbreviations
- GST
glutathione S-transferase
- HA
hemagglutinin
- DAPI
4′,6-iamidino-2-phenylindole
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nigg E A. BioEssays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 2.Whitmarsh A J, Davis R J. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 3.Karin M, Liu Z, Zandi E. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 4.Hunter T. Cell. 1998;92:141–143. doi: 10.1016/s0092-8674(00)80906-x. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X Z, Lu P J, Wulf G, Lu K P. Cell Mol Life Sci. 1999;56:788–806. doi: 10.1007/s000180050026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu K P, Hanes S D, Hunter T. Nature (London) 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 7.Yaffe M B, Schutkowski M, Shen M, Zhou X Z, Stukenberg P T, Rahfeld J, Xu J, Kuang J, Kirschner M W, Fischer G, et al. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 8.Ranganathan R, Lu K P, Hunter T, Noel J P. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 9.Lu P J, Zhou X Z, Shen M, Lu K P. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 10.Verdecia M A, Bowman M E, Lu K P, Hunter T, Noel J P. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 11.Shen M, Stukenberg P T, Kirschner M W, Lu K P. Genes Dev. 1998;12:706–720. doi: 10.1101/gad.12.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crenshaw D G, Yang J, Means A R, Kornbluth S. EMBO J. 1998;17:1315–1327. doi: 10.1093/emboj/17.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu P J, Wulf G, Zhou X Z, Davies P, Lu K P. Nature (London) 1999;399:784–788. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- 14.Patra D, Wang S X, Kumagai A, Dunphy W G. J Biol Chem. 1999;274:36839–36842. doi: 10.1074/jbc.274.52.36839. [DOI] [PubMed] [Google Scholar]
- 15.Albert A, Lavoie S, Vincent M. J Cell Sci. 1999;112:2493–2500. doi: 10.1242/jcs.112.15.2493. [DOI] [PubMed] [Google Scholar]
- 16.Morris D P, Phatnani H P, Greenleaf A L. J Biol Chem. 1999;274:31583–31587. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- 17.Wu X, Wilcox C B, Devasahayam G, Hackett R L, Arevalo-Rodriguez M, Cardenas M E, Heitman J, Hanes S D. EMBO J. 2000;19:3727–3738. doi: 10.1093/emboj/19.14.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerez L, Mohrmann K, va Raak M, Jongeneelen M, Zhou X Z, Lu K P, van der Sluijs P. Mol Biol Cell. 2000;11:2201–2211. doi: 10.1091/mbc.11.7.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stukenberg P T, Kirschner M W. Mol Cell. 2001;7:1071–1083. doi: 10.1016/s1097-2765(01)00245-3. [DOI] [PubMed] [Google Scholar]
- 20.Hsu T, McRackan D, Vincent T S, Gert De Couet H. Nat Cell Biol. 2001;3:538–543. doi: 10.1038/35078508. [DOI] [PubMed] [Google Scholar]
- 21.Wulf G M, Ryo A, Wulf G G, Lee S W, Niu T, Lu K P. EMBO J. 2001;20:3459–3472. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryo A, Nakamura N, Wulf G, Liou Y C, Lu K P. Nat Cell Biol. 2001;3:793–801. doi: 10.1038/ncb0901-793. [DOI] [PubMed] [Google Scholar]
- 23.Sicinski P, Donaher J L, Parker S B, Li T, Fazeli A, Gardner H, Haslam S Z, Bronson R T, Elledge S J, Weinberg R A. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 24.Fantl V, Stamp G, Andrews A, Rosewell I, Dickson C. Genes Dev. 1995;9:2364–2372. doi: 10.1101/gad.9.19.2364. [DOI] [PubMed] [Google Scholar]
- 25.Fujimori F, Takahashi K, Uchida C, Uchida T. Biochem Biophys Res Commun. 1999;265:658–663. doi: 10.1006/bbrc.1999.1736. [DOI] [PubMed] [Google Scholar]
- 26.Diehl J A, Zindy F, Sherr C J. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 27.Diehl J A, Cheng M, Roussel M F, Sherr C J. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alt J R, Cleveland J L, Hannink M, Diehl J A. Genes Dev. 2000;14:3102–3114. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, Counterman L J, Haslam S Z. Endocrinology. 1990;127:2183–2189. doi: 10.1210/endo-127-5-2183. [DOI] [PubMed] [Google Scholar]
- 30.Jochum W, Passegue E, Wagner E F. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- 31.Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Development (Cambridge, UK) 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 32.Oshima M, Dinchuk J E, Kargman S L, Oshima H, Hancock B, Kwong E, Trzaskos J M, Evans J F, Taketo M M. Cell. 1996;87:803–809. doi: 10.1016/s0092-8674(00)81988-1. [DOI] [PubMed] [Google Scholar]
- 33.Gillett C, Fantl V, Smith R, Fisher C, Bartek J, Dickson C, Barnes D, Peters G. Cancer Res. 1994;54:1812–1817. [PubMed] [Google Scholar]
- 34.Bartkova J, Lukas J, Muller H, Lutzhoft D, Strauss M, Bartek J. Int J Cancer. 1994;57:353–361. doi: 10.1002/ijc.2910570311. [DOI] [PubMed] [Google Scholar]
- 35.Hinds P W, Dowdy S F, Eaton E N, Arnold A, Weinberg R A. Proc Natl Acad Sci USA. 1994;91:709–713. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robles A I, Rodriguez-Puebla M L, Glick A B, Trempus C, Hansen L, Sicinski P, Tennant R W, Weinberg R A, Yuspa S H, Conti C J. Genes Dev. 1998;12:2469–2474. doi: 10.1101/gad.12.16.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Puebla M L, Robles A I, Conti C J. Mol Carcinog. 1999;24:1–6. [PubMed] [Google Scholar]
- 38.Yu Q, Geng Y, Sicinski P. Nature (London) 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 39.Sicinski P, Donaher J L, Geng Y, Parker S B, Gardner H, Park M Y, Robker R L, Richards J S, McGinnis L K, Biggers J D, et al. Nature (London) 1996;384:470–474. doi: 10.1038/384470a0. [DOI] [PubMed] [Google Scholar]
- 40.Zindy F, den Besten W, Chen B, Rehg J E, Latres E, Barbacid M, Pollard J W, Sherr C J, Cohen P E, Roussel M F. Mol Cell Biol. 2001;21:3244–3255. doi: 10.1128/MCB.21.9.3244-3255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zindy F, van Deursen J, Grosveld G, Sherr C J, Roussel M F. Mol Cell Biol. 2000;20:372–378. doi: 10.1128/mcb.20.1.372-378.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]