Abstract
Pentamer formation by Vp1, the major capsid protein of simian virus 40, requires an interdigitation of structural elements from the Vp1 monomers [Liddington, R. C., Yan, Y., Moulai, J., Sahli, R., Benjamin, T. L. & Harrison, S. C. (1991) Nature (London) 354, 278–284]. Our analyses reveal that disulfide-linked Vp1 homooligomers are present in the simian virus 40-infected cytoplasm and that they are derived from a 41-kDa monomeric intermediate containing an intrachain disulfide bond(s). The 41-kDa species, emerging within 5 min of pulse labeling with [35S]methionine, is converted into a 45-kDa, disulfide-free Vp1 monomer and disulfide-bonded dimers through pentamers. The covalent oligomer formation is blocked in the presence of a sulfhydryl-modifying reagent. We propose that there are two stages in this Vp1 disulfide bonding. First, the newly synthesized Vp1 monomers acquire intrachain bonds as they fold and begin to interact. Next, these bonds are replaced with intermolecular bonds as the monomers assemble into pentamers. This sequential appearance of transitory disulfide bonds is consistent with a role for sulfhydryl–disulfide redox reactions in the coordinate folding of Vp1 chains into pentamers. The cytoplasmic Vp1 does not colocalize with marker proteins of the endoplasmic reticulum. This paper demonstrates in vivo disulfide formations and exchanges coupled to the folding and oligomerization of a mammalian protein in the cytoplasm, outside the secretory pathway. Such disulfide dynamics may be a general phenomenon for other cysteine-bearing mammalian proteins that fold in the cytoplasm.
How proteins fold into functional, three-dimensional structures has been under intense study (1), and the folding pathways for a number of eukaryotic proteins have been characterized in vitro (2, 3) or in vivo (4–9). In the secretory pathway, protein folding is coupled to the formation and reshuffling of disulfide bonds. These redox conversions, leading to native, disulfide-bonded proteins, are catalyzed by prokaryotic Dsb proteins in the periplasm (10–12) and eukaryotic protein disulfide isomerase (PDI) in the endoplasmic reticulum (ER) (13–16). Proteins that fold and assemble in the reducing environment of the cytoplasm generally do not harbor native disulfides, owing to the activities of thioredoxins and glutaredoxins. Transitory disulfide bonding, though, is required for the folding of bacteriophage P22 tailspike protein in the cytoplasm (17, 18). Whether disulfide bond-coupled folding pathways exist for nonsecretory proteins in the mammalian cytoplasm is not known.
The structure of simian virus 40 (SV40), known at the atomic resolution, is determined by the major capsid protein Vp1 (19). Seventy-two pentamers of Vp1 form the outer shell of SV40, with each monomer making contact with its four intrapentamer neighbors via interdigitating secondary structural elements. The Vp1 pentamer is expected to form in the cytoplasm of SV40-infected cells during or soon after the monomers' synthesis (20, 21). There are seven cysteine residues in one Vp1 chain. No intrapentamer disulfide bridges, either between or within the monomers, are observed in the mature particle (22). Certain cysteine residues do lie in close proximity of one another, such as the Cys-49–Cys-87 and Cys-87–Cys-207 pairs within one monomer and the Cys-49–Cys-207 pair between two monomers within a pentamer (19). Each cysteine pair conceivably can become juxtaposed during the folding process and form a transient disulfide bond.
In this study, we show that in the virus-infected cytoplasm, the newly synthesized Vp1 chain is an intramolecularly disulfide-bonded monomer and is a precursor for intermolecularly disulfide-bonded Vp1 oligomers ranging from dimers to pentamers. We propose a model in which Vp1 achieves folding and oligomerization through transitory, disulfide-linked intermediates. Such sulfhydryl–disulfide redox dynamics conceivably may exist in the folding pathways of other nonsecretory mammalian proteins.
Materials and Methods
Cell Culture, Antibodies, Virus Infection, and Immunofluorescence.
The TC7 subline of African green monkey kidney cells (20) and rabbit preimmune and anti-Vp1 sera (23) have been described. Monoclonal anti-Vp1 antibody α597 is derived from the culture fluid of clone 597 and recognizes Vp1 in both immunoprecipitation and immunoblotting. Monoclonal mouse antibodies against PDI and calnexin were obtained from Affinity BioReagents (Golden, CO). SV40 infection (20) and immunofluorescence analysis (24) were performed as described.
Metabolic Radiolabeling, Dithiobis (Succinimidyl Propionate) (DTSP) Cross-Linking, and Subcellular Fractionation.
Cell labeling with [35S]methionine was performed essentially as described (21), with the following modifications. When indicated, the chase medium for pulse–chase contained an additional 2.5 mM N-ethylmaleimide (NEM), a reagent for sulfhydryl alkylation. Continuous labeling was for 1 h, and the cells were supplemented with 60 μM unlabeled methionine for the last 30 min of the hour. In situ cross-linking with DTSP (Pierce) was performed according to Lomant and Fairbanks (25). In brief, cells were washed and incubated for 10 min at room temperature with Dulbecco's PBS containing 1 mM CaCl2 and 1 mM MgCl2 (DPBSCM) and then incubated with 0.5 mM DTSP in DPBSCM at 4°C for 7 min. The reaction was terminated by adding lysine to 50 mM. Control non-DTSP-treated cells received 50 mM lysine together with DTSP in DPBSCM. The extent of protein cross-linking was equivalent to that reported by others (26, 27) (data not shown). Subcellular fractionation into an Nonidet P-40-soluble cytosolic fraction and a TWEEN 20 and sodium deoxycholate double-detergent-soluble cytoskeletal fraction was performed as described (20), except for the following changes in the lysis and homogenization buffers: Tris⋅Cl was substituted with Pipes (pH 7.2), 1 mM PMSF was added, and, when indicated, 1 mM NEM was added.
