Abstract
Phylogenetic analysis of 1.35 kb of mtDNA sequence from fossils revealed a previously unknown radiation of Hawaiian geese, of which only one representative remains alive (the endangered Hawaiian goose or nene, Branta sandvicensis). This radiation is nested phylogenetically within a living species, the Canada goose (Branta canadensis) and is related most closely to the large-bodied lineage within that species. The barnacle goose (Branta leucopsis) is also nested within the Canada goose species and is related most closely to the small-bodied lineage of Canada geese. The peripheral isolation of the barnacle goose in the Palearctic apparently allowed the evolution of its distinctive plumage pattern, whereas the two Nearctic lineages of Canada geese share a primitive plumage pattern. The Hawaiian lineage of Canada geese diverged more dramatically, splitting into at least three species that differ in body size, body proportions, and flight ability. One fossil species, limited to the island of Hawaii, was related closely to the nene but was over four times larger, flightless, heavy-bodied and had a much more robust cranium. Application of a rate calibration to levels of DNA divergence suggests that this species evolved on the island of Hawaii in less than 500,000 years. This date is consistent with the potassium/argon-based age of the island of Hawaii of 430,000–500,000 years. The giant Hawaii goose resembles the moa-nalos, a group of massive, extinct, flightless ducks that lived on older Hawaiian Islands and thus is an example of convergent evolution of similar morphologies in island ecosystems.
In continental ecosystems, feeding guilds of large herbivores usually are dominated by mammals. However, mammals have difficulty colonizing remote oceanic islands. In the extremely isolated Hawaiian islands, all the large native herbivores were waterfowl (1, 2). Based on the geographic distribution of the fossil species and the habits of the surviving species, these birds did not favor wetlands as most waterfowl do but instead occupied a broad range of terrestrial habitats. At least seven species occur in Holocene fossil faunas from the islands. Similar to island endemics elsewhere, Hawaii's large waterfowl were vulnerable to extinction, and all except the Hawaiian goose or nene (Branta sandvicensis) became extinct after human settlement of the islands ≈1,600 years ago (1, 2).
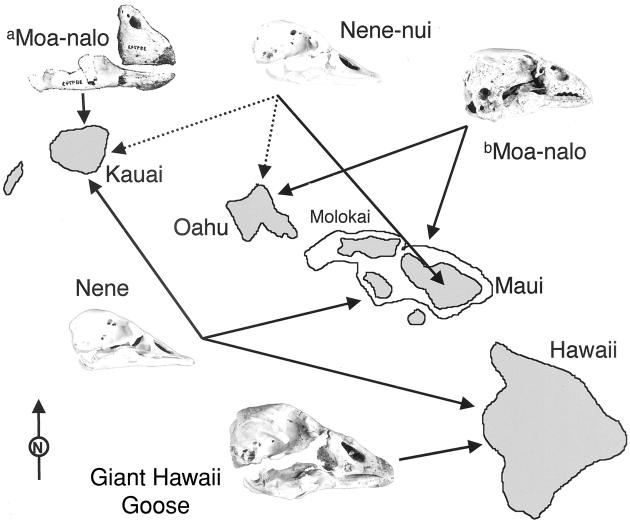
Hawaii's large waterfowl consist of two groups, the “moa-nalos” and the true geese (Fig. 1). The moa-nalos consist of four species of large flightless birds that are classified either with the dabbling ducks, tribe Anatini (1–3), or placed in a tribe of their own, the Thambetochenini (4). The true geese in Hawaii (tribe Anserini) comprise at least three species that exhibit a wide range of morphologies (Figs. 2 and 3). The nene is both the smallest and the only species capable of strong flight. Branta hylobadistes, the “nene-nui,” an extinct fossil species from Maui, is a heavier bird that was at best a weak flier (1, 2). The giant Hawaii goose, an as yet undescribed fossil species from the island of Hawaii (1, 2), was incapable of flight, was much larger in body size, and had a far more robust skull and bill (Figs. 1–3). Additional undescribed fossil geese not included in the molecular analyses presented here have been found on Kauai and Oahu (1).
Figure 1.
A map of the main Hawaiian Islands with skulls of two moa-nalos (a, Chelychelynechen quassus; b, Thambetochen chauliodous) and the three Hawaiian geese evaluated in this study [B. sandvicensis (nene), B. hylobadistes (nene-nui), and the undescribed giant Hawaii goose]. Fossils of the giant Hawaii goose have been found only on the island of Hawaii (1, 2). Moa-nalos were found only on the older islands from Maui to Kauai (1). Arrows show that the nene, B. sandvicensis, and nene-like (B. hylobadistes and relatives) geese were widely distributed among the main islands.
