Abstract
Little is known about how a morphogenetic rearrangement of a tissue is affected by individual cells. Starving Dictyostelium discoideum cells aggregate to form dendritic streams, which then break up into groups of ≈2 × 104 cells. Cell number is sensed at this developmental stage by using counting factor (CF), a secreted complex of polypeptides. A high extracellular concentration of CF indicates that there is a large number of cells, which then causes the aggregation stream to break up. Computer simulations indicated that stream breakup could be caused by CF decreasing cell–cell adhesion and/or increasing cell motility, and we observed that CF does indeed decrease cell–cell adhesion. We find here that CF increases cell motility. In Dictyostelium, motility is mediated by actin and myosin. CF increases the amounts of polymerized actin and the ABP-120 actin-crosslinking protein. Partially inhibiting motility by using drugs that interfere with actin polymerization reduces stream dissipation, resulting in fewer stream breaks and thus larger groups. CF also potentiates the phosphorylation and redistribution of myosin while repressing its basal level of assembly. The computer simulations indicated that a narrower distribution of group sizes results when a secreted factor modulates both adhesion and motility. CF thus seems to induce the morphogenesis of streams into evenly sized groups by increasing actin polymerization, ABP-120 levels, and myosin phosphorylation and decreasing adhesion and myosin polymerization.
How tissues regulate their size and morphology is poorly understood (1, 2). The simple eukaryote Dictyostelium discoideum lives as single amoebae and grows by fission. When starved, the cells begin secreting an 80-kDa glycoprotein called conditioned medium factor (3, 4). When there is a high density of starving cells, as indicated by a high concentration of conditioned medium factor, the cells aggregate by using relayed pulses of cAMP as a chemoattractant (5, 6). The aggregating cells form dendritic aggregation streams, which break up into groups of up to 105 cells, with our laboratory strains forming groups of ≈2 × 104 cells (7).
To explore how Dictyostelium aggregation streams are able to break up into groups of a fairly specific number of cells, we used shotgun antisense (a mutagenesis scheme where pooled cDNA is cloned into an antisense vector, and the resulting library is used to transform cells; after a transformant with an interesting phenotype is identified, PCR can be used to immediately isolate the antisense cDNA) to obtain smlAas, a transformant forming abnormally small fruiting bodies (8). smlA− cells, with smlA disrupted by homologous recombination, had the same phenotype as smlAas. These cells formed normal aggregation territories and streams, but the streams then broke up into many small groups. The exudate from smlA− cells caused wild-type cells to form small groups and fruiting bodies, suggesting that the smlA− phenotype was because of these cells oversecreting a factor (9). We purified the factor and found that it was a complex of polypeptides secreted by wild-type cells and oversecreted by smlA− cells. The counting factor (CF) has a net molecular mass of ≈450 kDa (10).
Disruption of the gene encoding countin, one of the components of CF, generated cells that secreted no detectable CF activity (10, 11). Countin− cells form normal aggregation territories and streams, but the streams do not break up, resulting in the formation of huge groups and fruiting bodies. Computer simulations indicated that if the ratio of the random cell motility force FM to the cell–cell adhesion force FA is low, the stream will stay intact. If, however, this ratio is high, the random motility of the cells will overcome the cells' adhesion to one another, and the stream will then dissipate; if the ratio then decreases, the dispersed cells will condense into groups (12). We found that CF negatively regulates cell–cell adhesion at the level of the expression of adhesion proteins, so if there are only a few cells in a stream, the low level of CF will permit a high adhesion and the stream will remain intact, whereas if there are a large number of cells in a stream, the high levels of CF decrease cell–cell adhesion, allowing the stream to break up (12).
In the above model, the breakup of the stream depends on the cells being motile. The motility of Dictyostelium cells depends on actin and myosin (13). Both proteins experience dynamic rearrangements after cAMP stimulation. The level of polymerized actin (F-actin) increases significantly within seconds of chemotactic stimulation. After a brief return to basal levels at ≈20–30 s, the F-actin level transiently increases again at ≈60 s (14–16). As the cell moves forward by extending a pseudopod (17), actin but not myosin II is present in the protruding pseudopod. The F-actin crosslinking protein ABP-120 then seems to play a major role in stabilizing the newly formed pseudopod (18, 19).
Myosin II is distributed uniformly, mainly as subunits, throughout the cytoplasm or is localized in the cell cortex as myosin filaments in nonmigrating cells, but upon cAMP stimulation, it transiently concentrates in the posterior end of migrating cells and in the tips of retracting pseudopods (20–23). The subcellular distribution of myosin II is regulated by the phosphorylation of the myosin heavy chain on three threonines, with phosphorylation causing myosin to disassemble, and dephosphorylation causing myosin to assemble and become active (24–27). The phosphorylation is necessary for the maintenance of cell shape and increases the ability of cells to chemotax (28, 29). In this article, we show that CF regulates this phosphorylation, myosin II localization and assembly, ABP-120 levels, and actin polymerization to regulate motility during stream formation and thus group size.
Materials and Methods
Cell Culture.