Immunoprecipitation and Western Blot Analysis.
Anti-Vp1 immunoprecipitation and elution under reducing conditions were performed as described (21), with the following modifications. First, protein samples were incubated at 25°C for 10 min in 25 mM Hepes (pH 6.8) and 2 mM NEM, clarified by centrifuging at 120,000 × g for 20 min at 4°C, diluted 5-fold with IP buffer (20 mM Tris⋅Cl, pH 7.5/150 mM NaCl/10 mM EDTA/0.5% Nonidet P-40/0.5% sodium deoxycholate/1 mM PMSF) with or without additional 0.2% SDS, before antibodies were added. Second, each reaction contained a 0.5–1 × 106 cell-equivalent sample in a total of 0.5 ml, and 7.5 μl of α597 monoclonal anti-Vp1 antibody, followed by 20 μl of rabbit anti-mouse-Ig serum, was used in place of anti-Vp1 serum for some of the reactions. Third, some of the immunoprecipitates were eluted under nonreducing conditions by incubating at 50°C for 30 min in 120 mM Hepes, pH 6.8/5% SDS/20% glycerol/0.01% bromophenol blue/4 mM NEM. Anti-Vp1 Western blot analysis was performed as described (20).
Sedimentation Analysis and SDS/PAGE.
For nondenaturing sedimentation, 100 μl of the cytosolic fraction was sedimented through a 4.2-ml, 5–20% continuous sucrose gradient on top of a 0.3-ml, 50% sucrose cushion, all in IP buffer, at 36,000 rpm at 20°C for 110 min in an SW51 rotor. Denaturing sedimentation was performed similarly, except the sucrose solutions contained additional 0.1% SDS and the cytosolic sample was incubated for 30 min at 50°C in 50 mM Hepes, pH 6.8/0.5% SDS/1% Nonidet P-40/2 mM NEM before loading. SDS/PAGE was performed by the method of Laemmli (28). For two-dimensional (2D) SDS/PAGE, proteins eluted under nonreducing conditions first were resolved on an 8%/12.5% step-gradient gel consisting of an 8% polyacrylamide layer above a 12.5% polyacrylamide layer. Then, individual lanes were cut from the gel, equilibrated with reducing solution (120 mM Tris⋅Cl, pH 6.8/1% SDS/50 mM DTT) at 37°C for 30 min and embedded on top of new 12.5% polyacrylamide gels in reducing solution that was warmed to 50°C and supplemented with 2 mM EDTA/0.001% bromophenol blue/5% glycerol/1% agarose. The mixture was left at room temperature for 60 min before electrophoresis.
Results
Vp1 Forms Disulfide-Bonded Oligomers in the Infected Cytoplasm.
We examined the oligomerization state of Vp1 in SV40-infected cytoplasm, in which the Vp1 concentration, estimated to be about 10−5 M at 48–50 h postinfection (20), is well above the 10−7 M monomeric concentration observed for Vp1 disulfide bond formation in vitro (29). At a multiplicity of infection of 5, less than 10% of the total physical particles is internalized in the cells (A.N., unpublished results). These input virions would amount to an intracellular Vp1 concentration of below 10−8 M, a negligible amount compared with that synthesized after infection.
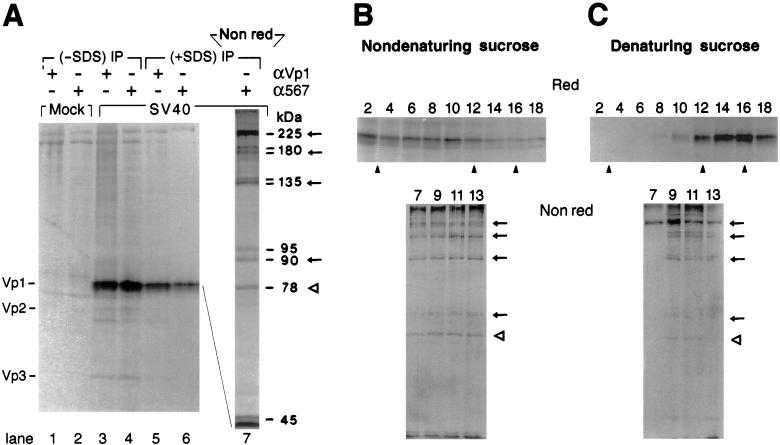
The infected cells were labeled for 1 h with [35S]methionine, and a cytosolic fraction soluble in nonionic detergent was prepared, treated with the sulfhydryl modifying reagent NEM, and analyzed by immunoprecipitation and SDS/PAGE. Both polyclonal anti-Vp1 serum (Fig. 1A, lanes 3 and 5) and monoclonal anti-Vp1 antibody α597 (lanes 4 and 6) precipitated a 45-kDa Vp1 under reducing conditions. Adding 0.2% SDS in the immune reaction only moderately decreased the amount of precipitated Vp1 (lanes 5 and 6, compared with lanes 3 and 4) but abolished the coprecipitation of minor capsid proteins Vp2 (37 kDa) and Vp3 (27 kDa), as expected of the noncovalent nature of Vp1's association with Vp2 and Vp3. Under nonreducing conditions, the α597 immunoprecipitate resolved into species with apparent molecular weights (Mr) of 225,000, 180,000, 135,000, 90,000/95,000, and 78,000 besides the 45-kDa monomer (lane 7). Except for the 78-kDa band, whose identity is not known at present (see also the 78-/80-kDa species described below), all other species have apparent masses that correspond to oligomers of Vp1. The 180-, 135-, and 90-/95-kDa bands migrated as doublets, and the 5,000–10,000 difference in Mr in each case could have arisen from posttranslational modifications (see references within refs. 30 and 31) or from the presence of additional intramolecular disulfide linkages. The possibility that the Vp1 oligomers are formed during immunoprecipitation after cell lysis was ruled out by a direct anti-Vp1 Western blot of the lysate. Under nonreducing conditions, distinct Vp1-containing bands are present (Fig. 2A, lane 1) whose Mr values correspond to the oligomers noted above.
