Abstract
Purpose
Accurate and robust segmentation of anatomical structures in brain MRI provides a crucial basis for the subsequent observation, analysis, and treatment planning of various brain diseases. Deep learning foundation models trained and designed on large-scale natural scene image datasets experience significant performance degradation when applied to subcortical brain structure segmentation in MRI, limiting their direct applicability in clinical diagnosis.
Methods
This paper proposes a subcortical brain structure segmentation algorithm based on Low-Rank Adaptation (LoRA) to fine-tune SAM (Segment Anything Model) by freezing SAM’s image encoder and applying LoRA to approximate low-rank matrix updates to the encoder’s training weights, while also fine-tuning SAM’s lightweight prompt encoder and mask decoder.
Results
The fine-tuned model’s learnable parameters (5.92 MB) occupy only 6.39% of the original model’s parameter size (92.61 MB). For training, model preheating is employed to stabilize the fine-tuning process. During inference, adaptive prompt learning with point or box prompts is introduced to enhance the model’s accuracy for arbitrary brain MRI segmentation.
Conclusion
This interactive prompt learning approach provides clinicians with a means of intelligent segmentation for deep brain structures, effectively addressing the challenges of limited data labels and high manual annotation costs in medical image segmentation. We use five MRI datasets of IBSR, MALC, LONI, LPBA, Hammers and CANDI for experiments across various segmentation scenarios, including cross-domain settings with inference samples from diverse MRI datasets and supervised fine-tuning settings, demonstrate the proposed segmentation algorithm’s generalization and effectiveness when compared to current mainstream and supervised segmentation algorithms.
Keywords: Subcortical structure segmentation, Cross-domain adaption, Fine-tuning SAM, Low-rank adaptation
Introduction
Subcortical brain structures, such as the caudate nucleus, putamen, pallidum, thalamus, amygdala, and hippocampus, are critical for regulating movement, emotion, memory, and learning [1]. Alterations in these structures have been associated with neuropsychiatric disorders, including schizophrenia, depression, and Alzheimer’s disease [2–4]. Accurate segmentation of these regions in brain MRI is crucial for diagnosis and treatment planning. However, challenges such as MR image noise, ambiguous boundaries, small structure sizes, and low grayscale contrast at edges pose significant obstacles for traditional segmentation algorithms. To overcome these limitations, various advanced segmentation algorithms and software tools have been developed [5–7].
Recent advances in deep learning, particularly through Deep Neural Networks (DNNs), have significantly advanced image classification [8–10] and hold promise for precise anatomical segmentation in medical imaging. However, models trained on specific datasets often face challenges in generalizing to new tasks or diverse medical images. General AI models such as ChatGPT [11], SegGPT [12], and SAM (Segment Anything Model) [13] offer potential solutions. SAM, a robust segmentation model, allows users to generate segmentation masks interactively or automatically, gaining attention for its zero-shot learning capabilities across various applications. While SAM’s generalization abilities have been explored in medical image processing [14–17], its effectiveness is constrained by domain-specific challenges, including variations in intensity, color, and texture in medical imaging.
Fine-tuning large models has proven effective in enhancing their adaptability for specialized tasks. Recent studies have adapted SAM for medical image segmentation through fine-tuning techniques. For example, SAM-Med2D [18] integrates adapter technology into SAM’s image encoder, tailoring it for the medical domain and demonstrating strong performance across CT, MRI, X-rays, and diverse medical targets. Similarly, MedSAM [19] fine-tunes SAM on over 200,000 masks across 11 modalities, employing Low-Rank Adaptation (LoRA) while freezing most of SAM’s parameters. Both SAM-Med2D and MedSAM exhibit robust adaptability to medical images but remain limited in segmenting brain structures with low contrast and ambiguous boundaries.
The full fine-tuning method requires training and adjusting all parameters of the entire model, consuming significant computational resources and time. Hu et al. [20] proposed the LoRA method, which is based on the idea of performing implicit low-rank transformations on the weight matrices of large models, making it a parameter-efficient fine-tuning technique. As illustrated in Fig. 1 (a), LoRA seeks to find a (linear) combination of a few dimensions in the original feature space (or matrix), approximating a high-dimensional matrix or dataset through a lower-dimensional matrix. Figures 1 (b) and (c) show the training methods for full parameter fine-tuning and LoRA fine-tuning, respectively; the blue snowflakes represent frozen pre-trained model parameters, while the red flames indicate the learnable parts. In the LoRA framework, two learnable structures, A and B, are added to the pre-trained model. The parameters of these structures are initialized to a Gaussian distribution and zero, respectively, meaning that the additional parameters start at zero during the initial phase of training. The input dimension of A and the output dimension of B are the same as the input and output dimensions of the original model, while the output dimension of A and the input dimension of B are significantly smaller than those of the original model, facilitating low-rank transformations. By freezing the high-dimensional information of the original model in the LoRA approach, only two low-rank matrices need to be trained to inject specific parameters into the original model, enabling low-rank adaptation fine-tuning. The LoRA fine-tuning method not only reduces computational demands and training resources but also retains the powerful performance of the original model.
Fig. 1.
Schematic diagram of low rank transformation process and LoRA fine-tuning
This study fine-tunes SAM with LoRA for segmenting deep brain structures in MRI, providing a foundational segmentation model and evaluation framework for researchers and clinicians involved in deep brain structure analysis. The main contributions of this paper are as follows:
The fine-tuned model follows SAM’s network architecture, including the image encoder, prompt encoder, and mask decoder, updating only a small set of parameters to adapt SAM for brain structure segmentation.
During training, the SAM image encoder’s weights are frozen, and LoRA fine-tuning is used for low-rank updates to the image encoder’s weights, while also fine-tuning SAM’s prompt encoder and mask decoder.
The learnable parameters (5.92 MB) are only 6.39% of the original model size (92.61 MB). Model warming stabilizes fine-tuning, and adaptive prompt learning with point or box prompts enables generalization to any MRI dataset.
The proposed SAM-based segmentation algorithm, trained on five brain MRI datasets (IBSR, MALC, LPBA, Hammers, and CANDI), is compared with mainstream and supervised segmentation algorithms. Results across various scenarios confirm the algorithm’s generalizability and effectiveness.
The rest of the paper is organized as follows: Section “Related work” covers related work. Section “Low-rank adaptation fine-tuning SAM for brain structure segmentation algorithm” presents the details of our proposed low-rank adapted fine-tuning SAM brain structure segmentation algorithm. Section “Experimental results and analysis” provides an analysis of the experimental results, and finally, Section “Conclusion” concludes our work.
Related work
In this section, we present some research related to this work.
SAM architecture
SAM is a foundational image segmentation model designed to generate masks interactively based on prompts like points, boxes, and masks, displaying strong zero-shot generalization due to its training on a large dataset. Built on a Transformer architecture [21], SAM includes an image encoder using Vision Transformer (ViT) [22] to extract embeddings, a prompt encoder for processing input prompts, and a mask decoder for segmentation mask outputs. SAM’s image encoder, adapted from the ViT-based Masked Auto-Encoder (MAE) [23], supports high-resolution images. The prompt encoder processes sparse (points and boxes) and dense (mask) prompts, while the mask decoder combines image and prompt embeddings through self-attention and cross-attention for segmentation.
Training involves focal and Dice loss to handle class imbalance and noise. SAM’s annotation tool enables semi-automatic mask generation, yielding a high-quality segmentation dataset with robust generalization.
Medical image segmentation with foundation models
Foundation models like ChatGPT [24, 25] and SAM [13] have propelled deep learning into a new era, leveraging extensive datasets and complex patterns for generalization and zero-shot learning [26–28]. SAM’s components image encoder, prompt encoder, and mask decoder enable it to generate segmentation masks in real-time, though SAM’s accuracy on medical images is limited due to domain differences like resolution, modality, and quality [14].
Fine-tuning foundation models enhances performance on specific tasks by updating select parameters. This can involve full or parameter-efficient fine-tuning (PEFT), as seen with models like MedSAM [16], which improved segmentation across modalities but still struggled with intricate structures. PEFT approaches such as LoRA, QLoRA, and adapter tuning have been explored to reduce computational costs [17, 18, 29]. For instance, Med-SA [17] fine-tunes SAM with LoRA modules, achieving better results on diverse medical tasks, while SAM-Med2D [18] created a multi-modal dataset for generalization, excelling in the MICCAI 2023 challenge.
While fine-tuning improves SAM’s generalization, deep brain structures pose challenges due to their low contrast and complex shapes. Current models perform well on defined targets but are limited in segmenting complex structures with unclear boundaries.
Low-rank adaptation fine-tuning SAM for brain structure segmentation algorithm
The goal of the proposed algorithm is to learn a brain structure segmentation model with strong cross-domain segmentation performance, suitable for segmentation scenarios where inference samples come from any MRI dataset. This model utilizes a variety of brain MRI datasets with different parameters and styles to perform low-rank adaptation fine-tuning on the foundational SAM model, enabling it to adapt to the deep brain structure segmentation task. The fine-tuning approach eliminates the need for training the model from scratch, reducing computational demands while retaining SAM’s robust segmentation performance.