Cell culture was done following Brock and Gomer (10). Green fluorescent protein (GFP)–myosin cells (22) and GFP–myosin heavy chain kinase cells (27) were cultured with 10 μg/ml G418. HS1 myosin null cells were cultured on SM/5 following Ruppel et al. (30). To deplete extracellular CF with anti-countin antibodies, cells were starved at 4 × 106 cells/ml in phosphate buffer with magnesium (20 mM KH2PO4/1 mM MgCl2/0.01 mM CaCl2, pH 6.1 with KOH; PBM) (10) on AABP04700 filters (Millipore) placed either on two layers of no. 3 paper or on three layers of no. 541 paper (Whatman) soaked with PBM. After 1 h, the filters were transferred to paper soaked with either 1:300 immune or preimmune anti-countin antisera in PBM. To measure motility, the cells were collected 4 h later and diluted to 3 × 105 cells/ml in PBM; 400 μl was placed in the well of a no. 155409 8-well chambered cover glass (Nalge). After 15 min, 200 μl of buffer was removed. Immune or preimmune antibodies were added to the well to a 1:300 dilution, and cells were videotaped 1 h after being placed in the well following Yuen et al. (4) by using a ×20 objective. Treatment of cells with 0.01 units/ml beef heart cAMP phosphodiesterase (PDE) or cAMP pulses, both starting at 4 h of starvation, followed Tang et al. (31). For other experiments, cells were collected 5 h after the filters were exposed to antibodies. To determine the effect of inhibiting actin-mediated motility, filters were transferred to paper soaked with latrunculin (Molecular Probes), cytochalasin A, or cytochalasin D (Sigma).
Actin Polymerization, Myosin Assembly, and Myosin Phosphorylation.
Preparation of crude cytoskeletons and gel electrophoresis to visualize the level of F-actin was done following Dharmawardhane et al. (15). Myosin assembly was assayed following Chung and Firtel (32). To examine myosin phosphorylation, cells starved in the presence of antibodies were washed with PBM twice, resuspended in PBM at 2 × 107 cells/ml, and stimulated with 1 μM cAMP. At each time point, stimulated cells were mixed with SDS/PAGE loading buffer containing protease inhibitor cocktail (Roche, Mannheim, Germany) and phosphatase inhibitors I and II (Sigma), boiled for 5 min, and stored at −70°C. Western blots were done as described (9) and were stained with 1:2,500 nu48 anti-myosin (a gift from Rex Chisolm, Northwestern University Medical School, Chicago) or 1:500 p-Thr(H-2) sc-5267 anti-phosphothreonine (Santa Cruz Biotechnology) antibodies. Bound antibody was detected with the ECL Western blotting kit (Amersham Pharmacia).
Microscopy.
For F-actin and ABP-120 localization, cells were starved as described for motility assays. After 6 h, cells were fixed and stained with phalloidin following Pang et al. (33), with the exception that PBM was used instead of HL5 medium, and Alexa 488 phalloidin (Molecular Probes) was used at a final concentration of 2 units/ml. For immunolocalization of ABP-120, cells were fixed and stained following Cox et al. (18). GFP–myosin or GFP–myosin heavy chain kinase cells were starved as described for the motility assays, and the live cells were imaged by using a Zeiss LSM 510 laser confocal microscope every 30 s.
Gel Electrophoresis.
Two-dimensional gel electrophoresis was performed essentially following the Bio-Rad two-dimensional gel instruction manual with 10-cm-long and 3-mm-diameter first-dimension tube gels. Samples were prepared by using the procedure of Morrissey et al. (34). One hundred microliters of lysate containing 5 × 107 cells was loaded onto the basic end of a first-dimension gel. Electrophoresis was at 500 V for 10 min and then 750 V for 13–16 h. For the second dimension 10% polyacrylamide gels, the upper chamber contained 1:10,000 thioglycolic acid (Sigma). Gels were stained with Coomassie blue R250, and spots of interest were excised, neutralized for 1 h with several changes of water, and then stored at −20°C. Trypsin digestion of the excised proteins and tandem electrospray MS of the peptides was performed by Richard Cook at the Baylor College of Medicine Protein core facility, Houston. Western blots were done following Roisin-Bouffay et al. (12).
Computer Simulations.
The computer simulations used the program described in Roisin-Bouffay et al. (12) with the modification that a calculated concentration of 100 ng/ml CF caused a 12.5% decrease in cell–cell adhesion, and the random motility force was increased proportionally to the concentration of CF so that at a calculated concentration of 100 ng/ml CF, the random motility force increased by a factor of 2.
Results
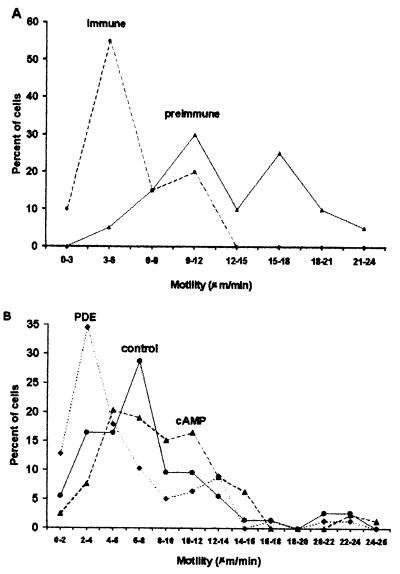
Cell motility during development can theoretically affect group size in Dictyostelium (12). We previously found that the levels of extracellular CF could be decreased by adding anti-countin antibodies, allowing experiments with different levels of CF to be performed on the same clone of cells. We tested the hypothesis that CF regulates motility by examining Ax4 cells developing in the presence of preimmune or immune anti-countin antibodies. As shown in Fig. 1A, cells tend to move slower in CF-depleted starvation medium than in control medium. The speed of cells in the presence of preimmune serum was in the range of speeds previously observed for wild-type cells (35–37). Similar assays with developing Ax4 cells incubated in conditioned medium for 15 min showed that the speed (in μm/min, mean ± SEM) was 6.8 ± 0.5 in countin−-conditioned medium and 8.8 ± 1.0 (P = 0.08) in smlA−-conditioned medium. The above data suggest that CF increases cell motility in developing Dictyostelium cells. A different secreted factor, conditioned medium factor, does not affect motility but does increase the rate of pseudopod extensions (4). There was no significant effect of CF on the number of pseudopods extended per minute (data not shown). A large number of similar experiments done over 2 years often (but not always) showed that smlA− had a higher motility than Ax4, which had a higher motility than countin−. We also observed that from one month to the next, the average speed of any given cell line tended to vary, suggesting that in addition to CF some aspect of the motility as measured by the translocation speed of a cell on a glass surface depends on unknown factors.
