Abstract
Background
Theileria annulata, a tick-borne protozoan that causes tropical theileriosis, poses a serious threat to livestock production in endemic regions. The emergence of resistance to buparvaquone, the primary chemotherapeutic treatment, has been attributed to acquired mutations in the cytochrome b (Cytb) gene, with identical resistance-associated polymorphisms observed in both laboratory-adapted strains and field isolates from China.
Methods
A dual probe-specific real-time polymerase chain reaction (PCR) assay was developed to detect point mutations in the Cytb gene. The specificity, sensitivity, and reproducibility of the assay were validated, and its field applicability was evaluated via cattle blood samples (n = 531) collected from five endemic Chinese provinces.
Results
Six point mutations were identified in the Cytb gene, and the developed dual probe-specific real-time PCR assay simultaneously detected T. annulata infection and distinguished between the buparvaquone-sensitive and buparvaquone-resistant genotypes. The assay demonstrated a detection limit of 1 × 101 copies/μl, high specificity, and satisfactory repeatability, with results consistent with those of Sanger sequencing. Field screening revealed a 21.7% (115/531) prevalence of T. annulata and a 4.3% (23/531) occurrence of resistant genotypes. Moreover, two-dimensional scatterplot visualization enabled clear genotype discrimination without post-PCR processing.
Conclusions
The developed dual probe-specific real-time PCR assay enables efficient detection of buparvaquone-resistant genotypes, providing important implications for guiding the treatment of tropical theileriosis and enhancing epidemiological surveillance of emerging resistance in endemic regions.
Graphic abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-025-06884-y.
Keywords: Buparvaquone resistance, Cytochrome b, Real-time PCR, SNP genotyping, Theileria annulata
Background
Theileria annulata is a highly pathogenic tick-borne protozoan parasite and the primary causative agent of bovine tropical theileriosis. This widespread disease poses serious threats to cattle health and results in substantial economic losses across Southern Europe, North Africa, and Asia, where approximately 250 million cattle are at risk [1]. The presence of multiple cattle breeds and buffaloes remains a major impediment to livestock development in many African and Asian regions [2, 3]; its complex epidemiology involves intricate interactions among hosts, pathogens, and vectors, which are further influenced by livestock management practices, agroecological factors, and sociopolitical conditions [4]. Buparvaquone is widely used to treat bovine tropical theileriosis owing to its high efficacy during the early stages of infection [5].
Currently, resistance to buparvaquone in T. annulata is attributed primarily to mutations in the TaPIN1 gene [6], which has also been widely used as a molecular target for detecting drug resistance. Several studies have reported treatment failure in T. annulata infections associated with these mutations, including a recent identification of a similar TaPIN1 mutation in a Chinese isolate [7]. A TaqMan probe-based real-time PCR assay was previously developed to investigate the prevalence of TaPIN1 mutations in Chinese isolates, providing a specific, sensitive, and reproducible method for detecting resistant TaPIN1 variants [8]. In addition to TaPIN1, mutations in the cytochrome b (Cytb) gene have emerged as key markers for buparvaquone resistance in T. annulata [9]. These resistance mechanisms are primarily linked to nonsynonymous Cytb mutations [10], such as methionine-to-isoleucine substitutions at position 128 [11], which disrupt drug binding within the mitochondrial electron transport chain and reduce treatment efficacy. Mutations at critical binding sites, particularly the Q01 and Q02 regions of Cytb, substantially diminish buparvaquone activity, thereby conferring resistance in infected cattle [7, 12].
To address this diagnostic challenge, advanced molecular techniques such as the TaqMan SNP real-time PCR assay utilizing minor groove binder (MGB) probes have been developed [13], offering increased sensitivity and specificity [14, 15]. TaqMan SNP probes have been successfully applied in various fields, including the identification of fluoroquinolone-resistant Mycoplasma bovis and oseltamivir-resistant influenza A, underscoring their versatility and precision in the detection of drug-resistant pathogens [16, 17].