Figure 1.
Discrete, disulfide-bonded Vp1 oligomers exist in the cytoplasm. (A) SV40-infected cells at 48–50 h postinfection (lanes 3–6) or mock-infected cells (lanes 1 and 2) were labeled for 1 h and fractionated as described in Materials and Methods. The cytosolic fraction from 2.5 × 105 cells was immunoprecipitated with anti-Vp1 serum (αVp1, lanes 1, 3, and 5) or monoclonal anti-Vp1 antibody α597 (lanes 2, 4, and 6) in the absence (lanes 1–4) or presence (lanes 5–7) of SDS, eluted under reducing (lanes 1–6) or nonreducing (lane 7) conditions, and analyzed by 12.5% (lanes 1–6) or 7% (lane 7) SDS/PAGE and autoradiography. The positions for Vp1, Vp2, and Vp3 (Left) and the apparent molecular masses of labeled bands in lane 7 (Right) are indicated. Arrows mark the positions for dimeric, trimeric, tetrameric, and pentameric Vp1s, and an empty arrowhead marks the position for the 78-kDa species. (B and C) The same cytosolic fraction as in A, prepared from 2.5 × 106 cells, was sedimented through a 5–20% sucrose gradient under nondenaturing (B) or denaturing (C) conditions. Eighteen fractions were collected from the bottom of the gradient, immunoprecipitated with anti-Vp1 serum in the absence of SDS (even fractions 2–18) or with α597 in the presence of SDS (odd fractions 7–13), and analyzed by reducing (Red) or nonreducing (Nonred) 7% SDS/PAGE, respectively, followed by autoradiography. The nonreducing gels represent a three-times-longer autoradiographic exposure relative to the reducing gels. Solid arrowheads point to positions for sedimentation markers determined in parallel gradients: (from left to right) 13S, β-galactosidase; 7S, goat IgG; and 5S, ovalbumin.
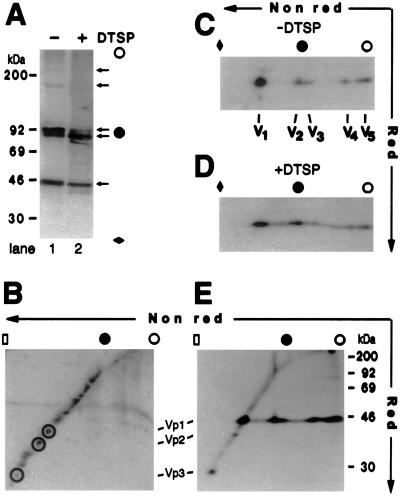
Figure 2.
Cytoplasmic Vp1 oligomers are homooligomers. (A) Anti-Vp1 Western blot of infected cytosolic fraction from 5 × 104 non-DTSP-treated (lane 1) or DTSP-treated (lane 2) cells. The cytosolic sample was incubated in 120 mM Hepes, pH 6.8/3% SDS/10% glycerol/4 mM NEM at 50°C for 30 min, clarified by centrifugation, and resolved by 8%/12.5% step-gradient SDS/PAGE before being electrotransferred to membrane and blotted as described (21). (B) Coomassie blue staining of cytosolic proteins from 2.5 × 105 non-DTSP-treated infected cells after 2D SDS/PAGE. Spots on the diagonal line corresponding to the presumed locations of Vp1, Vp2, and Vp3 are circled. (C and D) Anti-Vp1 Western blots of the same cytosolic fraction as in A, lane 1 (C) and lane 2 (D), after 2D SDS/PAGE. The positions for monomeric (V1), dimeric (V2), trimeric (V3), tetrameric (V4), and pentameric (V5) Vp1s are marked beneath respective spots. (E) The same cytosolic fraction as in Fig. 1A, lane 3, prepared from 8 × 105 cells, was immunoprecipitated with anti-Vp1 serum and analyzed by 2D SDS/PAGE and autoradiography. For A– E, an open circle or an open rectangle indicates the origin or the end, respectively, of the resolving gel; a solid diamond, a point two-thirds of the way down; and a solid circle, the boundary for the 8%/12.5% step. The positions for molecular mass standards of 30–200 kDa are indicated in A and E. Note that Vp1 dimers and trimers migrated at the boundary of the step gradient in A and C– E.