Figure 1.
CF and cAMP pulse size affect cell speed. (A) Anti-countin antibodies decrease cell speed. Ax4 cells were starved in the presence of preimmune or immune anti-countin antibodies. At 6 h of starvation, videomicroscopy was used to measure the speed of cell translocation. The average speed of cells exposed to preimmune serum was 13.0 ± 0.7 μm/min, and for immune serum 6.0 ± 0.3 μm/min (mean ± SEM). The averages are significantly different with P < 0.001 (t test), and the distributions are different with P < 0.001 (χ2). (B) Exposure to cAMP pulses increases cell speed. Cells were treated with either exogenous 30 nM cAMP pulses starting at 4 h of development to mimic large cAMP pulses, or PDE to decrease the endogenous cAMP pulses. The motility was measured at 6 h. The average speeds of cells exposed to cAMP pulses, buffer alone, and PDE were 8.9 ± 0.5, 7.7 ± 0.6, and 5.9 ± 0.5 μm/min (mean ± SEM). A Mann–Whitney rank sum test (the characteristics of the data precluded using t or χ2 tests) indicated that the speeds of cAMP-pulsed cells were different from control with P = 0.05, and the speeds of PDE-treated cells were different from control with P < 0.005. Because average motilities tend to vary from month to month, representative experiments are shown.
We previously found that CF potentiates cAMP pulse sizes and that large cAMP pulses decreased group size, whereas reducing cAMP pulse sizes by exposure of cells to PDE increased group size (31). Cells exposed to cAMP pulses moved faster than control cells, whereas cells treated with PDE moved slower (Fig. 1B). The results suggest that CF's effect on cell motility during development may be caused in part by its effect on cAMP pulses.
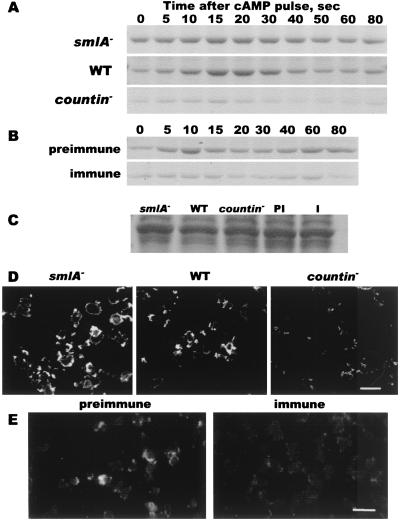
When a pulse of cAMP arrives at an aggregating cell, there is a transient increase in the amount of F-actin (15). To determine whether CF affects the level of F-actin, crude cytoskeletons were prepared from developing cells stimulated with extracellular cAMP and were then fractionated by SDS/gel electrophoresis. As previously observed, aggregating Ax4 parental cells contain F-actin, and within 10 s after cAMP stimulation the amount of F-actin increases, peaking at ≈10–20 s and then declining. A second peak of F-actin then appears at ≈60–80 s (ref. 15, Fig. 2A). A similar stimulation of smlA− cells showed that there is a higher basal level of F-actin in the cells, whereas countin− cells showed a lower level of F-actin compared with Ax4 cells (Fig. 2A). To verify that this difference was because of the level of extracellular CF, CF was depleted from developing Ax4 cells with exogenous anti-countin antibodies. As shown in Fig. 2B, the F-actin profile of Ax4 cells starved in the presence of preimmune antibodies was similar to the profile from untreated cells and to the previously observed profile (15). Cells starved in the presence of anti-countin antibodies showed a lower level of F-actin (Fig. 2B). Gels of the lysates used for the above experiments indicated that the total amount of actin in cells was similar in smlA−, wild-type, countin−, and preimmune- and anti-countin antiserum-treated cells (Fig. 2C). To determine whether CF affects the distribution of F-actin, cells were fixed, permeabilized, and stained with the F-actin specific dye phalloidin. The staining intensities observed by using both conventional and confocal fluorescence microscopy corresponded with the levels observed in the cytoskeletal preparations. There were no obvious differences in the distributions of F-actin (Fig. 2 D and E). Together, the data suggest that CF potentiates the amount of F-actin in streaming cells.
Figure 2.