Figure 2.
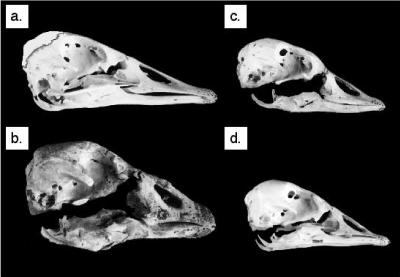
Skulls and mandibles in lateral view. (a) B. canadensis maxima (USNM 555497); (b) giant Hawaii goose (BPBM 179440); (c) B. hylobadistes (BPBM PPBH7); (d) B. sandvicensis (USNM 557998). The length of the giant Hawaii goose skull (b) is 127.6 mm.
Figure 3.
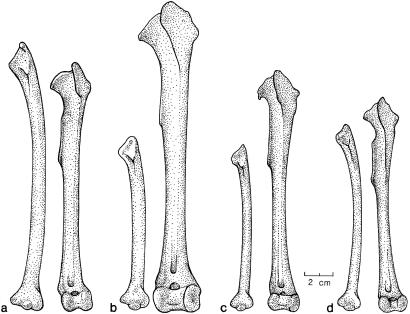
Left ulna and tibiotarsus. (a) B. canadensis maxima (USNM 555497); (b) giant Hawaii goose (BPBM 179440); (c) B. hylobadistes (BPBM PPBH7); (d) B. sandvicensis (USNM 557998).
Although most Hawaiian waterfowl are known only from bones, developing robust and well resolved hypotheses for their evolutionary relationships by studying osteological characters alone may not be possible, particularly for the flightless species such as the giant Hawaii goose. Some of the bones are highly modified compared with volant, continental waterfowl, making it difficult to identify shared character states (4). Also, the phylogenetic signal from osteological characters may be obscured by homoplastic evolution in Hawaiian birds that evolved in similar ecosystems on separate islands (Fig. 1). Therefore, we assessed the evolutionary relationships of Hawaii's large waterfowl by using genetic information (see ref. 3 for molecular phylogenetic analysis of the moa-nalos). Here we present phylogenetic analyses of ancient mtDNA sequences from three of the Hawaiian geese of the tribe Anserini.
Materials and Methods
Sampling and Radiocarbon Dating of Fossils.
We obtained DNA sequences from 13 bone samples representing the three Hawaiian geese collected in lava tube caves on the islands of Hawaii and Maui. Most samples were taken from associated skeletons cataloged at the National Museum of Natural History (USNM, Washington, DC) or the Bishop Museum (BPBM, Honolulu, HI). Three of the samples were taken from associated skeletons that are still in the caves in accordance with permit requirements. The sampled specimens include three individuals of the giant Hawaii goose [BPBM 179440, Gigo-1 (skeleton still in cave, bone powder from sample preserved at USNM), and Gigo-2 (skeleton still in cave)], three individuals of B. hylobadistes (USNM 519291, USNM 519295, and BPBM 183958), four individuals of nene from the island of Hawaii (USNM 519292, USNM 519293, USNM 519296, and Brsa-MPRC9 (skeleton still in cave, bone fragments from sample preserved at USNM), and three individuals of nene from Maui (USNM 519290, USNM 519294, and BPBM 183959).
Nine of the thirteen bones that yielded ancient DNA sequences were radiocarbon-dated from purified collagen by using accelerator mass spectrometry. Dates were obtained from Instaar Labs (University of Colorado, Boulder, CO), with the exception of two bones (Gigo-2 and BPBM 179440) that were obtained from Beta Analytic (Miami). The data reported are radiocarbon years before AD 1950, corrected for 13C fractionation. The dates range from 5,100 ± 50 to 510 ± 60 (giant Hawaii goose: Gigo-1, 870 ± 50; Gigo-2, 900 ± 60; BPBM 179440, 510 ± 60. B. sandvicensis: USNM 519296, 1,940 ± 50; Brsa-mprc9, 1,000 ± 50; USNM 519292, 2,010 ± 40; BPBM 183959, 3,190 ± 50. B. hylobadistes: 519291, 5,100 ± 50; BPBM 183958, 1,050 ± 50).
Taxonomic Sampling.
To place the Hawaiian geese in phylogenetic context, we obtained homologous sequences from 15 taxa of true geese (tribe Anserini). Because a close genetic relationship between the nene and the Canada goose has been reported previously (5), we sampled this polymorphic species more densely. Two lineages of Canada goose are recognized: a large-bodied lineage encompassing seven subspecies and a small-bodied lineage encompassing four extant subspecies (6). We sampled two subspecies from the large-bodied lineage (B. c. occidentalis and B. c. maxima) and three subspecies from the small-bodied lineage (B. c. canadensis, B. c. hutchinsii, and B. c. taverneri). Outgroups to Branta were six taxa of Anser (Fig. 4).