The size range of oligomerized Vp1 in the cytosolic fraction also was analyzed by sedimentation in sucrose gradients. In the absence of SDS, Vp1 was distributed throughout the gradient (Fig. 1B, fractions 2–18). When the anti-Vp1 immunocomplexes in the fractions expected for the oligomers, fractions 7–13, were resolved by nonreducing SDS/PAGE, the presence of the Vp1 oligomer species and of the 78-kDa band (Fig. 1A, lane 7) was confirmed (Fig. 1B). The broad sedimentation pattern is consistent with noncovalent association of monomeric and oligomeric Vp1s with many other cytoplasmic proteins (also see below). In contrast, when proteins in the cytosolic fraction were denatured with SDS to limit the noncovalent associations and then sedimented through a denaturing gradient, Vp1 was observed in fractions 10–18 (Fig. 1C), consistent with Vp1 existing as a monomer and covalent oligomers up to pentamer. Furthermore, the various Vp1 oligomers—225, 180, 135, and 90 kDa—are distributed in fractions 7–13 in a nonuniform manner, which roughly correlates with the respective sizes of the species (Fig. 1C). The combined results show that at the time of cell lysis, some of the cytoplasmic Vp1 molecules, including those synthesized during the labeling period, exist as intermolecularly disulfide-bonded dimers through pentamers and form native associations with cellular proteins.
Cytoplasmic Disulfide-Linked Vp1 Oligomers Are Homooligomers.
Whether the Vp1 oligomers detected above are disulfide-linked homooligomers was tested by a two-dimensional (2D) SDS/PAGE analysis of the cytosolic sample. After resolution in the nonreducing first dimension, non-disulfide-bonded proteins again would migrate according to their respective Mr values in the reducing second dimension, forming a diagonal line, whereas disulfide-linked protein complexes would separate into one or more spots of component proteins below the diagonal. The total infected cytosolic proteins detected by Coomassie blue staining resolved on the diagonal line (Fig. 2B), indicating that the majority of the proteins do not contain disulfide bonds. A distinctly different pattern is exhibited by Vp1 detected by Western blot (Fig. 2C). Whereas a major 45-kDa monomeric Vp1 (spot V1) falls on the diagonal line, the four oligomeric Vp1s (V2 through V5) were converted into 45-kDa spots below the diagonal after reduction in the second dimension. Quantitation of 125I radioactivity residing in excised spots of Fig. 2C shows that species V1, V2 plus V3, and V4 plus V5 constitute 42%, 26%, and 32% of the combined radiolabel, respectively. The Vp1 oligomers in [35S]methionine-labeled cytosolic fraction (Fig. 1A, lane 7) similarly resolved below the diagonal (Fig. 2E), whereas coprecipitated proteins, including Vp2 and Vp3 (Fig. 1A, lane 3), lie on the diagonal line (Fig. 2E). No off-diagonal spots of cytoplasmic proteins are detected above or below the reduced monomeric Vp1 spots, except for a faint band with a Mr of 80,000 near the right edge of the gel (Fig. 2E). The latter species is presumably a tightly associated protein similar or identical to the 78-kDa species observed in Fig. 1 (A– C), and its identity is not investigated further. These results indicate that a substantial fraction of cytoplasmic Vp1 exists as homooligomers containing intermolecular disulfide bonds.
To capture a full set of Vp1 oligomers in the cytoplasm, cells were treated with DTSP, a membrane-permeable, thiol-cleavable cross-linker (25), before lysis. This in situ cross-linking is expected to trap any proteins located in the immediate vicinity of the Vp1 monomers or disulfide-linked oligomers. Consistent with a heterogeneity of Vp1-containing complexes (Fig. 1B) and a predominantly disulfide-bonded nature of Vp1 homooligomers in the cytoplasm, the apparent effect of the cross-linking is a smearing of the four oligomeric Vp1 species toward one another (Fig. 2A, lane 2 compared with lane 1; Fig. 2 D compared with C). All results indicate that discrete Vp1 monomer and oligomer species are present in the infected cytoplasm and that a fraction of Vp1 monomers and oligomers is close to other cellular and viral (Vp2 and Vp3) cytoplasmic proteins. We conclude that the observed Vp1 species in the cytoplasm are genuine Vp1 monomers and homooligomers containing disulfide bonds.
Newly Synthesized Vp1 Monomers Are Intramolecularly Disulfide-Linked and Are Precursors for Vp1 Oligomers.
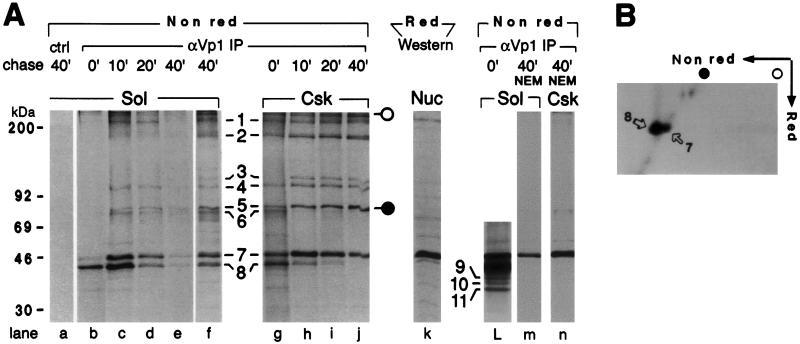
To demonstrate that the oligomers are derived kinetically from newly synthesized Vp1 and represent cytoplasmic assembly intermediates, a pulse–chase experiment was performed. The cytosolic and the double-detergent-soluble cytoskeletal fractions prepared from infected cells after 5-min [35S]methionine labeling and various times of chase were analyzed for Vp1 under nonreducing conditions. Unexpectedly, the newly synthesized Vp1, emerging during the pulse, appeared predominantly as a 41-kDa species under nonreducing conditions (band 8 in Fig. 3A, lanes b and g). The 41-kDa monomer was converted during the chase periods, in both cytosolic and cytoskeletal fractions, into the 45-kDa species (band 7 in lanes b–j), which is identical to the Vp1 in the nuclear fraction resolved by reducing SDS/PAGE (lane k). The 41- to 45-kDa conversion stalled by 20 min of chase in the cytosolic fraction (Fig. 3A, lanes c–e) but reached completion by 40 min in the cytoskeletal fraction (Fig. 3A, lanes h–j). A longer autoradiographic exposure of the pulse-labeled cytosolic fraction (lane b) shows three additional, monomer-like Vp1 species of 38, 36, and 34 kDa (bands 9, 10, and 11, respectively, in lane L) that were present at low abundance.