CF increases the amount of F-actin in cells. (A) Cytoskeletal actin in smlA−, Ax4, and countin− cells. After 6 h of starvation, cells were collected and lysed at the times indicated after cAMP stimulation. Triton-insoluble cytoskeletons were isolated, and a Coomassie blue-stained SDS-polyacrylamide gel from a typical experiment is shown. (B) Anti-countin antibodies decrease the level of cytoskeletal actin. Ax4 cells were starved in the presence of preimmune or immune anti-countin antibodies. At 6 h, the Triton-insoluble cytoskeletons were isolated at the times indicated after cAMP stimulation. (C) CF levels do not affect total actin levels. Shown is a Coomassie-stained gel of the 0-sec total cell lysates used for the experiments shown in A and B. The heavy band in A–C is at 42 kDa. (D) F-actin in smlA−, Ax4, and countin− cells. After 6 h of starvation, cells were fixed and stained for F-actin with phalloidin and visualized with a confocal microscope. (E) Anti-countin antibodies decrease the level of F-actin. Ax4 cells were starved in buffer containing preimmune or immune anti-countin antibodies. The cells were fixed and stained with phalloidin and visualized with conventional fluorescence microscopy. (Bars in C and D are 20 μm.)
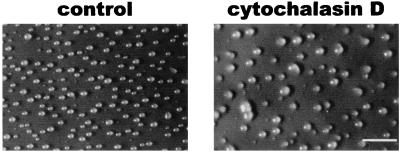
To test the hypothesis that decreasing cell motility while streams are condensing and breaking up can increase group size, we decreased motility by adding actin-destabilizing drugs such as latrunculin, cytochalasin A, or cytochalasin D to cells after streams had formed. Latrunculin (0.1–1 μM) depolymerizes actin filaments and inhibits cell movement (38–40); 1–100 μM cytochalasin A similarly inhibits motility (40–42), and 0.5–10 μM cytochalasin D also disrupts actin and inhibits motility (43, 44). Transferring filters with developing cells at 4 h after starvation (when streams had begun to form) onto pads soaked with 0.1 μM or higher latrunculin, 100 μM cytochalasin A, or 0.3 μM or higher cytochalasin D caused an increase in group size 4 h later, when streams had broken up (Fig. 3 and data not shown). Under these conditions and at the lowest drug concentrations, the motilities (in μm/min, mean ± SE) were control, 7.3 ± 0.3; latrunculin, 4.3 ± 0.2 (P < 0.001, compared with control); cytochalasin A, 3.0 ± 0.2 (P < 0.001); and cytochalasin D, 5.7 ± 0.3 (P < 0.001). This indicated that the drugs inhibited but did not abolish motility. Similar results were observed when cells were transferred at 5 or 6 h of development (data not shown). The above results suggest that inhibiting motility as streams are breaking up can increase group size.
Figure 3.
Inhibiting motility during stream formation increases group size. Wild-type Ax4 cells were starved on filters. At 4 h, the filters were transferred to pads soaked with buffer (control) or 0.3 μM cytochalasin D. The broken streams were photographed 4 h later. (Bar is 1 mm.)
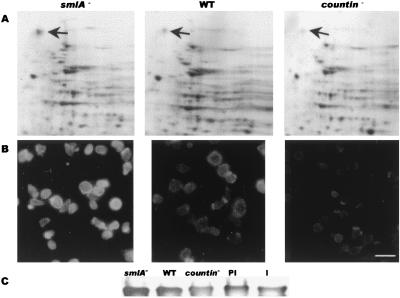
To determine whether CF affects motility and group size by regulating the expression of any major proteins, we examined two-dimensional gels of developing cells. As shown in Fig. 4A, there is a spot at ≈120 kDa that is visible in Ax4 cells and seems to be more prominent in smlA− cells and less prominent in countin− cells. This spot was excised and digested with trypsin, yielding peptides with the sequences LVGIGAEDIVDSQLK, VEVYGPGVEGGFVNK, and VFDNAPAEFTIFAVDTK. These were all exact matches to sequences in the 120-kDa ABP-120 actin crosslinking protein (45).
Figure 4.
CF increases the amount of the ABP-120 actin crosslinking protein in 6-h starved cells. (A) Two-dimensional gels of smlA−, Ax4, and countin− cells. The spot at ≈120 kDa (upper arrows) was excised, and the sequence of tryptic peptides indicated that it was ABP-120. The spot near the bottom of the gels that appears to be negatively regulated by CF is similar to aldose reductase. (B) The distribution of ABP-120 in smlA−, Ax4, and countin− cells. (Bar is 20 μm.) (C) A Western blot stained for ABP-120. PI and I indicate Ax4 cells treated with preimmune or immune anti-countin antibodies, respectively.
To verify that the level of ABP-120 protein is regulated by CF, we stained 6-h developing cells with anti-ABP-120 antibodies (a gift from John Condeelis, Albert Einstein College of Medicine, Bronx, NY). As shown in Fig. 4B, the Ax4 parental cells had a distribution of ABP-120 that was indistinguishable from previously published results (18). The smlA− cells had considerably brighter staining, whereas countin− cells showed staining that was much weaker than Ax4 cells. When anti-countin antibodies were used to deplete extracellular CF from developing Ax4 cells, the cells showed considerably less ABP-120 staining than cells developing in the presence of preimmune antibodies (data not shown). The different levels of ABP-120 seen by immunofluorescence were also seen in a Western blot stained for ABP-120 (Fig. 4C).