Figure 4.
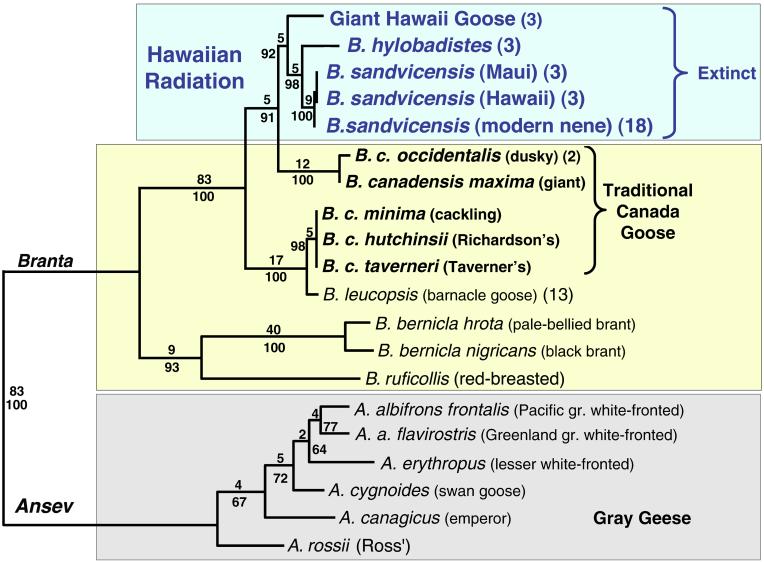
One of the two maximum parsimony (MP) phylograms found by using a heuristic search of 1,348 mitochondrial DNA sites in 20 taxa of true geese (Anserini). Group frequencies from a 1,000-replicate bootstrap (below branch) and Bremer decay indices (above branch) are indicated. Parenthetical numbers following taxon names indicate the number of individuals sequenced. Unweighted analysis tree statistics are: tree length = 529 steps, consistency index = 0.70, retention index = 0.85, and rescaled consistency index = 0.60. Support is strong in the MP analysis for a monophyletic Hawaiian clade arising from within the Canada goose and as sister taxon to the large-bodied subspecies (B. c. occidentalis and B. c. maxima). Maximum likelihood (ML) and minimum evolution (ME) analyses generally produced identical or nearly identical topologies (see Phylogenetic Inference).
DNA Amplification and Sequencing.
Measures were taken to detect and avoid contamination in the ancient DNA extractions and amplifications (7, 8). Fossil sequences were obtained before work with modern DNA samples. All fossil work was conducted in an ancient DNA laboratory established specifically for this task and located in a building separate from the main laboratory (7). For at least one specimen of each taxon, DNA isolations were replicated in temporally disjunct experiments to confirm results. For fossil material, PCR mixes were set up in a dedicated hood in the ancient DNA laboratory using appropriate contamination-control procedures and then brought to the main molecular genetics lab for thermocycling (7). PCR products were used in a few cases as templates in second-round amplifications. The same regions were amplified and sequenced for modern material under similar PCR conditions (7). For all ancient and modern reactions, amplification products were not detected in the negative extraction controls or PCR controls.
We amplified mitochondrial protein-coding regions including part of cytochrome b (307 bp) and all of ATPase subunit 8 (194 bp) and noncoding regions including hypervariable regions I (5′ end, 374 bp) and II (3′ end, 473 bp) of the mitochondrial control region (5′CR and 3′CR). To maximize the probability of successfully amplifying the fragmented and degraded template obtained from the fossil bones, internal PCR primers also were designed to flank short stretches (50–250 bp) of the mitochondrial regions of interest (7). Oligonucleotide primers used for PCR amplification and sequencing are listed by mitochondrial region. H and L refer to heavy and light strands of the mitochondrial genome, and the numbers refer to the position of the 3′ end of the primer on chicken sequence published in ref. 9. The PCR primers for the 5′CR were: Lnecr (L00078), 5′-tttggttatgcatattcgtg-3′; Hnecr (H00270), 5′-tgttatgtctggaagcattc-3′; C1R2 (H00390) and C1R (H00493), see ref. 3. The PCR primers for the 3′CR were: tPhe2 (H01249), 5′-cttcagtgccatgctttgtg-3′; GCDR (L01117), 5′-tattagagaaactccagtac-3′; CSB1 (L00887), 5′-tattggatgaatgctcgttg-3′; C2 (L00736), see ref. 3. The PCR primers for ATPase subunit 8 were: tLys(L9051), 5′-caccagcactagccttttaag-3′; birdsRus (H9241), 5′-tggtcgaagaagcttaggttca-3′. The PCR primers for cytochrome b are modified from ref. 10: Cytb1Kocher (L14990), 5′-tccaacatctccgcatgatgaaa-3′; Cytb2Kocher (H15298), 5′-tcagaatgatatttgtcctca-3′; Cytb3 (L15104), see ref. 3; Cytb4 (H15021), 5′-gtatgggtgaaatggaattttgtc-3′.