In this study, we aimed to construct a dual probe-specific real-time PCR assay employing TaqMan MGB probes to target single-nucleotide polymorphisms (SNPs) in the Cytb gene for the simultaneous detection of T. annulata infection and buparvaquone resistance. By leveraging the high specificity of MGB probes for key drug resistance mutations, this approach integrates pathogen detection and resistance profiling into a single, streamlined assay. This method not only improves diagnostic accuracy and efficiency but also provides a robust tool for managing bovine tropical theileriosis, offering substantial improvements in field diagnostics and enabling timely, evidence-based disease control strategies in endemic regions.
Methods
Cells and blood samples
TaNM1 and TaXJS, two bovine lymphocyte cell lines infected with T. annulata, were used in this study. Specifically, TaNM1 harbors a buparvaquone-susceptible strain (T. annulata-bss), whereas TaXJS harbors a buparvaquone-resistant strain (T. annulata-brs) [18]. The genomic DNA of T. sinensis, T. orientalis, B. bigemina, and B. bovis were provided by the Vector and Vector-borne Diseases (VVBD) Team, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (CAAS), China, to validate the specificity of the assay.
Field cattle blood samples (n = 531) were collected from five different provinces in China, and the DNA of the blood samples was extracted via the TIANamp blood DNA kit (Tiangen Biotech Co. Ltd; Beijing; China) according to the manufacturer’s protocol.
TaqMan-MGB probes and primers
The primers TaCytb-F/R used to amplify the full-length sequences of the Cytb gene (1092 bp) of TaNM1 and TaXJS were designed according to the standard protocol [19]. The published primer, Cytb-F/R, was used for the detection of T. annulata infection in the PCR assay (Additional File 1) [20]. The Ankara strain (XM949625.1) was used as reference sequence of the T. annulata Cytb gene for alignment with the sequenced TaNM1, TaXJS, and reported mutated strains [12, 21]. The 115-bp fragment of the T. annulata Cytb gene was amplified via primers (SNP-F/R) and TaqMan-MGB probes (Probe-NM1/XJS). Both the primers and probes were designed by SnapGene according to the standard protocol [22]. The probes were labeled with VIC/FAM dyes recognizing the ACT/ACC genotype, respectively. The details are presented as Table 1. The primers and probes were custom synthesized through outsourcing from Beijing Tsingke Biotech Co., Ltd.
Table 1.
Probes and primers used in the present study
| Primer/probe | Sequence (5′–3′) | Product size (bp) |
|---|---|---|
| TaCytb-F | ATGAATTTGTTTAACTCACATTTGC | 1092 bp |
| TaCytb-R | TTATGCACGAACTCTTGCAGAGTC | |
| Cytb-F | TGGTCTTGGTATTCTGGTGTT | 393 bp, Junlong et al. [20] |
| Cytb-R | GCCAATGGATTTGAACTTCC | |
| SNP-F | TGGTCTTGGTATTCTGGTGTT | 115 bp |
| SNP-R | AACCACCTATGACTGTAGCT | |
| Probe-NM1 | VIC-AGTATAGCAACTGCTT-MGB | |
| Probe-XJS | FAM-AGTATAGCAACCGCTT-MGB |
Construction of standard plasmids
The standard plasmid was constructed from the complete Cytb genes of T. annulata-bss and T. annulata-brs. The Cytb genes were amplified, cloned into the pGEM-T Easy vector (Progema, USA), and sequenced by Beijing Tsingke Biotech Co., Ltd. (Beijing, China), with the sequences subsequently validated [23]. The standard plasmids were labeled T-XJS and T-NM1, respectively (Additional File 2).
The T-XJS and T-NM1 plasmids were continuously diluted tenfold from 1 × 108 to 1 copies/μl. The copy number was calculated via the formula [copy number = recombinant plasmid concentration × Avogadro constant × 10–9/(660 bp × total size of the pGEM-T Easy vector)] [23].