Figure 3.
Kinetics of formation of disulfide-bonded Vp1 monomer and oligomers. (A) The Vp1 homooligomers are formed from intramolecularly disulfide-linked monomer in the cytoplasm. Infected cells were pulse-labeled for 5 min and then either harvested immediately (lanes b, g, and L) or chased in the absence of NEM for 10 min (lanes c and h), 20 min (lanes d and i), or 40 min (lanes a, e, f, and j) or chased in NEM-containing medium for 40 min (lanes m and n). The cytosolic fraction from 5 × 105 cells (Sol, lanes a–f and L and m) or the cytoskeletal fraction from 1 × 106 cells (Csk, lanes g–j and n) was immunoprecipitated in the presence of SDS with anti-Vp1 serum (αVp1 IP, lanes b–j and L–n) or with preimmune serum (ctrl, lane a), eluted under nonreducing conditions (Nonred), and analyzed by 8%/12.5% step-gradient SDS/PAGE and autoradiography. Lanes f and L are longer exposures of lanes e and b, respectively. In lane k, 2 × 104 infected, unlabeled nuclei (Nuc) were lysed with SDS and a reducing agent (Red) and analyzed by anti-Vp1 Western blot. The positions for Vp1 pentamer (band 1), tetramer (band 2), trimers (bands 3 and 4), dimers (bands 5 and 6), monomers (bands 7 and 8), and monomer-like species (bands 9–11, lane L) are marked, as are the positions for molecular mass standards of 30–200 kDa (Left). The apparent molecular masses of bands 7, 8, 9, 10, and 11 are 45, 41, 38, 36, and 34 kDa, respectively. (B) The 41-kDa Vp1 monomer is converted into the 45-kDa monomer upon reduction. The same cytosolic sample as in A, lane b, prepared from 1 × 106 cells, was immunoprecipitated with anti-Vp1 serum and analyzed by 2D SDS/PAGE and autoradiography. The 41-kDa Vp1 (spot 8) is positioned off the diagonal, whereas the 45-kDa Vp1 (spot 7) lies on the diagonal.
Whether the 41-kDa species is a full-length Vp1 monomer with intrachain disulfide linkage was determined by a 2D SDS/PAGE analysis. The 41-kDa species in the nonreducing first dimension (band 8 in Fig. 3A, lane b) acquired the full-length monomeric Mr of 45,000 (band 7) in the reducing second dimension, as shown by the horizontal alignment of the off-diagonal major spot 8 with the adjacent minor spot 7 that lies on the diagonal (Fig. 3B). Thus, the newly synthesized Vp1 chain contains at least one intramolecular disulfide linkage, and this linkage is soon reduced, giving rise to the extended 45-kDa monomer.
All four Vp1 oligomers appeared during the chase periods in the cytosolic fraction (Fig. 3A, bands 1–6, lanes c–f) and were found sooner, by the end of pulse, in the cytoskeletal fraction (lanes g–j). When the sulfhydryl modifying reagent NEM was present in the chase medium, no oligomers were detected and only the 45-kDa monomer remained in the cytosolic or the cytoskeletal fraction (lane m or n), indicating that the conversion of the 41-kDa species into the 45-kDa monomer preceded oligomer assembly. The steady decline of the total Vp1 radiolabel in the cytosolic fraction during the 40 min of chase (lanes c–e) is consistent with trafficking of the newly synthesized Vp1 within the cytoplasm or into the nucleus (20, 21). The oligomers constituted a greater proportion of the total Vp1 radiolabel in the cytoskeletal than in the cytosolic fraction by 40 min of chase (lane j compared with lane f), raising the possibility of the former fraction being a preferred site for Vp1 oligomerization. These findings demonstrate that interchain disulfide bonds are formed at the time of oligomerization and that the oligomers are intermediates of cytoplasmic Vp1 assembly. We conclude that a series of oxidation–reduction reactions accompanies cytoplasmic Vp1 assembly. The disulfide-linked Vp1 oligomers form from newly synthesized Vp1 monomers, which originally contain intramolecular disulfide linkages.
Redox-Coupled Vp1 Folding and Oligomerization Are Not Associated with the ER.
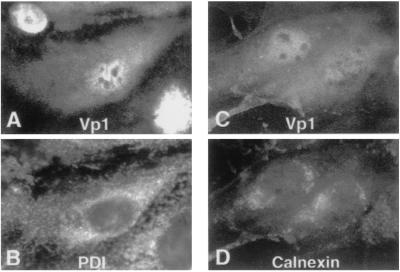
Vp1 is a soluble protein that folds and oligomerizes in the cytoplasm and then localizes to the nucleus. There is no evidence that it enters the ER-Golgi system, where disulfide formation and rearrangement are a common feature of secreted and membrane-bound proteins. In this study, we confirm the lack of association of Vp1 with the ER system by showing a distinct distribution pattern of Vp1 from those of two ER marker proteins, PDI and calnexin (Fig. 4). At 60 h postinfection, Vp1 showed nuclear and diffuse cytoplasmic staining (Fig. 4 A and C), whereas PDI had a broad, grainy perinuclear and cytoplasmic distribution (Fig. 4B) and calnexin localized primarily in perinuclear regions (Fig. 4D). The nonoverlapping Vp1 and ER marker distributions are confirmed by confocal microscopy (data not shown). We also assume that the Vp1 redox reactions observed above are catalyzed in vivo by cytoplasmic enzymes such as thioredoxins and glutaredoxins. The effective inhibition of disulfide-bonded oligomer formation by the presence of NEM in the chase medium (Fig. 3A, lanes m and n) is consistent with this interpretation. An alternative possibility, that the disulfides are catalyzed artifactually by the ER-resident PDI upon subcellular fractionation (32) and that this activity is inhibited by NEM, is unlikely, because the Vp1 oligomers also were detected (i) when fractions were treated with NEM and directly analyzed by immunoblotting (Fig. 2 A– C), (ii) in fractions prepared with lysis buffer containing NEM, and (iii) in a soluble cytoplasmic fraction prepared by hypotonic cell lysis (data not shown) that is not expected to contain PDI activity (32, 33). Therefore, Vp1 folding and oligomer assembly, accompanied by redox reactions, likely take place in the cytosol, outside the ER-Golgi system.