Overview
Figure 2 illustrates the overall framework of the proposed brain structure segmentation algorithm, which is based on LoRA fine-tuning of SAM. The framework comprises three main components: (1) a LoRA-based fine-tuned image encoder, (2) a prompt encoder capable of learning point or box prompts, and (3) a mask decoder that generates segmentation masks by fusing image and prompt embeddings.
Fig. 2.
Schematic diagram of the proposed algorithm
The proposed model builds upon the SAM architecture, inheriting all parameters of the SAM image encoder while introducing a trainable LoRA module into each Transformer module. The LoRA module first compresses the features output by the Transformer module into a low-rank space and then re-samples the compressed features to match the channel dimensions of the frozen output features. During training, only the default embedding from SAM is used as input to the mask decoder, while the prompt encoder and default embedding are fine-tuned automatically throughout the process.
During inference, when encountering samples from unseen datasets, the proposed model can incorporate interactive prompt information (points or boxes) to improve segmentation accuracy for arbitrary samples. In this case, the prompt encoder outputs both prompt embeddings and default embeddings, which are fed into the mask decoder. The mask decoder consists of a lightweight Transformer decoder that integrates features from the prompt encoder and image encoder, as well as a segmentation head that predicts the final brain structure segmentation results.
Although further fine-tuning of the Transformer module in the decoder using LoRA could significantly reduce the number of updated parameters, the proposed model applies LoRA fine-tuning only to the image encoder. This decision is based on the observation that fine-tuning the lightweight decoder may slightly degrade its performance. The segmentation head outputs multiple segmentation masks, each corresponding to a specific brain structure category. This design leverages prompt information more effectively compared to predicting multiple categories simultaneously, avoiding the challenges associated with interference from multiple prompt features in the segmentation head’s predictions. Finally, bilinear upsampling is applied to align the predicted masks with the input image.
LoRA fine-tuning
Unlike the SAM training process that updates all weight parameters, LoRA allows SAM to update only a small portion of parameters when training on brain MRI images. This not only significantly reduces computational demands but also decreases the complexity of deploying and storing the fine-tuned model, while retaining SAM’s robust segmentation performance. Figure 3 illustrates the schematic of fine-tuning the Transformer module in the image encoder using the LoRA module.
Fig. 3.
Schematic diagram of the transformer block and the LoRA block
The Transformer module includes multi-head self-attention (MHA), feedforward neural networks (FFN), and normalization, dropout, and residual components. The learnable parameters in the Transformer module are found in the QKV (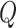 : query,
: query, 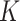 : key,
: key, 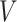 : value) mapping matrices. The self-attention module multiplies the input vectors by the learnable mapping matrices
: value) mapping matrices. The self-attention module multiplies the input vectors by the learnable mapping matrices  ,
, 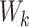 and
and 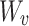 , and then performs the Scaled Dot-Product Attention calculation. Given an input embedding sequence
, and then performs the Scaled Dot-Product Attention calculation. Given an input embedding sequence 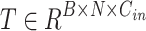 , the output embedding sequence
, the output embedding sequence 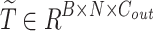 is computed using the mapping matrix
is computed using the mapping matrix 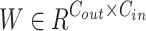 . LoRA assumes that during model adaptive fine-tuning, the updated parameter matrix has a lower intrinsic dimensionality, or low-rank representation, allowing for the fine-tuning of low-rank matrices instead of performing full parameter fine-tuning. For a pretrained model’s weight matrix
. LoRA assumes that during model adaptive fine-tuning, the updated parameter matrix has a lower intrinsic dimensionality, or low-rank representation, allowing for the fine-tuning of low-rank matrices instead of performing full parameter fine-tuning. For a pretrained model’s weight matrix 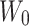 ,
, 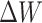 represents the updated weight matrix after fine-tuning for a specific task. Since
represents the updated weight matrix after fine-tuning for a specific task. Since  has a lower intrinsic dimensionality, it can be expressed as the product of two low-rank matrices
has a lower intrinsic dimensionality, it can be expressed as the product of two low-rank matrices 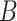 and
and 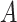 , meaning that only
, meaning that only 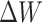 needs to be updated during fine-tuning.
needs to be updated during fine-tuning.
Based on the above analysis, we first freeze the Transformer module in the image encoder to keep the mapping matrix 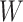 fixed, then add a LoRA bypass for low-rank approximation. As shown in Fig. 3, the LoRA module includes two linear layers,
fixed, then add a LoRA bypass for low-rank approximation. As shown in Fig. 3, the LoRA module includes two linear layers,  and
and  , where
, where 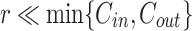 and
and 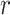 represents the rank of the LoRA module. Thus, the updated layer
represents the rank of the LoRA module. Thus, the updated layer  can be described as follows:
can be described as follows:
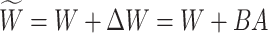 |
1 |
The multi-head self-attention mechanism obtains attention regions by calculating the similarity and weight distribution of matrices  ,
, 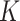 , and
, and 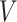 . The formula is as follows:
. The formula is as follows:
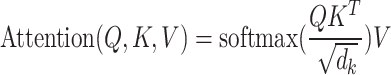 |
2 |
Here,  represents the number of columns in the matrices
represents the number of columns in the matrices 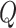 and
and 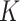 , which is the dimensionality of the vectors. Therefore, LoRA can be applied to the mapping layers of
, which is the dimensionality of the vectors. Therefore, LoRA can be applied to the mapping layers of 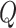 ,
,  , or
, or  to fine-tune the parameters of the Transformer module.
to fine-tune the parameters of the Transformer module.
We apply LoRA fine-tuning to the query 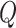 and value
and value  mapping layers; therefore, we can describe the computation of multi-head self-attention as follows:
mapping layers; therefore, we can describe the computation of multi-head self-attention as follows:
 |
3 |
where:
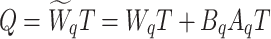 |
4 |
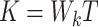 |
5 |
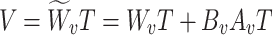 |
6 |
Here, 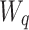 ,
, 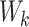 , and
, and 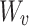 are the frozen parameters from the Transformer module in SAM, while
are the frozen parameters from the Transformer module in SAM, while 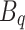 ,
, 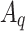 ,
, 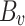 , and
, and  are the fine-tuned parameters of the LoRA module. Finally, the output embedding sequence of the Transformer layer after LoRA fine-tuning is computed as follows:
are the fine-tuned parameters of the LoRA module. Finally, the output embedding sequence of the Transformer layer after LoRA fine-tuning is computed as follows:
 |
7 |
During the fine-tuning training process, only the weight matrix 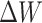 is updated, where
is updated, where 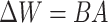 as shown in Eq. (1). Here,
as shown in Eq. (1). Here,  is obtained as the product of two low-rank matrices
is obtained as the product of two low-rank matrices  and
and 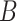 . This means that only
. This means that only  and
and 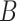 need to be updated during network fine-tuning. The updates are performed via backpropagation using the following gradient rules:
need to be updated during network fine-tuning. The updates are performed via backpropagation using the following gradient rules:
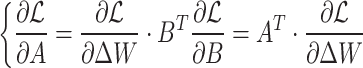 |
8 |
Here, 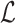 represents the network’s loss function. It is important to note that matrix
represents the network’s loss function. It is important to note that matrix 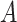 is initialized with random Gaussian values, while matrix
is initialized with random Gaussian values, while matrix 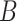 is initialized to zero. This setup ensures that the model starts from the pre-trained weights
is initialized to zero. This setup ensures that the model starts from the pre-trained weights  . During training, as matrix
. During training, as matrix 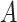 and
and 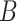 is updated, the adjustments introduced by LoRA accumulate progressively. This approach prevents excessive weight changes in the early training stages, mitigates potential gradient explosions or instability, and thereby stabilizes the model’s learning process.
is updated, the adjustments introduced by LoRA accumulate progressively. This approach prevents excessive weight changes in the early training stages, mitigates potential gradient explosions or instability, and thereby stabilizes the model’s learning process.
Mask decoder
The SAM mask decoder consists of a lightweight Transformer module and a dynamic prediction segmentation head, and we retain the structure of the SAM mask decoder in our brain structure segmentation algorithm. The mask decoder maps the image embeddings output and the prompt embeddings output to get the final segmentation mask. All embeddings continuously optimize their weights during the training process as the decoder updates.
To enhance the model’s segmentation accuracy for any unlearned MRI datasets, the proposed algorithm first employs multiple MRI datasets for supervised fine-tuning of the large model. During supervised fine-tuning, the prompt encoder section uses the default learnable prompt inputs from the SAM model, which are fed into the mask decoder. Sparse prompts employ learnable positional encodings, while dense prompts utilize convolutional encodings. Additionally, learned embeddings are incorporated for each prompt type, making them trainable throughout the fine-tuning process. During inference, adaptive prompt learning based on points or boxes is introduced, allowing the model to quickly locate deep brain structure regions by providing a few target points, background points, or bounding box prompts, effectively improving the speed and accuracy of segmentation. Ultimately, this results in an interactive large model suitable for deep brain structure segmentation.