Reaction conditions and optimization
The dual probe-specific real-time PCR assay was developed, and the final reaction mixtures consisted of 10 μl of 2 × ChemQ Geno-SNP Probe Master Mix (Vazyme, China), 1.8 μl of each primer (10 μM), 2 μl of DNA template, and 0.4 μl of each probe. For screening optimization, probe-NM1 and probe-XJS were systematically evaluated via stepwise concentration gradients (50–400 nM). Incremental titration at 50 nM intervals was applied to establish dose‒response relationships. Finally, nuclease-free water was added to a final volume of 20 μl. Amplification was performed on a QuantStudio 5 instrument (Applied Biosystems, USA), and the two-step protocol conditions were as follows: 30 s for DNA denaturation at 95 °C followed by 45 cycles of 95 °C for 10 s, 60 °C for 30 s. The fluorescence signal was collected at the end of each cycle.
Establishment of standard curves and repeatability of tests
Different concentrations of plasmid ranging from 1 × 108 to 1 × 103 copies/μl were used as templates for dual probe-specific real-time PCR to generate standard curves, on the basis of the calculated R2 (correlation coefficient), E value (amplification efficiency), and standard equation. Three independent tests were performed, and each reaction was repeated three times. The coefficient of variation (CV) of the Ct values was used to estimate repeatability.
Sensitivity and specificity tests
The sensitivity of the standardized method in this study was assessed by dual probe-specific real-time PCR amplification with plasmid templates ranging from 1 × 103 to 1 copy/μl. To assess the specificity of the test, the genomic DNA of T. sinensis, T. orientalis, B. bigemina, and B. bovis was used as a template.
Dual probe-specific real-time PCR assay for the detection of T. annulata in field blood samples
We collected 531 field cattle blood samples from five different provinces of China and applied our assay to detect T. annulata-bss/brs. Positive samples were sequenced and validated against sequences from the NCBI database. The comparison tests were performed via a previously published method to detect T. annulata-brs [8] and a method in which the Cytb gene was targeted to detect T. annulata [20].
Results
Detection of SNPs
Six point mutations located at positions 234, 348, 417, 753, 843, and 870 of the Cytb gene were identified (Fig. 1). The point mutations located at positions 234, 348, 417, and 870 have been demonstrated in previous studies [12, 21, 24], whereas positions 753 and 843 were newly identified in this study.
Fig. 1.
Nucleotide alignment of the T. annulata Cytb gene. The mutated nucleotides are marked in red
Primer probe specificity validation and concentration screening
A dual probe-specific real-time PCR genotyping assay was performed to validate the specificity of the primers and probes. Clear genotyping results were obtained, and no nonspecific amplification was observed (Fig. 2). On the basis of the Ct value (18.59) and analysis of the amplification curves, the optimal concentrations for probe-XJS and probe-NM1 were 150 nM and 200 nM, respectively (Fig. 3).
Fig. 2.
Primer probe specificity validation. The green dots represent the homozygous FAM/FAM, and the template was T-XJS. The blue dots represent the homozygous VIC/VIC, and the template was T-NM1. The red dots represent the heterozygous FAM/VIC, and the template was a mixture of T-XJS and T-NM1. The black X represents undetermined, and the template was nuclease-free water
Fig. 3.
Probe concentration screening. The numbers in the green and blue parts represent the concentrations of probe-XJS and probe-NM1, respectively
Establishment of standard curves and repeatability tests
The standard curves for T-XJS (y = − 3.4506x + 42.713, R2 = 0.9994, E value = 94.90%) and T-NM1 (y = − 3.5583x + 42.482, R2 = 0.9991, E value = 91.00%) demonstrated strong linear correlations between the Ct values and corresponding copy numbers (R2 > 0.999) of both amplifications (Fig. 4).
Fig. 4.