Figure 4.
Localization of Vp1 and ER marker proteins in infected cells. SV40 infected cells at 60 h postinfection were fixed, doubly stained either with rabbit anti-Vp1 (A) and mouse anti-PDI (B) antibodies or with rabbit anti-Vp1 (C) and mouse anti-calnexin (D) antibodies, followed by fluorescein-labeled (A and C) or rhodamine-labeled (B and D) secondary antibodies. The same cells are shown in A and B and in C and D.
Discussion
Our analysis has revealed naturally occurring, disulfide-bonded Vp1 species that are the assembly intermediates of Vp1 in the cytoplasm of SV40-infected cells. The noteworthy finding is that covalently linked Vp1 homooligomers from dimers to pentamers are formed from newly synthesized Vp1 monomers that initially contain an intrachain disulfide linkage(s). This transient, disulfide-linked monomer was detected as a pulse-labeled, 41-kDa Vp1 under nonreducing conditions that resolved into a 45-kDa species under reducing conditions. During chase periods, it was converted into the extended, 45-kDa Vp1 and then into the covalently linked homooligomers. The presence of NEM in the chase medium blocked the emergence of the disulfide-linked oligomers. The cytoplasmic Vp1 did not colocalize with ER marker proteins. We conclude that SV40 major structural protein Vp1 assembles into pentamers in the cytoplasm via a sulfhydryl–disulfide redox-coupled pathway. This paper demonstrates in vivo a mammalian viral protein that folds and oligomerizes through a series of oxidation–reduction reactions outside the ER-secretory pathway.
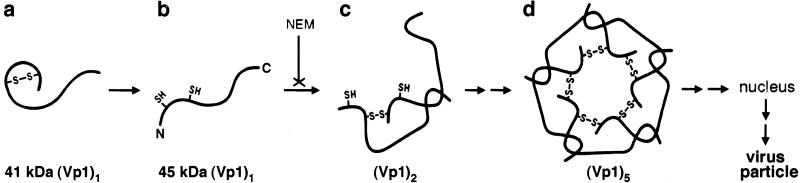
A model consistent with all results is illustrated in Fig. 5. The intramolecularly disulfide-bonded, 41-kDa Vp1 represents an intermediate (Fig. 5a) in the folding of a monomer. This intrachain linkage transiently stabilizes a local structure of Vp1 that facilitates the interdigitation of secondary structural elements between neighboring monomers. The intrachain disulfide is then broken (Fig. 5b), and interchain disulfides are forged to facilitate the formation of dimers (Fig. 5c) through pentamers (Fig. 5d). These interchain disulfide bonds are also transient, broken somewhere along the particle assembly and maturation process in the nucleus. In this model, Vp1 folding into a pentamer is guided by transitory disulfides in intermediates a and c through d in Fig. 5, and these disulfides necessarily are formed by at least two different cysteine residues. If these disulfides are essential for proper pentamer formation, mutating the cysteine pair(s) involved should disrupt normal Vp1 folding in the cytoplasm and eliminate the formation of the particle: the mutants should be nonviable. We previously have shown that viability is retained when the seven Vp1 cysteines are mutated individually (34). We have since tested a number of cysteine combination mutants and found a few double mutants to be nonviable. In particular, our preliminary results show that a Cys-49 → Ala/Cys-87 → Ala mutant Vp1 synthesized from the transfected mutant viral DNA is unstable, cannot enter the nucleus, and has a different profile of cytoplasmic intermediates as that observed for wild-type Vp1 in Fig. 3A (35), suggesting that the formation of the 41-kDa kinetic intermediate or of the Vp1 oligomers has been affected. The distinct viability difference between the pair mutant and the single mutants is consistent with an interpretation that Cys-49 and Cys-87 each can bond with other cysteine residues for Vp1 folding when the other is mutated but not when both are mutated. Which other cysteines may serve as the bonding partners for Cys-49 and Cys-87 remains to be identified. In an alternative model, the disulfides per se are not essential for Vp1 folding but are formed as a consequence of two cysteines being brought into close proximity by the folding of local structural elements. Mutating the two cysteines at the same time, but not mutating single cysteines, somehow could disrupt normal Vp1 folding. The murine polyomavirus particle contains an intrapentamer disulfide between Cys-19 and Cys-114 of Vp1 (36), so the linkage conceivably might be formed during pentamerization. Testing whether the major capsid protein pentamers of other papovaviruses, such as human JC virus and papillomaviruses (37), fold via transient disulfide bonding may reveal a general principle for the formation of these pentamers.
Figure 5.