In conjunction with actin, myosin II is involved in cell motility. To determine the effect of CF on myosin II, wild-type cells expressing GFP-tagged myosin II heavy chain (GFP–Myo cells) were mixed 1:9 with smlA−, wild-type, or countin− cells and allowed to develop in submerged culture. When mixed with wild-type cells, we observed the characteristic cytosolic/cell cortex localization of myosin, which transiently changed to a concentration at the rear of a cell (ref. 22, Fig. 5A). When mixed with smlA− cells, the GFP–Myo cells became very motile, and most of the GFP–myosin was condensed at the rear of the cells. When cells became less motile after encountering a cAMP pulse, the GFP–myosin stayed at the posterior end of the cells and did not relocate (Fig. 5A). When mixed with countin− cells, GFP–Myo cells became relatively immobile, and very few cells had a posterior accumulation of GFP–myosin. GFP–Myo cells treated with preimmune anti-countin serum behaved like wild-type cells, with normal myosin II localization dynamics, whereas GFP–Myo cells exposed to immune anti-countin serum became much less motile, with myosin II either distributed uniformly in the cytosol or concentrated in the peripheral cell cortex but seldom at the posterior end (Fig. 5B). For the preimmune antiserum-treated cells, 28 ± 3% (mean ± SEM) had a posterior concentration of GFP–myosin, 22 ± 4% had GFP–myosin around the entire cortex, and 50 ± 4% had myosin in what appeared to be an even distribution in the cytosol with no obvious concentration in the cortex. For the anti-countin-treated cells, 4 ± 1% had GFP myosin in the posterior, 32 ± 4% had a cortical distribution, and 64 ± 5% had GFP–myosin in the cytosol. The data thus suggest that CF affects the overall degree of cell polarization as measured by a posterior accumulation of myosin.
Figure 5.
CF regulates the distribution of GFP–myosin in developing cells. Cells expressing GFP–myosin were starved in a chambered cover glass slide. (A) The cells were mixed 1:9 with the indicated cell line. (B) The cells were mixed with wild-type cells, and preimmune or immune anti-countin antibodies were added 1 h after starvation. The distribution of GFP–myosin in the live cells was recorded 5 h later. (Bar is 10 μm.)
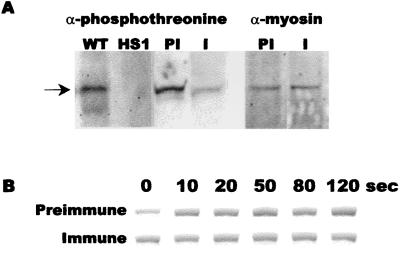
In addition to causing an increase in the amount of F-actin and a redistribution of myosin II, a pulse of cAMP causes a transient increase in myosin II phosphorylation and the amount of myosin in the cytoskeleton (26). Myosin phosphorylation can be detected by staining Western blots of cell proteins with anti-phosphothreonine antibodies (46). As shown in Fig. 6, anti-phosphothreonine antibodies stained a band at ≈210 kDa from Ax4 cells. There was no detectable staining of this band in HS1 cells, which have a disruption of the myosin II heavy chain (30). This indicated that the 210-kDa band stained by the anti-phosphothreonine antibodies is myosin II heavy chain. When Ax4 cells were starved in the presence of preimmune anti-countin antibodies or buffer alone (ref. 26 and data not shown), there was an observable basal level of myosin phosphorylation that transiently increased, with a peak at ≈60 s, after cAMP stimulation (Fig. 6A and data not shown). Treatment of cells with anti-countin antibodies caused a decrease in the basal level of myosin phosphorylation (Fig. 6A and data not shown). When identical blots were stained with anti-myosin antibodies, there was no detectable difference in the levels of myosin in cells treated with buffer, preimmune, or immune sera (Fig. 6A and data not shown). Treatment with anti-countin antibodies did not affect the distribution of GFP–myosin heavy chain kinase in cells expressing this protein (data not shown). Gel electrophoresis of cytoskeleton preparations indicated that cells treated with preimmune antisera had a cAMP-induced myosin polymerization that was similar to that previously observed for untreated cells (32), whereas cells treated with anti-countin antibodies had a higher basal level of myosin polymerization and a less prominent response to cAMP (Fig. 6B). Compared to wild type, smlA− cells had a lower basal level of myosin polymerization and a more pronounced response to cAMP, whereas countin− cells had a higher basal level of polymerized myosin and an apparently weaker response to cAMP (data not shown). These observations suggest that CF decreases the basal level of myosin phosphorylation and increases and somewhat stabilizes the level of myosin polymerization.
Figure 6.
Anti-countin antibodies decrease myosin phosphorylation and increase the basal level of myosin assembly. (A) Ax4 cells were starved in the presence of preimmune or anti-countin antibodies. Samples were collected at 6 h, and a Western blot of the samples was stained with anti-phosphothreonine or anti-myosin II heavy chain antibodies. WT, untreated Ax4 cells; HS1, untreated myosin II heavy chain null cells; PI, Ax4 cells exposed to preimmune antibodies; I, Ax4 cell exposed to anti-countin antibodies. Arrow indicates the 210-kDa myosin II heavy chain. (B) Ax4 cells were harvested at 6 h and stimulated with cAMP, and cytoskeletons were isolated at the times indicated in seconds after cAMP stimulation. For each treatment, the band shown is the 210-kDa myosin heavy chain from a Coomassie-stained 12% SDS-polyacrylamide gel of the cytoskeletons.