Figure 4 illustrates the mask decoder schematic, which comprises two Transformer modules and a segmentation layer. Initially, image embeddings from the encoder and prompt embeddings from the input prompts are fed into the Transformer modules for information reconstruction. Each Transformer module performs four steps: (1) prompt embeddings are input into a self-attention layer; (2) prompt embeddings (as queries in the self-attention mechanism) are passed through a cross-attention layer with image embeddings; (3) a point-wise MLP updates each prompt embedding vector; (4) image embeddings (as queries in the self-attention mechanism) go through a cross-attention layer with prompt embeddings, updating the image embeddings using prompt information in the final step. In the segmentation head, image embeddings processed by the Transformer modules are upsampled through two transposed convolution layers (Conv. trans). The prompt embeddings are used again to update the image embeddings by passing them through an MLP that outputs vectors matching the channel dimension of the updated image embeddings. Finally, spatial dot-product operations between the updated image embeddings and the MLP output embeddings yield the segmentation mask for each semantic class. Additionally, after processing through the MLP, the updated embeddings produce the corresponding segmentation confidence scores (IoU). Our model outputs 13 target class segmentation results, covering the background and 12 different target brain structures. Unlike SAM’s coarse predictions, the segmentation head of our algorithm performs accurate predictions for each semantic class, outputting the segmentation map 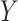 as follows:
as follows:
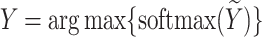 |
9 |
Fig. 4.
Schematic diagram of the mask decoder
Here, 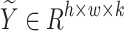 (where
(where 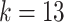 ) represents the semantic masks predicted by the model, corresponding to
) represents the semantic masks predicted by the model, corresponding to  semantic segmentation classes. The softmax and argmax operations are applied along the last channel dimension, respectively.
semantic segmentation classes. The softmax and argmax operations are applied along the last channel dimension, respectively.
Model training strategy
Model warmup
Learning rate warmup is a method proposed in ResNet that begins training with a small learning rate, allowing the model to stabilize gradually before switching to a predefined learning rate. This approach helps the model converge faster and achieve better performance. In Transformer architectures, where model size is significant, gradient vanishing or explosion can occur, making warmup strategies particularly beneficial in stabilizing training. Therefore, warmup is applied before training in this model to stabilize the process and allow early adaptation to brain MRI data. Additionally, inspired by the training strategies of TransUnet [30], an exponential learning rate decay is applied after warmup to facilitate gradual convergence during training.
The learning rate  is adjusted as follows during training:
is adjusted as follows during training:
 |
10 |
Here, 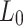 is the initial learning rate,
is the initial learning rate,  is the current iteration count,
is the current iteration count, 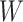 is the number of warmup iterations, and
is the number of warmup iterations, and 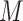 is the maximum number of iterations.
is the maximum number of iterations.
Loss function
Some recent works have shown the effectiveness of a hybrid loss function in various network models developed for image segmentation or augmentation precisely [31–33].
For semantic segmentation loss, we use a combined loss function of multi-class cross-entropy loss  and Dice loss
and Dice loss  to optimize the training process, calculated as follows:
to optimize the training process, calculated as follows:
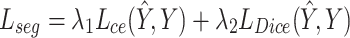 |
11 |
Here, 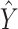 and
and 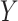 represent the model’s segmentation output and the ground truth, respectively. It is important to note that the labels need to undergo an additional downsampling operation to match the resolution of the model output.
represent the model’s segmentation output and the ground truth, respectively. It is important to note that the labels need to undergo an additional downsampling operation to match the resolution of the model output. 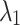 and
and 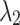 are loss weights used to balance the influence of each loss on the segmentation results, set to 0.2 and 0.8 in our experiments.
are loss weights used to balance the influence of each loss on the segmentation results, set to 0.2 and 0.8 in our experiments.
Optimizer
We selected the AdamW optimizer, based on the gradient descent algorithm, to train the fine-tuned network. AdamW is an improvement over the Adam optimizer. Adam is an adaptive learning rate optimization algorithm that adjusts the learning rate adaptively for each parameter based on its gradient and gradient history. AdamW combines Adam with weight decay, adding an additional L2 regularization term to the loss function to reduce the weight magnitudes. It also separates weight decay from gradient updates, applying it only to weight parameters rather than to bias parameters, effectively preventing adverse effects on bias terms and improving training stability. Compared to the SGD optimizer, which requires manual adjustment of learning rates, AdamW balances adaptive learning rates with regularization effects, helping to avoid local optima.
Experimental results and analysis
In this section, we conduct an experimental evaluation of our method and compare it with other approaches.
Datasets and data processing
We propose a segmentation algorithm for subcortical deep brain structures, focusing on the left and right thalamus, putamen, caudate nucleus, globus pallidus, hippocampus, and amygdala in the human brain, totaling 12 categories. We validate our proposed SAM-based brain structure segmentation algorithm, adapted through low-rank fine-tuning, on five public brain MR image datasets: the Internet Brain Segmentation Repository (IBSR)1, Multi-Atlas Labeling Challenge (MALC)2, LONI Probabilistic Brain Atlas (LPBA)3, Hammers4, and the Child and Adolescent NeuroDevelopment Initiative (CANDI).5 Table 1 provides the basic information for these five brain MR image datasets.
Table 1.
Information of the Datasets
| Dataset | Sample Size | Imaging Equipment | Modality | Age Range | Label Count | Image Size | Image Resolution ( ) ) |
|---|---|---|---|---|---|---|---|
| IBSR | 18 (14, 4) | 12: GE (1.5 T) 6: Siemens (1.5 T) | T1-w | 7–71 | 43 | 256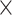 256 256 128 128 |
8: 0.94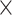 0.94 0.94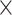 1.5, 6: 0.84 1.5, 6: 0.84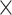 0.84 0.84 1.5, 4: 1.0 1.5, 4: 1.0 1.0 1.0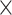 1.5 1.5 |
| MALC | 35 | — | T1-w | 19.3–39.5 | 135 | 256 256 256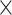 287 287 |
1.0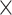 1.0 1.0 1.0 1.0 |
| LPBA | 40 (20, 20) | GE Signa (1.5 T) | T1-w | — | 56 | 256 256 256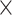 124 124 |
38: 0.86 0.86 0.86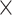 1.5, 2: 0.78 1.5, 2: 0.78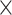 0.78 0.78 1.5 1.5 |
| Hammers | 30 (15, 15) | GE Signa (1.5 T) | T1-w | 20–54 | 83 | 192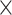 256 256 124 124 |
0.937 0.937 0.937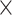 1.5 1.5 |
| CANDI | 103 (57, 46) | GE Signa (1.5 T) | T1-w | 4–17 | 62 | 192 256 256 124 124 |
— |
Before training, we first processed the 3D brain MR images from all datasets into a series of 2D slice images, resulting in a total of 15,815 coronal MRI slices from 226 cases of 3D brain MRI, with each slice cropped to a pixel size of 112 128. The experiments include cross-domain segmentation scenario validation (where test samples come from other MRI datasets not encountered during training) and supervised fine-tuning validation (where both the training and testing sets come from the same dataset). The cross-domain segmentation experiments are validated in two scenarios: (1) using MALC, LPBA, Hammers, and CANDI as the training set and IBSR as the testing set; and (2) using IBSR, LPBA, Hammers, and CANDI as the training set and MALC as the testing set. Supervised fine-tuning experiments are conducted on both the IBSR and MALC datasets.
128. The experiments include cross-domain segmentation scenario validation (where test samples come from other MRI datasets not encountered during training) and supervised fine-tuning validation (where both the training and testing sets come from the same dataset). The cross-domain segmentation experiments are validated in two scenarios: (1) using MALC, LPBA, Hammers, and CANDI as the training set and IBSR as the testing set; and (2) using IBSR, LPBA, Hammers, and CANDI as the training set and MALC as the testing set. Supervised fine-tuning experiments are conducted on both the IBSR and MALC datasets.
Evaluation metrics
In the experiments, two metrics are used to evaluate the segmentation performance of the proposed network: the pixel-overlap-based Dice Similarity Coefficient (DSC) and Hausdorff Distance (HD). The DSC describes the voxel overlap between the segmentation result and the ground truth, calculated as follows:
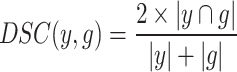 |
12 |
Here,  and
and 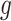 represent the final predicted segmentation result and the corresponding ground truth, respectively.
represent the final predicted segmentation result and the corresponding ground truth, respectively. 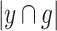 denotes the number of pixels in the overlapping area between
denotes the number of pixels in the overlapping area between 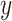 and
and 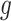 , while
, while  represents the total number of pixels in both regions. The DSC, which measures segmentation accuracy, typically ranges between 0 and 1.
represents the total number of pixels in both regions. The DSC, which measures segmentation accuracy, typically ranges between 0 and 1.