Model of cytoplasmic Vp1 folding and oligomerization. The newly synthesized Vp1 in the cytoplasm is an intrachain disulfide-linked monomer (a), which is converted into a disulfide-free monomer (b) and then into interchain disulfide-linked oligomers (c through d). The last species is transported into the nucleus, where it participates in virus particle assembly. A disulfide linkage is denoted by “S-S,” and a free sulfhydryl group of cysteine residue is denoted by “SH.”
Our study focuses on Vp1 folding and pentamer formation in the cytoplasm. Whether cysteine redox reactions are also involved in postpentamer virus assembly in the nucleus is not known. Postpentameric complexes have been observed in an in vitro translation system when at least one of Cys-9, Cys-104, and Cys-207 is intact (38). Disulfide linkage at Cys-9 and Cys-104 is also found to confer proteinase K resistance to virus-like particles (with no viral DNA encapsidated) assembled in Vp1-expressing insect cells (39). Because the triple mutant, Cys-9 → Ala/Cys-104 → Ala/Cys-207 → Ser, is viable (result to be reported elsewhere), it appears that the conditions inside the natural host cell are not reproduced by the cell-free and insect-cell systems. The Cys-104–Cys-104 disulfide exists between certain neighboring pentamers in mature SV40 (22). This disulfide is thought to form after virus assembly and contributes to capsid stability (22), yet it is dispensable for viability (34).
Besides the 41-kDa Vp1, the low-abundance 38-, 36-, and 34-kDa species also were present in the pulse-labeled cytosolic fraction and could represent additional intrachain intermediates; for example, monomers with progressively larger looping as a result of bonding between cysteines that are farther apart. Alternatively, those species could be incompletely synthesized or partly degraded Vp1 chains. The two possibilities remain unresolved because the amounts of these species were below the detection limit of the 2D gel analysis (Fig. 3B). Whether the intrachain linkage(s) in the 41-kDa species is formed cotranslationally or posttranslationally is not known, because our pulse-labeling period is longer than the expected time for the completion of chain synthesis.
The greater Vp1 oligomer-to-monomer ratio in the cytoskeletal fraction relative to the cytosolic fraction suggests that the cytoskeletal fraction is a preferred site for Vp1 oligomerization. Whether the cytoskeleton plays a role in Vp1 oligomerization and nuclear transport needs to be addressed by characterizing mutants that are defective in these processes. The absence of covalent Vp1 homooligomers larger than the pentamer in the cytosolic fraction (Fig. 1C) suggests that the pentamers are prevented from further assembly in the cytosol, perhaps through the involvement of chaperone proteins (40).
Our new finding on SV40 Vp1 raises the possibility that transient disulfide formation accompanies the folding of many cysteine-bearing mammalian proteins in the cytoplasm, whether or not native disulfides are present in these proteins. The recently determined structure for the complex of yeast superoxide dismutase with its metallochaperone (41) suggests that the two individually folded proteins undergo, during their interaction, conformational changes through disulfide rearrangements. This and our findings, analogous to that of phage P22 tailspike protein in the bacterial system (17, 18), hint that disulfide dynamics may be a more widely occurring phenomenon in the eukaryotic cytoplasm than previously assumed. It remains to be explored how general the involvement of sulfhydryl–disulfide redox is in cytoplasmic protein folding and what catalyzes these redox reactions in the cytoplasm.
Acknowledgments
We thank Dr. G. Fairbanks for advice on the use of the cross-linking reagent and Dr. E. Harlow for providing the monoclonal anti-Vp1 antibody. We also thank Ms. L. Perini for technical assistance. We are grateful to Drs. T. L. Benjamin, L. A. Bankston, S. G. Clarke, D. Eisenberg, and R. C. Liddington for suggestions, discussion, and critical reading. This work was supported by Public Health Service Grant CA50574 from the National Institutes of Health, a grant from the University of California, Los Angeles, Jonsson Comprehensive Cancer Center, and a grant from the University of California, Los Angeles, Academic Senate.