We sequenced ≈30 ng of amplification product for each 100 bp of sequence by using ABI Prism Ready-reaction dye/deoxy terminator chemistry (PE Biosystems). The reactions then were electrophoresed on an ABI 373 Stretch DNA sequencer. Sequences from ancient samples were unambiguous with one exception: although extraction and PCR controls detected no contamination, the cytochrome b sequence from one of the giant Hawaii goose extractions (Gigo-2) was contaminated with a second sequence and was excluded from these analyses.
Phylogenetic Analysis.
For phylogenetic inference, the sequences from the various gene fragments were combined for a total evidence analysis (11, 12). The CR sequences were aligned first by using the program MALIGN (13). In the final analyses, transversion substitutions and insertion/deletion events were weighted four times transition substitutions. However, different weighting schemes, including equal weights, yielded similar results. We also used the “optalign” feature of MALIGN (14) to find the shortest tree(s) without constructing a multiple alignment. The CR sequences then were concatenated to sequence fragments from protein-coding regions for a total of 1,348 nucleotide sites. Of these, 335 were variable and 250 were parsimony-informative.
To prevent apical rearrangements within species during tree searches, we included only one haplotype for each of 20 terminal taxa in the phylogenetic analysis [except the nene, for which we included the “modern” haplotype and fossil haplotypes from Maui and Hawaii islands (see ref. 7)]. Pairwise divergence within each Hawaiian taxon (B. sandvicensis, B. hylobadistes, and giant Hawaii goose) was low relative to among taxon distances [e.g., mean Hasegawa–Kishino–Yano distance within the giant Hawaii goose was 0.0024 and within B. sandvicensis was 0.0015 (calculated by using ref. 12)].
We conducted MP, ML, and ME analyses by using the program PAUP* (12). Parsimony analyses used unweighted and weighted heuristic searches (15) with 10 replicates and tree bisection-reconnection branch swapping. Alignment-generated gaps were treated as a fifth character state, and weighted searches used the same character weights as for the CR alignment. Support for nodes in the trees was estimated by bootstrap analysis with 1,000 replications (16) and calculation of Bremer decay indices (17). Support for alternative hypothesized topologies was assessed by nonparametric Templeton tests (18).
We conducted a heuristic search in an ME analysis by using a variety of evolutionary models (Fig. 4; ref. 12), both with and without variation in rates among sites as estimated by the Γ parameter (19, 20). We used two estimates of α: 0.496 calculated from the MP trees and 1.004 from the ML analysis below. We also bootstrapped the trees with 1,000 replications. We conducted a heuristic search by using an ML criterion and a general time-reversible model of sequence evolution that included estimation of the transition to transversion ratio, α from a Γ distribution (20), and empirical base frequencies. This tree was bootstrapped 100 times by using a stepwise addition of taxa, a fast method that may underestimate bootstrap support.
Results and Discussion
Phylogenetic Inference.
We obtained two optimal trees for the phylogeny of the tribe Anserini in both weighted and unweighted MP analyses (Fig. 4), differing only in the relationships among the small-bodied subspecies of Canada goose. Bootstrap analysis and Bremer decay indices (17) show strong support for nearly all nodes within Branta (Fig. 4). The removal of gap sites from the MP analysis results in the same topology and a similar level of support (results not shown). Support is strong for a monophyletic Hawaiian Branta lineage that is sister to the large-bodied Canada goose subspecies and embedded within the Canada goose (Fig. 4).
The ML heuristic search resulted in a single tree with a topology identical to that of the MP tree (results not shown). ME trees constructed without a Γ correction topologically match both the MP and ML trees (Fig. 4). However, with Γ, the Hawaiian clade is placed as a sister group to the small-bodied Canada goose lineage. This placement occurred despite a much closer, uncorrected sequence divergence between the Hawaiian clade and the large-bodied geese (3.0 versus 4.5%). Bootstrapping supports Hawaiian lineage monophyly (with frequencies of 92% for unweighted MP, 75% for ME, and 52% for ML) and placement within the Canada goose clade (100%). Sister-group status for the Hawaiian and large-bodied Canada geese lineages is supported by unweighted MP (91%) and ML (66%) but not by ME (47%) bootstraps.