The DSC focuses on the internal region of the segmentation mask and is less sensitive to boundaries. As a shape similarity measure, the HD describes the average boundary distance between the segmentation result and the ground truth, providing a valuable complement to DSC. The calculation methods are shown in Equations 13, 14, and 15.
 |
13 |
 |
14 |
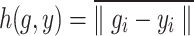 |
15 |
Here, 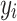 and
and  represent voxels in sets
represent voxels in sets  and
and 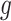 , respectively, and
, respectively, and 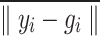 denotes the Euclidean distance. Therefore, the HD metric reflects the average distance from voxel
denotes the Euclidean distance. Therefore, the HD metric reflects the average distance from voxel 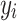 in set
in set 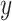 to voxel
to voxel 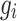 in set
in set 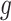 . Unlike DSC, a lower HD value indicates better segmentation performance.
. Unlike DSC, a lower HD value indicates better segmentation performance.
Experimental setup
To fairly compare the segmentation accuracy of our proposed algorithm with other state-of-the-art segmentation methods, all algorithms were implemented according to the authors’ publicly available code. All comparative experiments were conducted based on the PyTorch 1.10 framework and trained on 2  NVIDIA GTX 3090 Ti GPUs. For efficiency and performance optimization, LoRA was applied to fine-tune the
NVIDIA GTX 3090 Ti GPUs. For efficiency and performance optimization, LoRA was applied to fine-tune the 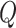 and
and 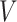 attention layers of the Transformer module, with the rank of the LoRA module set to 16. The initial learning rate for model warm-up was set to 0.005, and AdamW was used as the optimizer with betas set to (0.9, 0.999) and a weight decay of 0.1. During fine-tuning, the warm-up iteration count was set to 250, the total number of epochs to 200, and the batch size to 12.
attention layers of the Transformer module, with the rank of the LoRA module set to 16. The initial learning rate for model warm-up was set to 0.005, and AdamW was used as the optimizer with betas set to (0.9, 0.999) and a weight decay of 0.1. During fine-tuning, the warm-up iteration count was set to 250, the total number of epochs to 200, and the batch size to 12.
Experimental results and analysis in cross-domain scenarios
In this section, we conduct experimental validation for cross-domain segmentation scenarios where inference samples are derived from various MRI datasets. We compare our proposed method against current mainstream segmentation models, including the baseline segmentation model SAM [13] and the SAM-2D [18], a generalized medical image segmentation model based on fine-tuning. The experiments are performed with IBSR and MALC as the inference sample datasets, and results are averaged across all inference samples.
Experimental results for fine-tuning on other datasets → Inference on IBSR dataset
Table 2 shows the inference results on the IBSR dataset under point prompts (one, three, and five points) for our proposed method versus advanced algorithms, with the best scores marked in bold. The results indicate that all models achieve relatively low average DSC and HD values with point prompts. The SAM algorithm exhibits poor adaptability to medical image segmentation, and while SAM-2D serves as a generalized medical image segmentation algorithm, its accuracy remains low for deep brain structures with fuzzy boundaries. With an increase in the number of prompt points, segmentation metrics improve across all models. Our method achieves an average DSC of 60.92% with a single point prompt and 71.57% with five-point prompts, outperforming SAM and SAM-2D by 39.69% and 13.78%, respectively.
Table 2.
Comparison of inference results under the prompt of point
Figure 5 presents a visual comparison of inference results on two typical slices from the IBSR dataset under point-based prompts (including one, three, and five points) using our proposed algorithm and other advanced methods. In the figure, point prompts include red negative sample points (background) and green positive sample points (brain structures). Qualitative analysis shows that the segmentation performance of SAM and SAM-2D is relatively poor with point prompts, whereas our algorithm demonstrates significantly better segmentation performance under the same conditions.
Fig. 5.
Visual comparison of inference results under prompt of point
Table 3 presents the inference results of our proposed algorithm compared with other advanced methods on the IBSR dataset using bounding box prompts. The results show that with box prompts, the average DSC and HD metrics of all algorithms significantly improve compared to those under point prompts. For the average DSC across 12 brain structure classes, SAM achieves 52.73%, SAM-2D achieves 70.63%, and our algorithm achieves 81.11%. For the average HD metric, our algorithm achieves 0.640, representing improvements of 0.227 and 1.397 over SAM and SAM-2D, respectively. The visual comparison of inference results in Fig. 6 further illustrates that our algorithm achieves superior segmentation performance with bounding box prompts, accurately segmenting even complex or small brain structures. These quantitative and qualitative analyses indicate that our algorithm exhibits strong cross-domain segmentation performance with bounding box prompts, significantly outperforming existing advanced general medical image segmentation algorithms.
Table 3.
Comparison of inference results under the prompt of Bbox
| Models | SAM | SAM-2D | Ours | |||
|---|---|---|---|---|---|---|
| Segmentation Metrics | DSC(%) | HD | DSC(%) | HD | DSC(%) | HD |
| Left Thalamus | 60.08 | 1.734 | 73.39 | 0.774 | 86.68 | 0.491 |
| Right Thalamus | 59.76 | 1.749 | 80.07 | 0.527 | 88.84 | 0.467 |
| Left Caudate | 51.43 | 2.431 | 63.52 | 1.140 | 75.79 | 0.722 |
| Right Caudate | 44.74 | 3.051 | 63.06 | 1.310 | 74.78 | 0.745 |
| Left Putamen | 52.65 | 2.014 | 74.96 | 0.629 | 86.49 | 0.577 |
| Right Putamen | 53.15 | 2.031 | 70.80 | 0.849 | 85.60 | 0.632 |
| Left Pallidum | 47.74 | 1.905 | 67.55 | 0.921 | 72.69 | 0.814 |
| Right Pallidum | 52.25 | 1.925 | 67.20 | 0.960 | 73.85 | 0.804 |
| Left Hippocampus | 49.34 | 2.042 | 69.88 | 0.840 | 85.98 | 0.547 |
| Right Hippocampus | 49.68 | 1.885 | 69.84 | 0.864 | 84.73 | 0.588 |
| Left Amygdala | 51.51 | 2.083 | 67.30 | 1.130 | 80.86 | 0.598 |
| Right Amygdala | 59.08 | 1.494 | 71.55 | 0.922 | 76.99 | 0.693 |
| Average | 52.73 | 2.037 | 70.63 | 0.867 | 81.11 | 0.640 |
Fig. 6.
Visual comparison of inference results under prompt of Bbox
Fine-tuning on other datasets → Inference results on the MALC dataset
The histogram in Fig. 7 illustrates the average DSC and average HD values for segmenting 12 classes of deep brain structures using the proposed algorithm and other advanced algorithms on the MALC dataset under point and box prompt conditions. The experimental results indicate that all algorithms achieve optimal segmentation metrics under box prompts, followed by the five-point prompts. Under box prompts, the SAM algorithm yields an average DSC value of only 56.65%, while the SAM-2D algorithm achieves an average DSC value of 73.63%. In contrast, the proposed algorithm attains an average DSC value of 86.09%, surpassing the five-point prompt value by 8.81%. Additionally, similar performance is observed in the average HD metrics for segmenting all brain structures, with the SAM, SAM-2D, and proposed algorithms obtaining average HD values of 1.759, 0.867, and 0.357, respectively, under box prompts. The quantitative analysis on the MALC dataset demonstrates that the proposed algorithm effectively utilizes adaptive prompt learning during inference, enabling it to adapt well to the segmentation of deep brain structures across arbitrary MRI datasets. The visualization of inference results shown in Fig. 8 also reveals that the segmentation performance of the proposed algorithm, guided by interactive prompts, significantly outperforms existing advanced large segmentation models.
Fig. 7.
Comparison of inference results on MALC dataset
Fig. 8.
Visual comparison of inference results on MALC dataset
Supervised fine-tuning results and analysis
To further validate the segmentation performance of our proposed algorithm, we conduct experiments in a supervised fine-tuning setting, comparing our method with the latest supervised neural network segmentation algorithms. Notably, our approach requires no prompt information during inference. The comparative methods include: (1) the baseline segmentation algorithm U-Net [34]; (2) attention mechanism and transformer-based segmentation algorithms, AttenUnet [35] and TransUnet [30]; (3) the general medical image segmentation algorithm SAMed [19], which utilizes supervised fine-tuning. Supervised experiments are performed on the IBSR and MALC datasets. For the IBSR dataset, 12 brain MRI images are randomly selected as the training set, with the remaining 6 as the test set for each trial. For the MALC dataset, 20 brain MRI images are randomly selected as the training set, with the remaining 15 as the test set. The final prediction is obtained by averaging the results over 10 trials.