Standard curve established. Dual probe-specific real-time PCR amplification with serial dilutions of T-XJS (green color, FAM fluorescence channel) and T-NM1 (blue color, VIC fluorescence channel)
Repeatability of the real-time PCR assay
Repeatability was evaluated by testing different concentrations of T-XJS and T-NM1. The intra-assay and inter-assay CVs of the Ct values ranged from 0.10% to 0.92% and from 0.23% to 1.93%, respectively (Table 2). These results indicate that the method established in this study has satisfactory repeatability.
Table 2.
Repeatability test of the real-time PCR assay
| Pathogen | Concentration (copies/μL) | Intra-assay CV | Inter-assay CV | ||
|---|---|---|---|---|---|
| X ± SD | CV (%) | X ± SD | CV (%) | ||
| TaXJS | 108 | 14.93 ± 0.03 | 0.21 | 14.87 ± 0.12 | 0.80 |
| 107 | 18.55 ± 0.03 | 0.18 | 18.51 ± 0.08 | 0.44 | |
| 106 | 22.21 ± 0.03 | 0.15 | 22.19 ± 0.05 | 0.23 | |
| 105 | 25.52 ± 0.10 | 0.38 | 25.49 ± 0.10 | 0.41 | |
| 104 | 29.01 ± 0.10 | 0.35 | 28.93 ± 0.17 | 0.59 | |
| 103 | 32.15 ± 0.09 | 0.29 | 32.09 ± 0.14 | 0.43 | |
| TaNM1 | 108 | 14.05 ± 0.06 | 0.40 | 14.29 ± 0.25 | 1.78 |
| 107 | 17.50 ± 0.16 | 0.92 | 17.59 ± 0.20 | 1.16 | |
| 106 | 20.91 ± 0.09 | 0.42 | 21.15 ± 0.41 | 1.93 | |
| 105 | 25.00 ± 0.02 | 0.10 | 24.94 ± 0.11 | 0.46 | |
| 104 | 28.34 ± 0.05 | 0.19 | 28.43 ± 0.16 | 0.57 | |
| 103 | 31.64 ± 0.20 | 0.64 | 31.51 ± 0.28 | 0.90 | |
Sensitivity of the real-time PCR assay
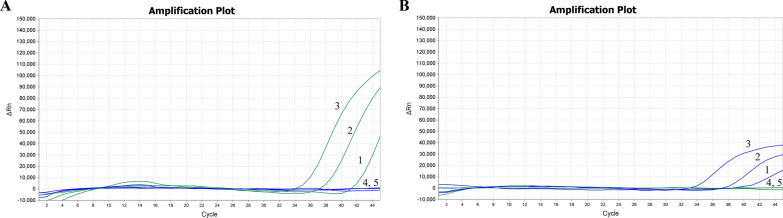
The results indicated that the method established in this study could detect T. annulata-bss and T. annulata-brs at concentrations as low as 1 × 101 copies/μl (Fig. 5).
Fig. 5.
Sensitivity test of the dual probe-specific real-time PCR assay. A T. annulata-brs, B T. annulata-bss, 1: 1 × 101 copies/μl, 2: 1 × 102 copies/μl, 3: 1 × 103 copies/μl, 4: 1 copies/μl, 5: negative control
Specificity of the real-time PCR assay
Templates of T. annulata-bss, T. annulata-brs, T. sinensis, T. orientalis, B. bigemina, and B. bovis were used in the dual probe-specific real-time PCR assay. FAM and VIC fluorescent signals were detected only when T. annulata-brs-positive clinical samples and T. annulata-bss-positive clinical samples were used as templates (Fig. 6). No fluorescent signals were observed for other pathogens that are common in bovine blood, viz. T. sinensis, T. orientalis, B. bigemina, and B. bovis. These results confirm the high specificity of the method developed in this study.
Fig. 6.