The most surprising outcome of our study is the placement of the giant Hawaii goose. A recent morphological study placed this species as sister to the genus Branta (4), yet on our trees the giant Hawaii goose is nested within the species B. canadensis. [Note that in ref. 4, the giant Hawaii goose is considered to be conspecific with Geochen rhuax (21). In our view, G. rhuax is a nomen dubium, and consequently, the giant Hawaii goose is an undescribed species.) Also unexpectedly, the giant Hawaii goose seems to be the sister group of a clade consisting of other Hawaiian geese. Our placement of the giant Hawaii goose is well supported (Fig. 4); moving its branch on our weighted MP trees to make it sister to the genus Branta, as Livezey (4) hypothesized, increases the tree length by 72 steps, a highly significant increase (nonparametric Templeton test: Z = −5.63, P < 0.0001). Our results imply that a single population of Canada goose became resident in the Hawaiian islands and gave rise to the diverse true geese of the islands, perhaps including the additional undescribed fossil forms from Kauai and Oahu (1). Thus, the endangered nene is the only surviving member of an overlooked radiation of Hawaiian Branta.
A sister-group relationship between the Hawaiian Branta and the two large-bodied subspecies of B. canadensis also is supported by our study (Fig. 4). In a previous study (5), restriction fragment and cytochrome b sequence data were interpreted as supporting a sister relationship between the nene and the Canada goose complex. However, the accompanying phylogenetic analysis showed unresolved relationships among the nene and both large- and small-bodied B. canadensis subspecies. Our trees show that the barnacle goose (Branta leucopsis) falls within the currently recognized species of Canada goose and is the sister group of the small-bodied lineage. Moving the barnacle goose outside of the Canada goose clade significantly increases tree length (Templeton test, Z = 3.91, P < 0.0001). Thus, five morphologically distinct species of geese are represented as lineages of B. canadensis. Clearly, the widespread and familiar Canada goose is a paraphyletic taxon.
Evolutionary Time Frame.
We used the molecular data to estimate the divergence time of the giant Hawaii goose from its closest relatives and the divergence time of the Hawaiian radiation from its Canada goose sister lineage. Distance (22) and ML approaches (23, 24) gave nearly identical divergence times and confidence intervals, and we report only the latter values here. A fossil-based estimate of the separation date of Anser and Branta was used to compute a rate of sequence divergence. The date of the Anser and Branta split is estimated from the presence in the fossil record of diagnosable remains of both up to 4–5 million years ago (mya; refs. 25 and 26), and thus we used 4.5 million years (Myr) as a date for the calibration. These are minimum ages of divergence for the taxa and thus estimate maximum rates of sequence evolution.
We used a likelihood tree topologically identical to that shown in Fig. 4 (constructed with gap sites excluded in PAUP* 4.0b8, ref. 12), and rooted with a sequence of Aythya americana (GenBank accession no. 000877). Date estimates were made by using RHINO, a modified version of the QDATE program (23) as used previously in ref. 24. This program allows for rate heterogeneity in different parts of the ML tree during the estimation procedure. The general time-reversible model of nucleotide substitution was applied along with a discrete γ (four rate categories) and invariable sites estimates. The invariant sites and γ values estimated were 0.523 and 1.004, respectively. Confidence intervals were obtained by using likelihood ratio testing as described in ref. 23. Phylogenetic uncertainty and molecular clock stochasticity both are incorporated into these error calculations. Only the error in the fossil date is unaccounted for. Rates calculated from the distance analyses (22) were typical of avian mtDNA sequences [e.g., using all sites, k = 0.030 (95% confidence interval = 0.026–0.035), and using protein-coding sites only, k = 0.021 (0.016–0.027); refs. 27–29 and references within].
The ML estimates of divergence represent the coalescence of the mtDNA sequences, which should predate the divergence of the taxa (30, 31). The difference between the two can be estimated empirically. The simplest method is subtraction of the length of time corresponding to the mean within taxon distance for the ancestral taxon (30, 31). For bird mtDNA, these values average ≈0.175 Myr (31). Accounting for this error associated with lineage sorting, the estimated time of divergence of B. sandvicensis and the giant Hawaii goose is 0.566 Myr (95% consistency index = 0.395–0.880 Myr). Thus, the estimated lower confidence limit on the divergence time is consistent with an origin of the giant Hawaii goose on the island of Hawaii, an island whose earliest subaerial magmas (Kohala volcano) are dated ≈0.43 mya (32). If the adjacent and now-submerged volcano of Mahukona had been connected to Kohala, as is suggested in ref. 32, the date of possible colonization would extend back to ≈0.50 mya.