Supervised experiment results on the IBSR dataset
Table 4 presents quantitative results for our proposed algorithm compared to other advanced methods on the IBSR dataset, with the best scores highlighted in bold. The results indicate that our method achieves superior DSC scores across eight brain structures, while U-Net achieves the highest DSC scores for the left thalamus, left caudate nucleus, and right caudate nucleus, and SAMed attains the highest DSC scores for the right putamen and left amygdala. Additionally, our method achieves the highest average DSC score of 87.52% across 12 brain structures, representing improvements of 0.74%, 0.91%, 0.62%, and 0.56% compared to U-Net, AttenUnet, TransUnet, and SAMed, respectively. In terms of HD scores, our algorithm records the best scores in ten brain structures, followed by U-Net with the best score for the left thalamus and SAMed for the left amygdala. Our method achieves a best average HD score of 0.497 across 12 structures, improving by 5.7%, 7.1%, 3.8%, and 3.6% over U-Net, AttenUnet, TransUnet, and SAMed, respectively.
Table 4.
Comparison of supervised experimental results on IBSR dataset
| Models | U-Net [34] | AttenUnet [35] | TransUnet [30] | SAMed [19] | nnSAM [36] | Ours | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Segmentation metrics | DSC% | HD | DSC% | HD | DSC% | HD | DSC% | HD | DSC% | HD | DSC% | HD |
| Left Thalamus | 91.66 | 0.551 | 91.40 | 0.567 | 91.14 | 0.593 | 91.38 | 0.577 | 91.35 | 0.569 | 91.45 | 0.553 |
| Right Thalamus | 91.88 | 0.547 | 91.51 | 0.557 | 91.32 | 0.553 | 91.91 | 0.545 | 91.87 | 0.541 | 91.92 | 0.513 |
| Left Caudate | 89.69 | 0.424 | 89.67 | 0.423 | 89.18 | 0.431 | 89.60 | 0.417 | 89.64 | 0.409 | 89.69 | 0.391 |
| Right Caudate | 90.24 | 0.422 | 89.71 | 0.418 | 89.27 | 0.454 | 89.51 | 0.434 | 89.56 | 0.429 | 89.68 | 0.414 |
| Left Putamen | 90.68 | 0.490 | 90.92 | 0.472 | 90.71 | 0.475 | 91.04 | 0.482 | 91.07 | 0.478 | 91.31 | 0.435 |
| Right Putamen | 91.17 | 0.469 | 91.34 | 0.454 | 90.62 | 0.486 | 91.37 | 0.452 | 91.34 | 0.450 | 91.21 | 0.445 |
| Left Pallidum | 84.25 | 0.614 | 85.54 | 0.631 | 85.73 | 0.509 | 85.62 | 0.544 | 85.63 | 0.538 | 86.23 | 0.469 |
| Right Pallidum | 85.02 | 0.571 | 85.62 | 0.563 | 85.97 | 0.541 | 85.61 | 0.523 | 85.80 | 0.517 | 86.40 | 0.501 |
| Left Hippocampus | 84.94 | 0.567 | 84.74 | 0.561 | 85.47 | 0.521 | 84.91 | 0.532 | 84.96 | 0.527 | 85.91 | 0.481 |
| Right Hippocampus | 85.42 | 0.551 | 85.35 | 0.532 | 86.65 | 0.467 | 86.06 | 0.494 | 86.06 | 0.488 | 87.25 | 0.427 |
| Left Amygdala | 78.24 | 0.711 | 77.21 | 0.843 | 78.24 | 0.722 | 78.72 | 0.669 | 78.75 | 0.676 | 78.60 | 0.682 |
| Right Amygdala | 78.14 | 0.732 | 76.31 | 0.791 | 78.47 | 0.671 | 77.81 | 0.723 | 77.84 | 0.716 | 80.63 | 0.651 |
| Average | 86.78 | 0.554 | 86.61 | 0.568 | 86.90 | 0.535 | 86.96 | 0.533 | 86.99 | 0.528 | 87.52 | 0.497 |
Figure 9 shows a visualization of supervised experiment results on the IBSR dataset, with segmentation outcomes and error maps for 12 brain structures on three representative slices (red indicates error regions and green indicates correct regions). White arrows highlight areas of significant errors in the results from the comparison algorithms. Observing the highlighted areas, our method shows the fewest segmentation errors, followed by TransUnet and SAMed, while U-Net and AttenUnet exhibit the most errors. Both quantitative and qualitative results on the IBSR dataset demonstrate that our proposed method outperforms the compared advanced supervised algorithms in the supervised fine-tuning scenario.
Fig. 9.
Visual comparison of supervised experimental results on IBSR dataset
Supervised experiment results on the MALC dataset
Table 5 presents the supervised experimental results of our proposed method and other advanced segmentation algorithms on the MALC dataset, with best scores highlighted in bold. The results indicate that our method achieves optimal segmentation metrics for most brain structures, obtaining eight highest DSC scores and nine highest HD scores. SAMed achieved the best DSC scores for the right thalamus, left caudate, and right caudate, while U-Net obtained the highest DSC for the right hippocampus. Additionally, our method achieved the highest average DSC score of 89.35%, surpassing SAMed and TransUnet by 0.63% and 0.75%, respectively. In terms of average HD, our method outperformed U-Net, AttenUnet, TransUnet, and SAMed by 8.6%, 8.7%, 6.2%, and 4.4%, respectively.
Table 5.
Comparison of supervised experimental results on MALC dataset
| Models | U-Net [34] | AttenUnet [35] | TransUnet [30] | SAMed [19] | nnSAM [36] | Ours | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Segmentation metrics | DSC% | HD | DSC% | HD | DSC% | HD | DSC% | HD | DSC% | HD | DSC% | HD |
| Left Thalamus | 92.31 | 0.575 | 92.24 | 0.574 | 92.32 | 0.534 | 92.42 | 0.516 | 92.53 | 0.507 | 92.54 | 0.364 |
| Right Thalamus | 92.18 | 0.574 | 92.21 | 0.569 | 92.38 | 0.521 | 92.49 | 0.503 | 92.75 | 0.496 | 92.46 | 0.372 |
| Left Caudate | 90.43 | 0.429 | 90.44 | 0.429 | 91.44 | 0.381 | 91.47 | 0.365 | 90.56 | 0.361 | 90.52 | 0.392 |
| Right Caudate | 90.78 | 0.416 | 90.71 | 0.424 | 91.51 | 0.374 | 91.57 | 0.356 | 90.55 | 0.355 | 90.43 | 0.409 |
| Left Putamen | 91.74 | 0.432 | 91.81 | 0.471 | 91.92 | 0.436 | 91.98 | 0.427 | 92.26 | 0.424 | 93.15 | 0.355 |
| Right Putamen | 91.92 | 0.414 | 92.01 | 0.427 | 91.98 | 0.429 | 92.23 | 0.371 | 91.93 | 0.374 | 92.80 | 0.375 |
| Left Pallidum | 86.88 | 0.568 | 86.75 | 0.573 | 86.95 | 0.586 | 87.05 | 0.584 | 87.97 | 0.576 | 88.92 | 0.494 |
| Right Pallidum | 87.15 | 0.552 | 86.91 | 0.547 | 86.97 | 0.578 | 87.07 | 0.579 | 88.17 | 0.573 | 89.30 | 0.485 |
| Left Hippocampus | 86.76 | 0.551 | 86.91 | 0.535 | 86.93 | 0.527 | 87.04 | 0.512 | 86.96 | 0.508 | 87.42 | 0.487 |
| Right Hippocampus | 87.55 | 0.534 | 87.74 | 0.537 | 86.88 | 0.531 | 87.18 | 0.501 | 87.18 | 0.504 | 87.31 | 0.496 |
| Left Amygdala | 81.33 | 0.659 | 80.84 | 0.649 | 81.98 | 0.595 | 82.04 | 0.585 | 82.15 | 0.573 | 83.51 | 0.547 |
| Right Amygdala | 80.77 | 0.670 | 80.71 | 0.653 | 81.95 | 0.591 | 82.15 | 0.57 | 82.13 | 0.569 | 83.82 | 0.562 |
| Average | 88.32 | 0.531 | 88.27 | 0.532 | 88.60 | 0.507 | 88.72 | 0.489 | 88.77 | 0.485 | 89.35 | 0.445 |
Figure 10 provides a visual comparison of the supervised experimental results on the MALC dataset, showing the segmentation results and error maps (with red indicating error regions and green indicating correct regions) for three typical slices across 12 brain structures. White arrows highlight notable areas where other algorithms made segmentation errors. Observing the highlighted regions, our method’s results closely match the ground truth, accurately segmenting even smaller or boundary-blurred structures. In contrast, other advanced medical image segmentation algorithms showed poor performance in distinguishing challenging brain structures or misidentified them entirely. The quantitative and qualitative results on the MALC dataset demonstrate that our method outperforms the compared supervised algorithms in segmentation performance.
Fig. 10.
Visual comparison of supervised experimental results on MALC dataset
DSC and HD metrics are commonly used evaluation indicators in brain structure segmentation, so this paper primarily utilizes them for performance analysis. To demonstrate the effectiveness of our method in segmenting small structures and those with ambiguous boundaries, we further compare it with mainstream methods on the MALC dataset using Intersection over Union (IoU) and Average Symmetric Surface Distance (ASSD). The comparative results are presented in Table 6.