Specificity of the dual probe-specific real-time PCR assay. A T. annulata-brs (1), other pathogens and negative control (2–7), B T. annulata-bss (1), and other pathogens and negative control (2–7)
Field sample detection
The results revealed that 92 samples were positive for T. annulata-bss and that 23 samples were positive for T. annulata-brs (Table 3, Fig. 7.), with no coinfection detected. The overall infection rate of T. annulata was 21.7% (115/531), of which T. annulata-brs accounted for 20% (23/115) of the total number of infections. A subset of positive samples was randomly selected for validation via Sanger sequencing, and the results were consistent with those obtained via the dual probe-specific assay. Comparison test results revealed an equivalent detection rate for T. annulata-brs, whereas the overall detection rate for T. annulata was higher than that reported using a previously described method (infection rate = 17.1%, 91/531) [8, 20
Table 3.
Results of field sample detection
| Province | Number of samples | Number of T. annulata -bss |
Number of T. annulata -brs |
Number of T. annulata-brs Previous method Su et al. [8] |
Number of T. annulata Previous method Junlong et al. [20] |
|---|---|---|---|---|---|
| Present study | |||||
| Xinjiang | 227 | 59 | 23 | 23 | 71 |
| Ningxia | 33 | 4 | 0 | 0 | 0 |
| Qinghai | 103 | 11 | 0 | 0 | 11 |
| Gansu | 65 | 16 | 0 | 0 | 9 |
| Inner Mongolia | 53 | 2 | 0 | 0 | 0 |
| Total | 531 | 92 | 23 | 23 | 91 |
Fig. 7.
Scatter plot of SNP genotyping results from field sample detection and Sanger sequencing. The green dots (homozygous FAM/FAM) represent T. annulata-brs positive, the blue dots (homozygous VIC/VIC) represent T. annulata-bss positive, the red dots (heterozygous FAM/VIC) represent coinfection T. annulata-bss and T. annulata-brs, and the black X (undetermined) represents T. annulata-bss and T. annulata-brs negative
Discussion
Although buparvaquone remains a frequently used treatment for bovine tropical theileriosis caused by T. annulata, recent genomic surveillance has revealed alarming therapeutic failures in endemic regions. The emergence of point mutations in the Cytb and/or TaPIN1 genes is thought to contribute to the development of buparvaquone resistance [7]. These dual-target mutations likely interfere with buparvaquone binding through steric hindrance, representing a concerning mechanism underlying the evolution of drug resistance.
Although Sanger sequencing remains the gold standard for detecting resistance-associated mutations, its reliance on post-PCR processing is time-consuming and laborious [25]. Recent advances have led to the development of a TaPIN1-targeted real-time PCR assay with a detection limit of 2.72 × 101 copies/μl (higher than our method) for identifying T. annulata -brs [8]. While this previous method achieved a comparable detection rate in field-collected cattle blood samples (likely because of the high load of T. annulata in samples, which is above both assays’ sensitivity thresholds), it has a critical limitation: the inability to distinguish coinfections, which is a common epidemiological challenge in endemic areas. The Cytb gene has been shown in previous studies to be a highly specific and sensitive target [26] that is widely used in phylogenetic studies [27], and TaqMan real-time PCR is considered to be one of the most convenient and accurate approaches for SNP genotyping [28]. In this study, we identified six point mutations in the Cytb gene in T. annulata-brs strains and developed a dual probe-specific real-time PCR assay targeting a 115-bp fragment of the T. annulata Cytb gene. The assay employs TaqMan-MGB probes for rapid, one-step differentiation between T. annulata-bss (genotype: ACT) and T. annulata-brs (genotype: ACC). Probes were labelled at the 5′ end with VIC (T. annulata-bss)/FAM (T. annulata-brs) dyes and nonfluorescent quenchers (MGB) at the 3′ end to improve sensitivity and specificity. This design enables both genotypes to be detected in a single reaction, making the assay simple to perform and suitable for large-scale genotyping applications.