We also estimated the divergence time of the Hawaiian Branta lineage and the large Canada goose lineage to be 0.890 Myr (95% consistency index = 0.577–1.294 Myr). Thus, the Branta lineage in Hawaii seems to have resulted from a fairly recent colonization and radiation. In contrast, Sorenson et al. (3) estimate that the moa-nalos colonized the archipelago ≈3.6 mya.
Morphological Evolution.
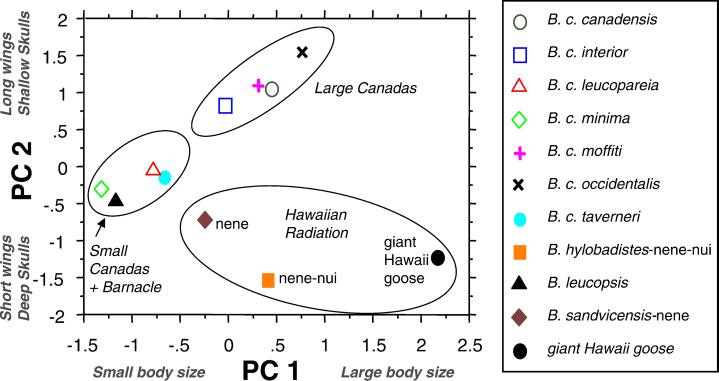
Body proportions of Hawaiian Branta can be compared with those of mainland Canada geese to shed light on morphological changes that have occurred in the Hawaiian lineage after it split from its continental ancestor. We used principal components analysis of 10 log-transformed osteological measurements to examine changes in the cranium, wing, and hind limb (skull depth; maxilla length and depth; lengths of the humerus, ulna, carpometacarpus, femur, tibiotarsus, and tarsometatarsus; and tibiotarsus circumference). One individual was measured representing each of four large subspecies of Canada goose, three small subspecies of Canada goose, the barnacle goose, and the three Hawaiian species. Principal components analysis in STATVIEW (Version 4.5) with a correlation matrix and orthogonal rotation of axes found two major components with eigenvalues of 6.74 (67% of the variance) and 2.87 (28% of the variance). The first component mostly represents relative size, with high positive factor loadings on the skull and hind limb measurements (0.92–0.98) and lower positive loadings on the wing measurements (humerus, 0.56; ulna, 0.32; carpometacarpus, 0.20). Factor two reflects shape variance, contrasting the wing measurements, which receive high positive factor loadings (carpometacarpus, 0.98; ulna, 0.94; humerus, 0.82), primarily with maxilla depth, skull depth, and tibiotarsus circumference (factor loadings −0.31, −0.23, and −0.29).
A plot of factor scores for the taxa on the first two components (Fig. 5) reveals an allometric relationship among the Canada goose subspecies, in which larger birds have relatively long wings and shallow bills and skulls. The barnacle goose conforms to this pattern. However, the three Hawaiian species plot on a different area of the graph. Examination of the factor loadings reveals that the Hawaiian birds have short wings and deep skulls and bills in comparison with continental geese.
Figure 5.
Plot of principal component scores derived from osteological measurements for Branta species and subspecies (see Morphological Evolution for details).
The broad spread of factor scores for the Hawaiian species on component one reflects evolutionary changes in body size within the Hawaiian lineage (Fig. 5). Because this component gives only a relative indication of body size, we obtained a direct estimate of body mass in the giant Hawaii goose by using published regression equations for estimating body mass from tibiotarsus circumference (33). Two equations (one based on all birds and one based on waterfowl only) gave an average mass estimate of 8.6 kg for seven individuals of the giant Hawaii goose. Comparing this figure with body masses of extant waterfowl (6), we estimate that the giant Hawaii goose was on average 1.4 times heavier than the largest subspecies of continental Canada goose (B. canadensis maxima) and 4.3 times heavier than its closest living relative, the nene (6). We can conclude with a high degree of confidence that the giant Hawaii goose evolved large body size within the islands. More tentatively, our study suggests that the nene evolved to become smaller in body size within the islands. The MP analysis indicates with 91% bootstrap confidence that the Hawaiian lineage is sister to the large-bodied subspecies of Canada goose. If the ancestor of the Hawaiian lineage had a body mass within the range of the large subspecies of Canada goose, then the nene has evolved to become at least 24% lighter in body mass (in comparison with B. c. parvipes, the smallest subspecies of the large-bodied lineage of Canada goose, ref. 6).