Table 6.
Comparison of IoU and ASSD on the MALC Dataset
| Models | U-Net [34] | AttenUnet [35] | TransUnet [30] | SAMed [19] | nnSAM [36] | Ours | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Segmentation metrics | IoU% | ASSD | IoU% | ASSD | IoU% | ASSD | IoU% | ASSD | IoU% | ASSD | IoU% | ASSD |
| Left Thalamus | 91.78 | 0.725 | 91.71 | 0.721 | 91.81 | 0.678 | 92.02 | 0.655 | 92.08 | 0.605 | 92.11 | 0.482 |
| Right Thalamus | 91.65 | 0.722 | 91.67 | 0.713 | 91.85 | 0.665 | 92.05 | 0.644 | 92.06 | 0.617 | 92.05 | 0.490 |
| Left Caudate | 89.91 | 0.579 | 89.93 | 0.576 | 90.93 | 0.524 | 90.17 | 0.511 | 90.14 | 0.505 | 90.16 | 0.503 |
| Right Caudate | 90.01 | 0.567 | 90.11 | 0.572 | 89.97 | 0.518 | 90.14 | 0.508 | 90.11 | 0.519 | 90.25 | 0.511 |
| Left Putamen | 91.22 | 0.582 | 91.25 | 0.612 | 91.41 | 0.578 | 91.58 | 0.566 | 91.83 | 0.554 | 92.72 | 0.475 |
| Right Putamen | 91.38 | 0.566 | 91.43 | 0.576 | 91.44 | 0.573 | 91.83 | 0.512 | 91.53 | 0.545 | 92.37 | 0.485 |
| Left Pallidum | 86.35 | 0.719 | 86.21 | 0.717 | 86.41 | 0.729 | 86.65 | 0.722 | 87.57 | 0.706 | 88.49 | 0.612 |
| Right Pallidum | 86.61 | 0.702 | 86.39 | 0.699 | 86.46 | 0.723 | 86.67 | 0.718 | 87.75 | 0.704 | 88.87 | 0.607 |
| Left Hippocampus | 86.22 | 0.701 | 86.39 | 0.681 | 86.42 | 0.672 | 86.64 | 0.653 | 86.56 | 0.637 | 86.99 | 0.609 |
| Right Hippocampus | 86.77 | 0.683 | 86.91 | 0.686 | 86.34 | 0.677 | 86.78 | 0.644 | 86.77 | 0.633 | 86.93 | 0.613 |
| Left Amygdala | 80.81 | 0.809 | 80.33 | 0.792 | 81.49 | 0.739 | 81.64 | 0.726 | 81.74 | 0.704 | 83.43 | 0.669 |
| Right Amygdala | 80.24 | 0.821 | 80.17 | 0.799 | 81.41 | 0.733 | 81.75 | 0.713 | 81.72 | 0.698 | 83.51 | 0.677 |
| Average | 87.75 | 0.681 | 87.71 | 0.679 | 88.00 | 0.651 | 88.16 | 0.631 | 88.32 | 0.619 | 88.99 | 0.561 |
As can be seen, our method achieves the best segmentation metrics for the three small or boundary-ambiguous structures of the Pallidum, Hippocampus, and Amygdala, demonstrating the superior capability of our approach in segmenting subtle anatomical structures within brain imaging data.
Ablation study results and analysis
To evaluate the effectiveness of the LoRA fine-tuning parameters and training strategies used in our algorithm, this section conducts an ablation study on the IBSR dataset. The ablation experiments include assessments of model preheating and optimizer selection (SGD, AdamW) in the training strategy, LoRA fine-tuning component ablation (encoder and decoder fine-tuning options), self-attention layer selection for LoRA fine-tuning ( ,
, 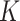 ,
, 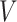 attention layers), and rank size selection for the LoRA module.
attention layers), and rank size selection for the LoRA module.
Model training strategy
Figure 11 presents the loss curves of the model using different training strategies. The comparison indicates that model preheating facilitates faster convergence, leads to more stable training, and enables the model to converge to a lower loss value. Furthermore, the AdamW optimizer significantly reduces the final training loss compared to the SGD optimizer. This demonstrates that model preheating and the selection of an appropriate optimizer play a crucial role in the performance of the LoRA fine-tuning algorithm.
Fig. 11.

Comparison of loss curves for different training strategies
Ablation of the LoRA module
Ablation of LoRA Fine-Tuning Components
In our proposed algorithm, LoRA is used to fine-tune the Transformer module of the image encoder in the SAM model. Since SAM is primarily trained on natural images, and there is a significant domain distribution difference between medical images and natural images, specific fine-tuning of the image encoder helps the model extract useful features from medical images, which also benefits the mask decoder for subsequent segmentation tasks. On the other hand, considering that the SAM mask decoder also contains two lightweight Transformer modules for decoding the extracted image embeddings, this section conducts ablation experiments to compare the segmentation performance when fine-tuning the Transformer modules in the image encoder and the mask decoder separately.
Figure 12 illustrates the ablation experiment results for separately fine-tuning the mask decoder, the image encoder, and simultaneously fine-tuning both the encoder and decoder. Comparing the segmentation DSC values of the three LoRA fine-tuning models for various brain structures reveals that fine-tuning only the encoder yields the best segmentation performance, followed by simultaneous fine-tuning of both the encoder and decoder. Conversely, the experiments that only fine-tune the decoder perform poorly, indicating that solely fine-tuning the decoder does not enable the model to learn the specific features of brain MR images. Simultaneous fine-tuning of the encoder and decoder may lead to feature overfitting during the training process, resulting in poorer model performance compared to fine-tuning the encoder alone.
Fig. 12.
Ablation studies for LoRA fine-tuning components
Table 7 presents a comparison of the learnable parameters and training times for models fine-tuned using LoRA for the encoder and decoder. It can be observed that the LoRA fine-tuning of the decoder requires the fewest learnable parameters and results in the shortest inference time. In our proposed algorithm, which fine-tunes only the encoder, the number of learnable parameters is 5.92 MB, and the inference time per slice is 0.135 seconds, which is only 0.006 seconds longer than the time taken when only fine-tuning the decoder, indicating that our algorithm achieves a fast inference speed. Additionally, when only the encoder is fine-tuned, it is necessary to learn all parameters of the decoder; therefore, the total learnable parameters when simultaneously fine-tuning both the encoder and decoder are fewer than those when fine-tuning the encoder alone.
-
(2)
Ablation Study of LoRA Fine-Tuning on Self-Attention Layers
Table 7.
Comparison of learnable parameters and training time
| LoRA Fine-tuning block | Decoder | Encoder | Encoder + Decoder |
|---|---|---|---|
| Learnable Parameters (MB) | 2.13 | 5.92 | 2.72 |
| Frozen Parameters (MB) | 90.57 | 87.28 | 90.57 |
| Inference Time per Slice (s) | 0.129 | 0.135 | 0.149 |
We investigated the segmentation performance of the LoRA module integrated into different self-attention layers within the Transformer module. The experimental results are presented in Fig. 1, where the segmentation DSC values indicate that applying the LoRA module to the  and
and 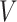 attention layers is a reasonable choice, yielding the best DSC values for all brain structures. However, using the LoRA module across all attention layers (
attention layers is a reasonable choice, yielding the best DSC values for all brain structures. However, using the LoRA module across all attention layers (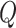 ,
, 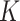 , and
, and 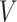 ) results in a significant decline in performance. This finding also corroborates the previous analysis: excessive parameter fine-tuning with LoRA may disrupt the original image segmentation capabilities of the SAM large model, introducing training interference into the model fine-tuning process.
) results in a significant decline in performance. This finding also corroborates the previous analysis: excessive parameter fine-tuning with LoRA may disrupt the original image segmentation capabilities of the SAM large model, introducing training interference into the model fine-tuning process.
Fig. 13.
Ablation studies for LoRA fine-tuning self-attention layers
-
(3)
Ablation Study of LoRA Rank
Table 8 presents the learnable parameters and training time of the LoRA module at different rank sizes. It can be observed that the model with a rank of 16 has a similar number of learnable parameters compared to the model with a rank of 8, with the inference time only increasing by 0.001 seconds when using a rank of 16.
Table 8.
Comparison of learnable parameters and training time
| LoRA Rank | r(LoRA) = 8 | r(LoRA) = 16 | r(LoRA) = 32 |
|---|---|---|---|
| Learnable Parameters (MB) | 5.62 | 5.92 | 6.51 |
| Frozen Parameters (MB) | 87.28 | 87.28 | 87.28 |
| Inference Time per Slice (s) | 0.134 | 0.135 | 0.138 |
Table 9 presents the segmentation performance of the LoRA module at different ranks. It can be observed from the table that, within a certain range, a higher rank leads to better model performance; however, when the rank is excessively high, the model performance significantly declines. This indicates that the LoRA module requires a specific number of parameters to effectively adapt to the fine-tuning of the brain MRI dataset. As the rank of LoRA increases, the number of trainable parameters in the fine-tuned model also increases, but too many parameters may impair the retained performance of the original SAM model in the fine-tuning process, thereby increasing the difficulty of training. Experimental results show that a rank setting of 16 for the LoRA module in this algorithm achieves optimal segmentation accuracy and speed.