Molecular surveillance data have revealed that T. annulata is the predominant and clinically significant piroplasm species in Chinese cattle and cocirculates with T. sinensis and T. orientalis along transnational transmission corridors [29]. In this study, we identified a geographically clustered emergence of T. annulata-brs in Xinjiang, a strategically linked area of China bordering eight countries, including Pakistan, Kazakhstan, and Tajikistan, through the Tianshan Border Trade Corridor. This epidemiological pattern has emerged despite the absence of buparvaquone use in local livestock, suggesting potential resistance gene introgression through two key mechanisms: transboundary movement of asymptomatic carrier cattle via the Central Asian livestock trade network [30, 31] and vector-mediated spread by Hyalomma tick species migrating along seasonal pastures. The 100% concordance between our TaqMan-MGB assay and Sanger sequencing results confirms the assay’s diagnostic robustness. These findings not only validate the assay’s suitability for large-scale resistance monitoring but also raise a critical alert that the expanding presence of drug-resistant T. annulata genotypes along the northwestern frontier of China may signal broader antimicrobial resistance proliferation in piroplasmosis-endemic regions across Eurasia.
This multiplex detection platform represents a significant advance over workflows for transboundary resistance surveillance; it enables one-hour single-tube dual-channel detection (48 h for Sanger sequencing) and simultaneous discrimination of wild-type (ACT_ (T. annulata-bss)) and mutant (ACC_ (T. annulata-brs)) haplotypes at sensitivity thresholds ≤ 101 copies/μl. This operational advantage facilitates real-time geospatial monitoring of resistance alleles, a capability that is particularly critical along China’s 5700-km border with Central Asia.
Conclusions
We identified six point mutations in the Cytb gene and developed a dual probe-specific TaqMan SNP real-time PCR assay for the rapid detection of Cytb mutations associated with buparvaquone resistance in T. annulata. This assay provides implications for guiding the treatment of tropical theileriosis and for enhancing the epidemiological surveillance of emerging buparvaquone resistance in endemic regions.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- SNP
Single-nucleotide polymorphism
- Cytb
Cytochrome b
- MGB
Minor groove binder
- TaPIN1
T. annulata prolyl isomerase I gene
- NCBI
National Center for Biotechnology Information
- CAAS
Chinese Academy of Agricultural Sciences
- VVBD
Vector and Vector-borne Diseases
Author contributions
W.L., H.Y., and J.L. planned the research. S.Z. and J.W. collected the samples. J.C. conducted the experiments. J.C. and Y.C. drafted the manuscript. H.Y. and G.G. revised the manuscript. All authors read and approved the final manuscript.
Funding
The study was financially supported by the Science Fund for Creative Research Groups (22JR5RA024), Special Project (22CX8NA011) and Natural Science Foundation (22JR5RA031) of Gansu Province, National Key Research and Development Program of China (2022YFD1800200), NSFC (№31972701), the Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2021-LVRI), NBCITS (CARS-37), and National Parasitic Resources Center (NPRC-2019–194-30).
Availability of data and materials
The data supporting the conclusions of this article are included within the article.
Declarations
Ethics approval and consent to participate
No human subjects were involved in this study. In the present study, all of the animal experiments were approved by the Animal Ethics Committee of the Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences. All experimental animals used were dealt with according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China (SYXK2010-0001).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jin Che and Yijun Chai contributed equally to this work.
Contributor Information
Guiquan Guan, Email: guanguiquan@caas.cn.
Hong Yin, Email: yinhong@caas.cn.
Wei Li, Email: neaulw@gmail.com.