Most of these evolutionary changes in morphology are evident from direct comparisons of the birds' bones (Figs. 2 and 3). Compared with the skull of B. canadensis maxima (a large-bodied subspecies), the bills of B. sandvicensis and B. hylobadistes are shorter and relatively deep (Fig. 2 a–d). The bill of the giant Hawaii goose is much larger and deeper, and the entire skull is much larger and more robust (Fig. 2b). As can be visualized by comparing relative lengths of the tibiotarsus and ulna, the continental B. canadensis maxima has a relatively long wing with ulna length greater than tibiotarsus length, which is a reflection of the well developed pectoral girdle needed for migratory flights (Fig. 3a). B. sandvicensis, a nonmigratory island endemic that still is capable of strong flight, has a relatively short wing with ulna length less than tibiotarsus length (Fig. 3d). Reduction of the wing relative to the leg has progressed even farther in B. hylobadistes, in which the ulna is markedly shorter than the tibiotarsus (Fig. 3c); this species was flightless or at most capable of weak flight. Finally, in the flightless giant Hawaii goose, there has been a dramatic increase in overall size and further reduction of the wing (Fig. 3b).
Conclusions.
The giant Hawaii goose is restricted to an island that is only ≈430,000–500,000 years old (32) and is separated from its closest relatives by small genetic distances. Application of a local rate calibration suggests that it split from an ancestor with the nene less than ≈0.5 mya. This result supports the hypothesis that its gigantism, flightlessness, and robust cranial morphology evolved during that time frame on the island of Hawaii. Although extreme morphological change occurred on the island of Hawaii, geese from the same ancestral clade on the older Hawaiian islands underwent more modest changes. The older islands were occupied already by a lineage of terrestrial dabbling ducks (the moa-nalos) that had evolved similar body proportions to the giant Hawaii goose (Fig. 1; refs. 1–3). The independent evolution of flightlessness, gigantism, and deep bills and skulls in the distantly related giant Hawaii goose on the island of Hawaii and the moa-nalos on the older main islands suggests that these changes occurred in response to similar selective factors on different islands in the chain. The most likely explanation is that in the absence of mammalian herbivores, selection on the waterfowl favored adaptation for terrestrial herbivory (34).
Our data also indicate that both the radiation of Hawaiian Branta and the Palearctic barnacle goose are nested within a paraphyletic Canada goose clade. The large- and small-bodied lineages of Canada goose have long been viewed as a single species because of their shared plumage pattern, which now appears to be plesiomorphic, and their parapatric breeding and wintering distributions in the Nearctic. However, a morphological and molecular study suggested that these lineages are separate species (5, 35), and our results could be interpreted as supporting this interpretation. Alternatively, the mtDNA divergence between large- and small-bodied subspecies could reflect long-term retention of divergent mtDNA clades from which two morphologically divergent lineages subsequently arose. In this case the Canada goose could be envisioned as a “living ancestor” of a diverse goose radiation.
Until now, the sister relationship of the barnacle goose with the small-bodied lineage of Canada goose was obscured by its apomorphic plumage pattern and discontinuous distribution in the Palearctic. Although the barnacle goose is peripherally isolated, it inhabits a similar environment to Canada geese. Its body proportions are similar to its sister taxon, suggesting that these two lineages have experienced stabilizing selection on body form under similar ecological conditions.
On the other hand, the ancestor of the Hawaiian radiation colonized a very different ecosystem. This ancestor underwent two important changes in life history: (i) loss of migration and (ii) a niche shift from mainly wetland to mainly terrestrial habitats. These changes may underlie two morphological traits shared by all members of the radiation: reduction in wing length (probably because of loss of migration) and increase in the depth of the skull and bill (probably because of dietary shift). Interrupted gene flow between sedentary populations on different islands of the chain permitted speciation and diversification of the lineage, leading to dramatic differences in body size and proportions.
Acknowledgments
We thank J. Giffin, W. Cuccaro, A. Cooper, P. Fiene-Severns, W. Covington, M. Severns, B. Schaefer, and C. Kishinami for field assistance. We are very grateful to C. Kishinami (Bishop Museum, BPBM) and M. and A. Lubbock, who provided samples for analysis. A. Cooper and A. Rambaut provided valuable advice on laboratory and statistical analyses and Mary Parrish provided the stipple art for Fig. 3. Kula Forest Reserve and Pohakuloa Military Installation provided permission to sample specimens. This research was supported in part by the Scholarly Studies Program, Walcott and Radiocarbon Funds of the Smithsonian Institution, Friends of the National Zoo, and the National Geographic Society.