Abbreviations
- SV40
simian virus 40
- PDI
protein disulfide isomerase
- ER
endoplasmic reticulum
- NEM
N-ethylmaleimide
- DTSP
dithiobis (succinimidyl propionate)
- 2D
two-dimensional
References
- 1.Yon J M. Braz J Med Biol Res. 2001;34:419–435. doi: 10.1590/s0100-879x2001000400001. [DOI] [PubMed] [Google Scholar]
- 2.Creighton T E. Biol Chem. 1997;378:731–744. doi: 10.1515/bchm.1997.378.8.731. [DOI] [PubMed] [Google Scholar]
- 3.Wedemeyer W J, Welker E, Narayan M, Scheraga H A. Biochemistry. 2000;39:4207–4216. doi: 10.1021/bi992922o. [DOI] [PubMed] [Google Scholar]
- 4.Bergman L W, Kuehl W M. J Biol Chem. 1979;254:5690–5694. [PubMed] [Google Scholar]
- 5.Bergman L W, Kuehl W M. J Biol Chem. 1979;254:8869–8876. [PubMed] [Google Scholar]
- 6.Braakman I, Hoover-Litty H, Wagner K R, Helenius A. J Cell Biol. 1991;114:401–411. doi: 10.1083/jcb.114.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huth J R, Mountjoy K, Perini F, Ruddon R W. J Biol Chem. 1992;267:8870–8879. [PubMed] [Google Scholar]
- 8.Carleton M, Brown D T. J Virol. 1996;70:5541–5547. doi: 10.1128/jvi.70.8.5541-5547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirazimi A, Svensson L. J Virol. 2000;74:8048–8052. doi: 10.1128/jvi.74.17.8048-8052.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raina S, Missiakas D. Annu Rev Microbiol. 1997;51:179–202. doi: 10.1146/annurev.micro.51.1.179. [DOI] [PubMed] [Google Scholar]
- 11.Rietsch A, Beckwith J. Annu Rev Genet. 1998;32:163–184. doi: 10.1146/annurev.genet.32.1.163. [DOI] [PubMed] [Google Scholar]
- 12.Fabianek R A, Hennecke H, Thony-Meyer L. FEMS Microbiol Rev. 2000;24:303–316. doi: 10.1111/j.1574-6976.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert H F. J Biol Chem. 1997;272:29399–29402. doi: 10.1074/jbc.272.47.29399. [DOI] [PubMed] [Google Scholar]
- 14.Ruddon R W, Bedows E. J Biol Chem. 1997;272:3125–3128. doi: 10.1074/jbc.272.6.3125. [DOI] [PubMed] [Google Scholar]
- 15.Huppa J B, Ploegh H L. Cell. 1998;92:145–148. doi: 10.1016/s0092-8674(00)80907-1. [DOI] [PubMed] [Google Scholar]
- 16.Frand A R, Cuozzo J W, Kaiser C A. Trends Cell Biol. 2000;10:203–210. doi: 10.1016/s0962-8924(00)01745-1. [DOI] [PubMed] [Google Scholar]
- 17.Robinson A S, King J. Nat Struct Biol. 1997;4:450–455. doi: 10.1038/nsb0697-450. [DOI] [PubMed] [Google Scholar]
- 18.Haase-Pettingell C, Betts S, Raso S W, Stuart L, Robinson A, King J. Protein Sci. 2001;10:397–410. doi: 10.1110/ps.34701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liddington R C, Yan Y, Moulai J, Sahli R, Benjamin T L, Harrison S C. Nature (London) 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 20.Lin W, Hata T, Kasamatsu H. J Virol. 1984;50:363–371. doi: 10.1128/jvi.50.2.363-371.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin W, Shurgot J L, Kasamatsu H. Virology. 1986;154:108–120. doi: 10.1016/0042-6822(86)90434-4. [DOI] [PubMed] [Google Scholar]
- 22.Stehle T, Gamblin S J, Yan Y, Harrison S C. Structure (London) 1996;4:165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 23.Kasamatsu H, Nehorayan A. J Virol. 1979;32:648–660. doi: 10.1128/jvi.32.2.648-660.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clever J, Kasamatsu H. Virology. 1991;181:78–90. doi: 10.1016/0042-6822(91)90472-n. [DOI] [PubMed] [Google Scholar]
- 25.Lomant A J, Fairbanks G. J Mol Biol. 1976;104:243–261. doi: 10.1016/0022-2836(76)90011-5. [DOI] [PubMed] [Google Scholar]
- 26.Peters K, Richards F M. Annu Rev Biochem. 1977;46:523–551. doi: 10.1146/annurev.bi.46.070177.002515. [DOI] [PubMed] [Google Scholar]
- 27.Schweizer E, Angst W, Lutz H U. Biochemistry. 1982;21:6807–6818. doi: 10.1021/bi00269a029. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Gharakhanian E, Takahashi J, Clever J, Kasamatsu H. Proc Natl Acad Sci USA. 1988;85:6607–6611. doi: 10.1073/pnas.85.18.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Garcea R L. J Virol. 1994;68:320–327. doi: 10.1128/jvi.68.1.320-327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burton K S, Consigli R A. Virus Res. 1996;40:141–147. doi: 10.1016/0168-1702(95)01266-4. [DOI] [PubMed] [Google Scholar]
- 32.Kaderbhai M A, Austen B M. Eur J Biochem. 1985;153:167–178. doi: 10.1111/j.1432-1033.1985.tb09283.x. [DOI] [PubMed] [Google Scholar]
- 33.Lambert N, Freedman R B. Biochem J. 1985;228:635–645. doi: 10.1042/bj2280635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li P P, Nakanishi A, Tran M A, Salazar A M, Liddington R C, Kasamatsu H. J Virol. 2000;74:11388–11393. doi: 10.1128/jvi.74.23.11388-11393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li P P. Ph.D. dissertation. Los Angeles: University of California; 2001. [Google Scholar]
- 36.Stehle T, Yan Y, Benjamin T L, Harrison S C. Nature (London) 1994;369:160–163. doi: 10.1038/369160a0. [DOI] [PubMed] [Google Scholar]
- 37.Chen X S, Garcea R L, Goldberg I, Casini G, Harrison S C. Mol Cell. 2000;5:557–567. doi: 10.1016/s1097-2765(00)80449-9. [DOI] [PubMed] [Google Scholar]
- 38.Jao C C, Weidman M K, Perez A R, Gharakhanian E. J Gen Virol. 1999;80:2481–2489. doi: 10.1099/0022-1317-80-9-2481. [DOI] [PubMed] [Google Scholar]
- 39.Ishizu K I, Watanabe H, Han S I, Kanesashi S N, Hoque M, Yajima H, Kataoka K, Handa H. J Virol. 2001;75:61–72. doi: 10.1128/JVI.75.1.61-72.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cripe T P, Delos S E, Estes P A, Garcea R L. J Virol. 1995;69:7807–7813. doi: 10.1128/jvi.69.12.7807-7813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamb A L, Torres A S, O'Halloran T V, Rosenzweig A C. Nat Struct Biol. 2001;8:751–755. doi: 10.1038/nsb0901-751. [DOI] [PubMed] [Google Scholar]