References
- 1.Liu J, Guan G, Yin H. Theileriaannulata. Trends Parasitol. 2022;38:265–6. [DOI] [PubMed] [Google Scholar]
- 2.El-Alfy ES, Abbas I, Elseadawy R, Saleh S, Elmishmishy B, El-Sayed S, et al. Global prevalence and species diversity of tick-borne pathogens in buffaloes worldwide: a systematic review and meta-analysis. Parasit Vectors. 2023;16:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu Z, Wang H, Wang T, Sun W, Yang X, Liu J. Tick-borne pathogens and the vector potential of ticks in China. Parasit Vectors. 2015;8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gharbi M, Darghouth MA, Elati K, Al-Hosary A, Ayadi O, Salih DA, et al. Current status of tropical theileriosis in Northern Africa: a review of recent epidemiological investigations and implications for control. Transbound Emerg Dis. 2020;67:8–25. [DOI] [PubMed] [Google Scholar]
- 5.Hashemi-Fesharki R. Chemotherapeutic value of parvaquone and buparvaquone against Theileria annulata infection of cattle. Res Vet Sci. 1991;50:204–7. [DOI] [PubMed] [Google Scholar]
- 6.Salim B, Chatanga E, Jannot G, Mossaad E, Nakao R, Weitzman JB. Mutations in the TaPIN1 peptidyl prolyl isomerase gene in Theileria annulata parasites isolated in Sudan. Int J Parasitol Drugs Drug Resist. 2019;11:101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rashid M, Hayat MH, Zahra N, Khan MS, Nadeem M, et al. Systematic review on buparvaquone resistance associated with non-synonymous mutation in drug binding genes site of Theileria annulate. Vet Parasitol. 2024;332:110321. [DOI] [PubMed] [Google Scholar]
- 8.Su S, Zhao S, Liu J, Zhang C, Zhu H, Guan G, et al. Establishment and application of TaqMan real-time PCR method for detection of Theileria annulata resistant to buparvaquone. Vet Parasitol. 2024;328:110183. [DOI] [PubMed] [Google Scholar]
- 9.Ali Q, Zahid O, Mhadhbi M, Jones B, Darghouth MA, Raynes G, et al. Genetic characterisation of the Theileria annulata cytochrome b locus and its impact on buparvaquone resistance in bovine. Int J Parasitol Drugs Drug Resist. 2022;20:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharifiyazdi H, Namazi F, Oryan A, Shahriari R, Razavi M. Point mutations in the Theileria annulata cytochrome b gene is associated with buparvaquone treatment failure. Vet Parasitol. 2012;187:431–5. [DOI] [PubMed] [Google Scholar]
- 11.Tajeri S, Chattopadhyay D, Langsley G, Nijhof AM. A Theileria annulata parasite with a single mutation, methionine 128 to isoleucine (M128I), in cytochrome B is resistant to buparvaquone. PLoS ONE. 2024;19:e0299002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacilarlioglu S, Bilgic HB, Bakirci S, Tait A, Weir W, Shiels B, et al. Selection of genotypes harbouring mutations in the cytochrome b gene of Theileria annulata is associated with resistance to buparvaquone. PLoS ONE. 2023;18:e0279925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Zhang S, Dang S, Gao L, Li G, Cheng D, et al. Establishment of an A/T-rich specifically MGB probe digital droplet PCR assays based on SNP for Brucella wild strains and vaccine strains. Diagn Microbiol Infect Dis. 2024;110:116432. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, He J, Liu J, You Y, Yuan L, Wen Y. Nested PCR and the TaqMan SNP genotyping assay enhanced the sensitivity of drug resistance testing of Mycobacterium leprae using clinical specimens of leprosy patients. PLoS Negl Trop Dis. 2019;13:e0007946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wadhwa A, Wilkins K, Gao J, Condori CR, Gigante CM, Zhao H, et al. A Pan-Lyssavirus TaqMan real-time RT-PCR assay for the detection of highly variable Rabies virus and other Lyssaviruses. PLoS Negl Trop Dis. 2017;11:e0005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben SM, Mikula I, Gerchman I, Lysnyansky I. Development and evaluation of a novel single-nucleotide-polymorphism real-time PCR assay for rapid detection of fluoroquinolone-resistant Mycoplasma bovis. J Clin Microbiol. 