Table 9.
Ablation study on the rank size of the LoRA block
| LoRA Rank |  |
 |
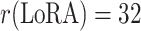 |
|||
|---|---|---|---|---|---|---|
| Segmentation Metrics | DSC (%) | HD | DSC (%) | HD | DSC (%) | HD |
| Left Thalamus | 90.59 | 0.623 | 91.45 | 0.553 | 81.78 | 1.114 |
| Right Thalamus | 90.62 | 0.596 | 91.92 | 0.513 | 81.13 | 1.149 |
| Left Caudate | 89.15 | 0.429 | 89.69 | 0.391 | 75.07 | 0.973 |
| Right Caudate | 89.17 | 0.462 | 89.68 | 0.414 | 72.39 | 1.061 |
| Left Putamen | 90.85 | 0.478 | 91.31 | 0.435 | 79.91 | 0.986 |
| Right Putamen | 90.93 | 0.461 | 91.21 | 0.445 | 78.97 | 1.020 |
| Left Pallidum | 85.94 | 0.521 | 86.23 | 0.469 | 70.10 | 1.123 |
| Right Pallidum | 85.67 | 0.526 | 86.40 | 0.501 | 71.44 | 1.072 |
| Left Hippocampus | 83.72 | 0.590 | 85.91 | 0.481 | 61.79 | 1.336 |
| Right Hippocampus | 85.17 | 0.545 | 87.25 | 0.427 | 64.36 | 1.239 |
| Left Amygdala | 77.17 | 0.775 | 78.60 | 0.682 | 61.07 | 1.403 |
| Right Amygdala | 78.48 | 0.735 | 80.63 | 0.651 | 67.04 | 1.275 |
| Average | 86.46 | 0.562 | 87.52 | 0.497 | 72.09 | 1.146 |
Influence study on hyperparameters of and
The paper involves two key hyperparameters: 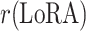 , which was analyzed in Section “Experimental results and analysis”, and
, which was analyzed in Section “Experimental results and analysis”, and 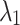 and
and 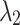 in Eq. (11). For brain image segmentation, the Dice loss plays a more significant role; thus, assigning higher
in Eq. (11). For brain image segmentation, the Dice loss plays a more significant role; thus, assigning higher 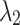 weights in these parameters facilitates model convergence and leads to more stable results. Here, we investigate the impact of the hyperparameters
weights in these parameters facilitates model convergence and leads to more stable results. Here, we investigate the impact of the hyperparameters  and
and 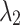 on segmentation performance. With
on segmentation performance. With 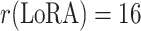 , the comparative segmentation results are summarized in Table 10.
, the comparative segmentation results are summarized in Table 10.
Table 10.
Influence study on hyperparameters of  and
and  r(LoRA) = 16
r(LoRA) = 16
| Parameter Settings |
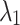 =0.5 =0.5 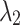 =0.5 =0.5 |
 =0.4 =0.4 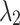 =0.6 =0.6 |
 =0.3 =0.3 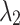 =0.7 =0.7 |
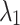 =0.2 =0.2 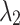 =0.8 =0.8 |
||||
|---|---|---|---|---|---|---|---|---|
| Segmentation Metrics | DSC (%) | HD | DSC (%) | HD | DSC (%) | HD | DSC (%) | HD |
| Average | 87.22 | 0.531 | 87.38 | 0.516 | 87.47 | 0.507 | 87.52 | 0.497 |
As shown in Table 10, the hyperparameters 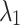 and
and 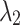 exhibit minimal sensitivity to segmentation performance under sufficient training epochs. Based on this observation, we empirically set
exhibit minimal sensitivity to segmentation performance under sufficient training epochs. Based on this observation, we empirically set 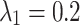 and
and 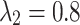 in our experiments.
in our experiments.
Limitation and discussion
The effectiveness of the proposed segmentation has been evaluated only for segmentation of brain structures. Its performance has not been assessed for segmentation of other tissues and organs from MRIs. This can be considered a limitation of this study. Particularly, segmentation and labeling of vascular structures (e.g., liver vessels) and organs (e.g., kidneys, liver) from abdominal MRIs is very challenging, and although different approaches based on level sets, probabilistic atlas and Gaussian model fitting [37], their effectiveness is not always sufficient. The proposed segmentation can be modified and its effectiveness can be evaluated as a future work.
The segmentation of deep brain structures in brain MR images is a pivotal task in the study of brain diseases, as well as in applications such as intelligent medical screening and diagnosis [38, 39]. Due to the complexity of segmenting subcortical deep brain structures, the difficulty in obtaining high-quality medical images, and the challenges in acquiring expert-annotated labels, the precise segmentation of deep brain structures in MR images remains a challenging research endeavor. The following issues should be further addressed on real-world clinical applications:
There may be complex noise and artifacts superimposed in actual clinical data. Therefore, how to extend this algorithm to more realistic brain MR image segmentation tasks under noise and artifact interference should be considered.
This paper fine tunes the existing semantic segmentation model SAM with low rank adaptation, transferring the powerful segmentation performance of SAM for natural image targets to deep brain structure segmentation tasks in brain MR images, eliminating the complex process of training the network from scratch. However, the drawback of fine-tuning methods is that models fine tuned based on specific datasets may have relatively low generalization when applied to other datasets of the same type but with different distributions. Further fine-tuning is needed on diverse brain MRI data to enhance the model’s generalization.
Develop appropriate data augmentation techniques and utilize additional data annotated by prompt learning for data augmentation, in order to achieve a more versatile brain structure segmentation algorithm.
During our experiments, we encountered an issue where large models lost their generalizability after applying certain fine-tuning methods, a phenomenon known as catastrophic forgetting. This occurs because the model primarily learns and adapts to the data characteristics of specific tasks or domains during fine-tuning, while neglecting the broader knowledge acquired during pretraining. To mitigate this loss of generalizability, several strategies can be employed, such as multi-task learning, transfer learning, reinforcement learning, or specialized fine-tuning techniques. In this paper, we adopted LoRA for fine-tuning, which alleviated the degradation of generalizability to some extent and reduced the impairment of the model’s original capabilities.
Conclusion
We investigated the segmentation method for deep brain structures in MRI images within cross-domain scenarios, proposing a brain structure segmentation algorithm based on LoRA to fine-tune SAM. This addresses the issue that current general semantic segmentation models like SAM and other common medical image segmentation algorithms perform well on clearly defined targets but exhibit lower segmentation accuracy for ambiguously defined deep brain structures. The proposed algorithm utilizes parameter-efficient fine-tuning techniques to transfer the large SAM model from the domain of natural image segmentation to the task of deep brain structure segmentation in MRI images. The model freezes all weights of the SAM image encoder and adds a trainable low-rank adaptation module to each Transformer module within the encoder. This module fine-tunes only a small number of parameters by training a low-rank approximation matrix of the original model weights, allowing the model to adapt to the complex task of brain structure segmentation. The fine-tuned model follows the architecture of SAM, including the image encoder, prompt encoder, and mask decoder, and adjusts only a small portion of the weight parameters during the training process. Specifically, the model applies LoRA fine-tuning strategies to approximate the low-rank matrix updates of the training weights in the image encoder while also fine-tuning SAM’s lightweight prompt encoder and mask decoder. The size of the learnable parameters in the fine-tuned model (5.92 M) constitutes only 6.39% of the original model’s parameter size (92.61 M). In terms of training strategy, model warming is employed to stabilize the fine-tuning process. During inference, adaptive prompt learning using point or box prompts is introduced to enhance the model’s accuracy in segmenting arbitrary brain MR images. This interactive prompt learning approach provides clinicians with an intelligent segmentation tool for deep brain structures, effectively alleviating the challenges of limited data labeling and the high costs associated with manual annotations in the current medical image segmentation field. Finally, comparisons are made with current mainstream general segmentation algorithms and supervised segmentation algorithms in cross-domain scenarios, where inference samples are drawn from arbitrary MRI datasets and in supervised fine-tuning scenarios. The proposed algorithm demonstrates superior segmentation accuracy compared to the advanced general segmentation algorithms and mainstream supervised medical image segmentation algorithms. Experimental results across multiple segmentation scenarios confirm the generalization and effectiveness of the proposed segmentation algorithm.
The current mainstream paradigm in medical imaging adopts the ‘pre-training + lightweight fine-tuning’ approach, balancing the generalization capabilities of large models like SAM with precise domain-specific control. Our solution achieves a triangular optimization among small data, high performance, and low latency through LoRA-based fine-tuning, demonstrating significant practical advantages in brain disease diagnosis scenarios.
Acknowledgements
Not applicable.