Discussion
Computer simulations had suggested that the key parameter for breaking a column or stream of cells into discrete groups is the ratio r = FM/FA, where FM is the random motility force and FA is the total adhesion force holding a cell in place (12). If r > 1, then the random motility force will tend to cause cells to tear themselves away from the cells they are touching, and this would then cause a stream to break up and thus form groups. If r < 1, then the adhesion forces will be greater than the random motility forces and the cells will remain in contact, and the stream will remain intact. Using the parameters of how much CF is secreted by a cell, the diffusion coefficient of CF, and the quantitative effect of CF concentration on cell–cell adhesion in the computer simulation, we showed that a roughly even group size could be obtained with CF modulating only cell–cell adhesion. Under these conditions, the variance in group size (the standard deviation in number of cells per group divided by the average number of cells per group) was 0.46. When we modified the program to have the calculated concentration of CF increase the motility in addition to decreasing the adhesion, the variance became 0.28. This difference was significant by using a one-tail test with P < 0.05. Thus, a possible explanation for CF regulating both motility and adhesion rather than regulating adhesion alone is that this dual regulation allows a more precise regulation of group size.
We previously found that CF negatively regulates the expression of the gp24 adhesion molecule and cell–cell adhesion (12). There is a close relationship between adhesion, the actin cytoskeleton, and motility (47), and there is thus a possibility that the observed effects of CF on motility are an indirect result of the effects of CF on adhesion, and the effects of actin-destabilizing drugs on group size is because of altered adhesion. However, the regulation by CF of four different aspects of motility (actin and myosin polymerization, myosin phosphorylation, and ABP-120 expression) and the ability of a 1-min exposure of cells to recombinant countin to increase the basal level of F-actin (T.G. and R.H.G., unpublished results) suggest that CF directly regulates some properties of the cytoskeleton and motility.
We consistently found that anti-countin antibodies decrease motility and that recombinant countin increases motility (L.T., T.G., and R.H.G., unpublished results). For motility, actin polymerization, ABP-120 expression, and myosin localization, the effects of anti-countin antibodies on wild-type cells were also seen comparing countin− to parental cells, suggesting the effects were not an artifact of using the antibodies, such as the antibodies binding to cells and impeding their movement. We also found a consistent pattern in the effect of CF on group size, actin polymerization, ABP-120 levels, myosin distribution, and myosin phosphorylation. However, we observe that the average motility of wild-type cells, as measured by translocation speed on tissue culture-treated glass slides, tends to vary from month to month. This suggests that either properties of the glass tend to vary or that some unknown physiological factors affect cell speed. The breakup of a stream of cells into groups requires that cells break their adhesion to other cells. To cause that rupture, the force of cell motility is more important than cell speed. We thus hypothesize that CF might increase the force of cell motility and that this increased force is manifested by an increased cell speed.
Motility of cells requires that the cells become polarized (32, 48). Our observations on cells expressing GFP–myosin suggest that a major effect of CF is to increase the extent to which cells form a polarized distribution of myosin. We previously observed that CF does not affect the extent of cytosolic regulator of adenylyl cyclase (CRAC)–GFP relocalization to the leading edge of cells during endogenous cAMP waves (31), suggesting that the effect of CF on GFP–myosin polarization occurs downstream of CRAC polarization.
Our data on the gross effect of altering cAMP pulse sizes suggest that some aspect of the cAMP signal transduction pathway mediates the effect of CF on motility and group size. CF increases the size of cAMP pulses and motility and decreases adhesion and group size. We found that large cAMP pulses increase motility and decrease group size, but actually slightly increase adhesion (12, 31). This then suggests that the ability of large cAMP pulses to decrease group size is mediated by their effect on motility and not adhesion and that with respect to regulating stream breakup and altering group size, the effect of cAMP pulses on motility overrides their effect on adhesion.
We previously found that CF regulates the cAMP-induced cGMP pulse but does not affect basal cGMP levels (31). Other workers have found that cGMP affects myosin phosphorylation (49–53). We see here that CF increases the basal level of myosin phosphorylation and decreases the basal level of myosin in the cytoskeleton. Because CF does not affect basal cGMP levels, a strong possibility is that CF regulates this basal level of myosin phosphorylation and localization through a pathway that does not involve cGMP. Our observations suggest that when CF concentrations are low, the basal level of myosin phosphorylation is low, causing a high level of myosin to polymerize, resulting in many cells having myosin in a relatively rigid and inflexible cytoskeleton around the entire cortex, reducing their motility and keeping the stream intact. When there is a large number of cells in a stream, the high CF concentration causes a high basal level of myosin phosphorylation, decreasing the amount of polymerized myosin, leaving regions in the cortex able to extend pseudopods, increasing motility and the consequent possibilities of cells moving apart from one another, and leading to breaks in the stream.
Cells lacking the myosin II heavy chain, or with a modified myosin lacking all three of the threonine phosphorylation sites, have reduced motilities compared with wild-type cells, but tend to change direction more frequently (28, 29, 54–56). In our model of stream breakup, a slower motility would tend to keep a stream intact and lead to larger groups, whereas turning more frequently or more drastically would tend to disrupt the orderly progression of a stream and lead to ruptures and smaller groups. The cells with no or an unphosphorylatable myosin heavy chain are able to form aggregation streams that break into groups that are similar in size to wild-type groups. We hypothesize that this is because of the effect of the slower speeds being counteracted by the effect of more frequent turns, leading to roughly normal group sizes.
Our results indicate that having CF regulating actin polymerization, myosin II phosphorylation, myosin distribution, and thus motility in a stream of Dictyostelium cells can affect the breakup of the stream into groups and regulate the size of these groups. In other systems, there exist secreted factors, mechanisms that can use the concentration of an extracellular signal to regulate gene expression, actin, and actin-crosslinking proteins. It is thus possible that in other systems, the morphogenesis of a column of cells into groups, and the initial number and size of these groups, could be regulated by using similar mechanisms.