2010;48:2909–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong S, Pabbaraju K, Wong A, Fonseca K, Drews SJ. Development of a real-time RT-PCR assay for detection of resistance to oseltamivir in influenza A pandemic (H1N1) 2009 virus using single nucleotide polymorphism probes. J Virol Methods. 2011;173:259–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rashid M, Guan G, Luo J, Zhao S, Wang X, Rashid MI, et al. Establishment and expression of cytokines in a Theileria annulata-infected bovine B cell line. Genes (Basel). 2019;10:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dieffenbach CW, Lowe TM, Dveksler GS. General concepts for PCR primer design. PCR Methods Appl. 1993;3:S30–7. [DOI] [PubMed] [Google Scholar]
- 20.Junlong L, Li Y, Liu A, Guan G, Xie J, Yin H, et al. Development of a multiplex PCR assay for detection and discrimination of Theileria annulata and Theileria sergenti in cattle. Parasitol Res. 2015;114:2715–21. [DOI] [PubMed] [Google Scholar]
- 21.Afshari A, Tavassoli M, Esmaeilnejad B, Habibi G, Esmaeilnia K. Molecular Characterization and phylogenetic analysis of pathogenic Theileria spp. isolated from cattle and sheep based on cytochrome b gene in Iran. Arch Razi Inst. 2021;76:243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez A, Rodriguez M, Cordoba JJ, Andrade MJ. Design of primers and probes for quantitative real-time PCR methods. Methods Mol Biol. 2015;1275:31–56. [DOI] [PubMed] [Google Scholar]
- 23.Cao T, Liu J, Li Z, Shi K, Shi M, Li Y, et al. Establishment and application of a qPCR diagnostic method for Theileria annulata. Parasitol Res. 2022;121:973–80. [DOI] [PubMed] [Google Scholar]
- 24.Chatanga E, Mosssad E, Abdo AH, Amin AS, Katakura K, Nakao R, et al. Evidence of multiple point mutations in Theileria annulata cytochrome b gene incriminated in buparvaquone treatment failure. Acta Trop. 2019;191:128–32. [DOI] [PubMed] [Google Scholar]
- 25.Malnati MS, Biswas P, Ugolotti E, Di Marco E, Sironi F, Parolini F, et al. A fast and reliable method for detecting SNP rs67384697 (Hsa-miR-148a binding site) by a single run of allele-specific real-time PCR. HLA. 2020;96:312–22. [DOI] [PubMed] [Google Scholar]
- 26.Bilgic HB, Karagenc T, Shiels B, Tait A, Eren H, Weir W. Evaluation of cytochrome b as a sensitive target for PCR based detection of T. annulata carrier animals. Vet Parasitol. 2010;174:341–7. [DOI] [PubMed] [Google Scholar]
- 27.Meyer A. Shortcomings of the cytochrome b gene as a molecular marker. Trends Ecol Evol. 1994;9:278–80. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Cui G, Li Z, Wang H, Ding H, Wang DW. Comparison of high-resolution melting analysis, TaqMan Allelic discrimination assay, and sanger sequencing for clopidogrel efficacy genotyping in routine molecular diagnostics. J Mol Diagn. 2013;15:600–6. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Chen YY, Liu G, Lyu C, Hu Y, An Q, et al. Prevalence of Theileria in cattle in China: a systematic review and meta-analysis. Microb Pathog. 2022;162:105369. [DOI] [PubMed] [Google Scholar]
- 30.Gebrekidan H, Abbas T, Wajid M, Ali A, Gasser RB, Jabbar A. Molecular characterisation of Theileria orientalis in imported and native bovines from Pakistan. Infect Genet Evol. 2017;47:19–25. [DOI] [PubMed] [Google Scholar]
- 31.Hassan MA, Liu J, Rashid M, Iqbal N, Guan G, Yin H, et al. Molecular survey of piroplasm species from selected areas of China and Pakistan. Parasit Vectors. 2018;11:457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the conclusions of this article are included within the article.