Author contributions
Yuan Sui: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Resources; Software; Validation; Visualization; Writing – original draft; Writing – review & editing. Qian Hu: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Resources; Software; Validation; Visualization; Writing – original draft; Writing – review & editing. Yujie Zhang: Conceptualization; Formal analysis; Funding acquisition; Methodology; Project administration; Resources; Supervision; Validation; Writing – review & editing.
Funding
This work was supported by the National Natural Science Foundation of China under Grant U22A2025 and 62,441,231, Key R & D projects of Liaoning Province, China (Grant No. 2024JH2/102,500,015), Fundamental Research Funds for the Central Universities of Ministry of Education (Grant No.N25BSS034).
Data availability
This paper uses publicly available datasets. The link to the dataset used in Section is as follows: http://www.neuromorphometrics.com/2012_MICCAI_Challenge_Data.html
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
http://masi.vuse.vanderbilt.edu/workshop2012/index.php
www.loni.usc.edu/research/atlases
https://soundray.org/hammers-n30r95
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.BrainFacts/SfN, Mapping the brain, https://www.brainfacts.org/brainanatomy-and-function/anatomy/2012/mapping-the-brain. (1 april 2012). (2012).
- 2.Teipel SJ, Grothe M, Lista S, Toschi N, Garaci FG, Hampel H. Relevance of magnetic resonance imaging for early detection and diagnosis of alzheimer disease. Med Clin. 2013;97(3):399–424. [DOI] [PubMed] [Google Scholar]
- 3.Debernard L, Melzer TR, Alla S, Eagle J, Van Stockum S, Graham C, Osborne JR, Dalrymple-Alford JC, Miller DH, Mason DF. Deep grey matter mri abnormalities and cognitive function in relapsing-remitting multiple sclerosis. Psychiatry Res Neuroimaging. 2015;234(3):352–61. [DOI] [PubMed] [Google Scholar]
- 4.Mak E, Bergsland N, Dwyer M, Zivadinov R, Kandiah N. Subcortical atrophy is associated with cognitive impairment in mild parkinson disease: a combined investigation of volumetric changes, cortical thickness, and vertex-based shape analysis. AJNR Am J Neuroradiol. 2014;35(12):2257–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischl B, Freesurfer. Neuroimage. 2012;62(2):774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56(3):907–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coupé P, Manjón JV, Fonov V, Pruessner J, Robles M, Collins DL. Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation. NeuroImage. 2011;54(2):940–54. [DOI] [PubMed] [Google Scholar]
- 8.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Adv Neural Inf Process. 2012;25.
- 9.He K, Zhang X, Ren S, Sun J, Deep residual learning for image recognition, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–78.
- 10.Minaee S, Boykov Y, Porikli F, Plaza A, Kehtarnavaz N, Terzopoulos D. Image segmentation using deep learning: a survey. IEEE Trans Pattern Anal Mach Intell. 2021;44(7):3523–42. [DOI] [PubMed] [Google Scholar]
- 11.Achiam J, Adler S, Agarwal S, Ahmad L, Akkaya I, Aleman FL, Almeida D, Altenschmidt J, Altman S, Anadkat S, et al, Gpt-4 technical report, arXiv preprint arXiv:2303.08774 (2023).
- 12.Wang X, Zhang X, Cao Y, Wang W, Shen C, Huang T. SegGPT: Towards segmenting everything in context. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. 2023. p. 1130–40.
- 13.Kirillov A, Mintun E, Ravi N, Mao H, Rolland C, Gustafson L, Xiao T, Whitehead S, Berg AC, Lo C, et al. Segment anything. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. 2023. p. 4015–26.
- 14.Zhang Y, Shen Z, Jiao R. Segment anything model for medical image segmentation: Current applications and future directions. Comput Biol Med. 2024:108238. [DOI] [PubMed]
- 15.Hu M, Li Y, Yang X, Skinsam: Empowering skin cancer segmentation with segment anything model. arXiv preprint arXiv:2304.13973. (2023).
- 16.Ma J, He Y, Li F, Han L, You C, Wang B. Segment anything in medical images. Nat Commun. 2024;15(1):654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J, Ji W, Liu Y, Fu H, Xu M, Xu Y, Jin Y, Medical sam adapter: Adapting segment anything model for medical image segmentation, arXiv preprint arXiv:2304.12620 (2023). [DOI] [PubMed]
- 18.Cheng J, Ye J, Deng Z, Chen J, Li T, Wang H, Su Y, Huang Z, Chen J, Jiang L, et al., Sam-med2d, arXiv preprint arXiv:2308.16184 (2023).
- 19.Zhang K, Liu D, Customized segment anything model for medical image segmentation, arXiv preprint arXiv:2304.13785 (2023).
- 20.Hu EJ, Shen Y, Wallis P, Allen-Zhu Z, Li Y, Wang S, Wang L, Chen W, Lora: low-rank adaptation of large language models, arXiv preprint arXiv:2106.09685 (2021).
- 21.Vaswani A. Attention is all you need. Adv Neural Inf Process Syst. 2017.
- 22.Dosovitskiy A, An image is worth 16×16 words: Transformers for image recognition at scale, arXiv preprint arXiv:2010.11929 (2020).
- 23.He K, Chen X, Xie S, Li Y, Dollár P, Girshick R, Masked autoencoders are scalable vision learners, in: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 2022, pp. 16000–09.
- 24.Brown TB, Language models are few-shot learners, arXiv preprint arXiv:2005.14165 (2020).
- 25.Ouyang L, Wu J, Jiang X, Almeida D, Wainwright C, Mishkin P, Zhang C, Agarwal S, Slama K, Ray A, et al. Training language models to follow instructions with human feedback. Adv Neural Inf Process Syst. 2022;35:27730–44. [Google Scholar]
- 26.Awais M, Naseer M, Khan S, Anwer RM, Cholakkal H, Shah M, Yang MH, Khan FS, Foundational models defining a new era in vision: A survey and outlook, arXiv preprint arXiv:2307.13721 (2023). [DOI] [PubMed]
- 27.Liang PP, Zadeh A, Morency LP. Foundations and trends in multimodal machine learning: Principles, challenges, and open questions. arXiv preprint arXiv:2209.03430. (2022).
- 28.Wang X, Chen G, Qian G, Gao P, Wei X–Y, Wang Y, Tian Y, Gao W. Large-scale multi-modal pre-trained models: a comprehensive survey. Mach Intell Res. 2023;20(4):447–82. [Google Scholar]
- 29.Feng W, Zhu L, Yu L, Cheap lunch for medical image segmentation by fine-tuning sam on few exemplars, arXiv preprint arXiv:2308.14133 (2023).
- 30.Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y, Transunet: Transformers make strong encoders for medical image segmentation, arXiv preprint arXiv:2102.04306 (2021).
- 31.Goceri E. Polyp segmentation using a hybrid vision transformer and a hybrid loss function. J Imaging Inform Med. 2024;37(2):851–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goceri E. Nuclei segmentation using attention aware and adversarial networks. Neurocomputing. 2024; 579:127445. [Google Scholar]
- 33.Goceri E. Gan based augmentation using a hybrid loss function for dermoscopy images. Artif Intell Rev. 2024;57(9):234. [Google Scholar]
- 34.Ronneberger O, Fischer P, Brox T, U-net: Convolutional networks for biomedical image segmentation, in: Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, Munich, Germany, October 5-9, 2015, proceedings, part III 18, Springer, 2015, pp. 234–41.
- 35.Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B, et al., Attention u-net: Learning where to look for the pancreas, arXiv preprint arXiv:1804.03999 (2018).
- 36.Li Y, Jing B, Li Z, Wang J, Zhang Y. Plug-and-play segment anything model improves nnunet performance. Med Phys. 2025;52(2):899–912. arXiv:https://aapm.onlinelibrary.wiley.com/doi/pdf/10.1002/mp.17481,10.1002/mp.17481. URL. https://aapm.onlinelibrary.wiley.com/doi/abs/10.1002/mp.17481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dura E, Domingo J, Göçeri E, Mart-Bonmat L. A method for liver segmentation in perfusion mr images using probabilistic atlases and viscous reconstruction. Pattern Anal Appll. 2018;21(4):1083–95. [Google Scholar]
- 38.Paithane P, Kakarwal S. Lmns-net: Lightweight multiscale novel semantic-net deep learning approach used for automatic pancreas image segmentation in ct scan images, expert syst. Appl. Dec 2023;234(C). 10.1016/j.eswa.2023.121064. 10.1016/j.eswa.2023.121064.
- 39.Paithane P. Optimize multiscale feature hybrid-net deep learning approach used for automatic pancreas image segmentation, Mach. Vision Appl. Oct 2024;35(6). 10.1007/s00138-024-01619-y. 10.1007/s00138-024-01619-y.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This paper uses publicly available datasets. The link to the dataset used in Section is as follows: http://www.neuromorphometrics.com/2012_MICCAI_Challenge_Data.html