Acknowledgments
We thank Hamam Alrabaa for assistance with two-dimensional gels, Richard Cook for protein sequencing, Diane Hatton for assistance with sequence analysis, Tom Egelhoff and Jim Spudich for their gifts of cells, Rex Chisolm and John Condeelis for their gifts of antibodies, Gadi Shaulsky for the use of lab space, and Rex Chisolm, Tom Egelhoff, Mike Gustin, Dave Knecht, and Mike Stern for advice. R.H.G. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- CF
counting factor
- GFP
green fluorescent protein
- PDE
phosphodiesterase
- PBM
phosphate buffer with magnesium
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Conlon I, Raff M. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 2.Gomer R H. Nat Rev. 2001;2:48–54. doi: 10.1038/35048058. [DOI] [PubMed] [Google Scholar]
- 3.Jain R, Yuen I S, Taphouse C R, Gomer R H. Genes Dev. 1992;6:390–400. doi: 10.1101/gad.6.3.390. [DOI] [PubMed] [Google Scholar]
- 4.Yuen I S, Jain R, Bishop J D, Lindsey D F, Deery W J, Van Haastert P J M, Gomer R H. J Cell Biol. 1995;129:1251–1262. doi: 10.1083/jcb.129.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devreotes P. Science. 1989;245:1054–1058. doi: 10.1126/science.2672337. [DOI] [PubMed] [Google Scholar]
- 6.Firtel R A. Genes Dev. 1995;9:1427–1444. doi: 10.1101/gad.9.12.1427. [DOI] [PubMed] [Google Scholar]
- 7.Shaffer B M. Q J Microb Sci. 1957;98:393–405. [Google Scholar]
- 8.Spann T, Brock D A, Lindsey D F, Wood S A, Gomer R. Proc Natl Acad Sci USA. 1996;93:5003–5007. doi: 10.1073/pnas.93.10.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brock D A, Buczynski F, Spann T P, Wood S A, Cardelli J, Gomer R H. Development (Cambridge, UK) 1996;122:2569–2578. doi: 10.1242/dev.122.9.2569. [DOI] [PubMed] [Google Scholar]
- 10.Brock D A, Gomer R H. Genes Dev. 1999;13:1960–1969. doi: 10.1101/gad.13.15.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown J M, Firtel R A. Trends Genet. 2000;16:191–193. doi: 10.1016/s0168-9525(00)01977-6. [DOI] [PubMed] [Google Scholar]
- 12.Roisin-Bouffay C, Jang W, Gomer R H. Mol Cell. 2000;6:953–959. [PubMed] [Google Scholar]
- 13.Noegel A A, Schleicher M. J Cell Sci. 2000;113:759–766. doi: 10.1242/jcs.113.5.759. [DOI] [PubMed] [Google Scholar]
- 14.McRobbie S J, Newell P C. Biochem Biophys Res Commun. 1983;115:351–359. doi: 10.1016/0006-291x(83)91011-2. [DOI] [PubMed] [Google Scholar]
- 15.Dharmawardhane S, Warren V, Hall A L, Condeelis J. Cell Motil Cytoskeleton. 1989;13:57–63. doi: 10.1002/cm.970130107. [DOI] [PubMed] [Google Scholar]
- 16.Noegel A A, Luna J E. Experientia. 1995;51:1135–1143. doi: 10.1007/BF01944731. [DOI] [PubMed] [Google Scholar]
- 17.Condeelis J. Cell Motil Cytoskeleton. 1992;22:1–6. doi: 10.1002/cm.970220102. [DOI] [PubMed] [Google Scholar]
- 18.Cox D, Condeelis J, Wessels D, Soll D, Kern H, Knecht D A. J Cell Biol. 1992;116:943–955. doi: 10.1083/jcb.116.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukui Y, de Hostos E, Yumura S, Kitanishi-Yumura T, Inoue S. Exp Cell Res. 1999;249:33–45. doi: 10.1006/excr.1999.4445. [DOI] [PubMed] [Google Scholar]
- 20.Ogihara S, Carboni J, Condeelis J. Dev Genet (Amsterdam) 1988;9:505–520. doi: 10.1002/dvg.1020090427. [DOI] [PubMed] [Google Scholar]
- 21.Nachmias V T, Fukui Y, Spudich J A. Cell Motil Cytoskeleton. 1989;13:158–169. doi: 10.1002/cm.970130304. [DOI] [PubMed] [Google Scholar]
- 22.Moores S, Sabry J, Spudich J. Proc Natl Acad Sci USA. 1996;93:443–446. doi: 10.1073/pnas.93.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clow P A, McNally J G. Mol Biol Cell. 1999;10:1309–1323. doi: 10.1091/mbc.10.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luck-Vielmetter D, Schleicher M, Grabatin B, Wippler J, Gerisch G. FEBS Lett. 1990;269:239–243. doi: 10.1016/0014-5793(90)81163-i. [DOI] [PubMed] [Google Scholar]
- 25.Sabry J H, Moores S L, Ryan S, Zang J H, Spudich J A. Mol Biol Cell. 1997;8:2605–2615. doi: 10.1091/mbc.8.12.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berlot C H, Spudich J A, Devreotes P N. Cell. 1985;43:307–314. doi: 10.1016/0092-8674(85)90036-4. [DOI] [PubMed] [Google Scholar]
- 27.Steimle P, Yumura S, Cote G, Medley Q, Polyakov M, Leppert B, Egelhoff T. Curr Biol. 2001;11:708–713. doi: 10.1016/s0960-9822(01)00182-8. [DOI] [PubMed] [Google Scholar]
- 28.Egelhoff T T, Lee R J, Spudich J A. Cell. 1993;75:363–371. doi: 10.1016/0092-8674(93)80077-r. [DOI] [PubMed] [Google Scholar]
- 29.Stites J, Wessels D, Uhl A, Egelhoff T, Shutt D, Soll D R. Cell Motil Cytoskeleton. 1998;39:31–51. doi: 10.1002/(SICI)1097-0169(1998)39:1<31::AID-CM4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 30.Ruppel K M, Uyeda T Q P, Spudich J A. J Biol Chem. 1994;269:18773–18780. [PubMed] [Google Scholar]
- 31.Tang L, Ammann R, Gao T, Gomer R H. J Biol Chem. 2001;276:27663–27669. doi: 10.1074/jbc.M102205200. [DOI] [PubMed] [Google Scholar]
- 32.Chung C Y, Firtel R A. J Cell Biol. 1999;147:559–576. doi: 10.1083/jcb.147.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pang K M, Lee E, Knecht D A. Curr Biol. 1998;8:405–408. doi: 10.1016/s0960-9822(98)70159-9. [DOI] [PubMed] [Google Scholar]
- 34.Morrissey J H, Devine K M, Loomis W F. Dev Biol. 1984;103:414–424. doi: 10.1016/0012-1606(84)90329-4. [DOI] [PubMed] [Google Scholar]
- 35.Schwarz E C, Neuhaus E M, Kistler C, Henkel A W, Soldati T. J Cell Sci. 2000;113:621–633. doi: 10.1242/jcs.113.4.621. [DOI] [PubMed] [Google Scholar]
- 36.Chen T L, Wolf W A, Chisholm R L. Development (Cambridge, UK) 1998;125:3895–3903. doi: 10.1242/dev.125.19.3895. [DOI] [PubMed] [Google Scholar]
- 37.Wilkins A, Khosla M, Fraser D J, Spiegelman G B, Fisher P R, Weeks G, Insall R H. Genes Dev. 2000;14:1407–1413. [PMC free article] [PubMed] [Google Scholar]
- 38.Ayscough K. Methods Enzymol. 1998;298:18–25. doi: 10.1016/s0076-6879(98)98004-1. [DOI] [PubMed] [Google Scholar]
- 39.Konzok A, Weber I, Simmeth E, Hacker U, Maniak M, Muller-Taubenberger A. J Cell Biol. 1999;146:453–464. doi: 10.1083/jcb.146.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parent C A, Blacklock B J, Froehlich W M, Murphy D B, Devreotes P N. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 41.Maniak M, Rauchenberger R, Albrecht R, Murphy J, Gerisch G. Cell. 1995;83:915–924. doi: 10.1016/0092-8674(95)90207-4. [DOI] [PubMed] [Google Scholar]
- 42.Hacker U, Albrecht R, Maniak M. J Cell Sci. 1997;110:105–112. doi: 10.1242/jcs.110.2.105. [DOI] [PubMed] [Google Scholar]
- 43.Hall A L, Schlein A, Condeelis J. J Cell Biochem. 1988;37:285–299. doi: 10.1002/jcb.240370304. [DOI] [PubMed] [Google Scholar]
- 44.Chu Q A, Fukui Y. Cell Motil Cytoskeleton. 1996;35:254–268. doi: 10.1002/(SICI)1097-0169(1996)35:3<254::AID-CM7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 45.Noegel A A, Rapp S, Lottspeich F, Schleicher M, Weeds A G. J Cell Biol. 1989;109:607–618. doi: 10.1083/jcb.109.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Leeuwen F, van Delft S, Kain H, van der Kammen R, Collard J. Nat Cell Biol. 1999;1:242–248. doi: 10.1038/12068. [DOI] [PubMed] [Google Scholar]
- 47.Angst B, Marcozzi C, Magee A. J Cell Sci. 2001;114:629–641. doi: 10.1242/jcs.114.4.629. [DOI] [PubMed] [Google Scholar]
- 48.Parent C A, Devreotes P N. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 49.Liu G, Newell P C. J Cell Sci. 1991;98:483–490. doi: 10.1242/jcs.98.4.483. [DOI] [PubMed] [Google Scholar]
- 50.Liu G, Newell P C. J Cell Sci. 1994;107:1737–1743. doi: 10.1242/jcs.107.7.1737. [DOI] [PubMed] [Google Scholar]
- 51.Kuwayama H, Ecke M, Gerisch G, Van Haastert P J. Science. 1996;271:207–209. doi: 10.1126/science.271.5246.207. [DOI] [PubMed] [Google Scholar]
- 52.Dembinsky A, Rubin H, Ravid S. J Cell Biol. 1996;134:911–921. doi: 10.1083/jcb.134.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silveira L A, Smith J L, Tan J L, Spudich J A. Proc Natl Acad Sci USA. 1998;95:13000–13005. doi: 10.1073/pnas.95.22.13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Lozanne A, Spudich J A. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 55.Wessels D, Soll D R, Knecht D, Loomis W F, De Lozanne A, Spudich J. Dev Biol. 1988;128:164–177. doi: 10.1016/0012-1606(88)90279-5. [DOI] [PubMed] [Google Scholar]
- 56.Manstein D J, Ruppel K M, Spudich J A. Science. 1989;246:656–658. doi: 10.1126/science.2530629. [DOI] [PubMed] [Google Scholar]