Abbreviations
- MP
maximum parsimony
- CR
control region
- ML
maximum likelihood
- ME
minimum evolution
- mya
million years ago
- Myr
million years
Footnotes
References
- 1.Olson S L, James H F. Ornithol Monogr. 1991;45:1–88. [Google Scholar]
- 2.James H F. In: Ecological Studies. Vitousek P, editor. Vol. 115. Berlin: Springer; 1995. pp. 87–102. [Google Scholar]
- 3.Sorenson M D, Cooper A, Paxinos E E, Quinn T W, James H F, Olson S L, Fleischer R C. Proc R Soc London Ser B. 1999;266:2187–2194. doi: 10.1098/rspb.1999.0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livezey B. Syst Biol. 1996;45:415–450. [Google Scholar]
- 5.Quinn T W, Shields G F, Wilson A C. Auk. 1991;108:585–593. [Google Scholar]
- 6.Todd F S. Natural History of the Waterfowl. Vista, CA: Ibis; 1996. [Google Scholar]
- 7.Paxinos E E. Doctoral Dissertation. Providence, RI: Brown University; 1998. [Google Scholar]
- 8.Handt O, Hoss M, Krings M, Pääbo S. Experientia. 1994;50:524–529. doi: 10.1007/BF01921720. [DOI] [PubMed] [Google Scholar]
- 9.Desjardins P, Morais R. J Mol Biol. 1990;212:599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- 10.Kocher T D, Thomas W K, Meyer A, Edwards S V, Pääbo S, Villablanca F X, Wilson A C. Proc Natl Acad Sci USA. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kluge A G. Syst Zool. 1989;38:7–25. [Google Scholar]
- 12.Swofford D L. paup*: Phylogenetic Analyses Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 2000. , Version 4.0d65. [Google Scholar]
- 13.Wheeler W C, Gladstein D G. malign. New York: The American Museum of Natural History; 1992. [Google Scholar]
- 14.Wheeler W C. Cladistics. 1996;12:1–9. [Google Scholar]
- 15.Farris J S. In: Advances in Cladistics. Platnick N I, Funk V A, editors. Vol. 2. New York: Columbia Univ. Press; 1983. pp. 7–36. [Google Scholar]
- 16.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 17.Bremer K. Evolution (Lawrence, Kans) 1988;42:795–803. doi: 10.1111/j.1558-5646.1988.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 18.Templeton A R. Evolution (Lawrence, Kans) 1983;37:221–244. doi: 10.1111/j.1558-5646.1983.tb05533.x. [DOI] [PubMed] [Google Scholar]
- 19.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;21:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan J, Holsinger K E, Simon C. Mol Biol Evol. 1995;12:988–1001. doi: 10.1093/oxfordjournals.molbev.a040292. [DOI] [PubMed] [Google Scholar]
- 21.Wetmore A. The Condor. 1943;45:146–148. [Google Scholar]
- 22.Kumar S, Tamura K, Jakobsen I, Nei M. mega: Molecular Evolutionary Genetic Analysis. Tempe, AZ: Arizona State University; 2001. , Version 2.1. [Google Scholar]
- 23.Rambaut A, Bromham L. Mol Biol Evol. 1998;15:442–448. doi: 10.1093/oxfordjournals.molbev.a025940. [DOI] [PubMed] [Google Scholar]
- 24.Cooper A, Lalueza-Fox C, Anderson S, Rambaut A, Austin J, Ward R. Nature (London) 2001;409:704–707. doi: 10.1038/35055536. [DOI] [PubMed] [Google Scholar]
- 25.Olson S L, Rasmussen P C. Smithson Contrib Paleobiology. 2001;90:233–365. [Google Scholar]
- 26.Bickart K J. Ornithol Monogr. 1990;44:1–72. [Google Scholar]
- 27.Shields G F, Wilson A C. J Mol Evol. 1987;24:212–217. doi: 10.1007/BF02111234. [DOI] [PubMed] [Google Scholar]
- 28.Klicka J, Zink R M. Science. 1997;277:1666–1669. [Google Scholar]
- 29.Fleischer R C, McIntosh C E, Tarr C T. Mol Ecol. 1998;7:533–545. doi: 10.1046/j.1365-294x.1998.00364.x. [DOI] [PubMed] [Google Scholar]
- 30.Moore W S. Evolution (Lawrence, Kans) 1995;49:718–726. doi: 10.1111/j.1558-5646.1995.tb02308.x. [DOI] [PubMed] [Google Scholar]
- 31.Edwards S V, Beerli P. Evolution (Lawrence, Kans) 2000;54:1839–1854. doi: 10.1111/j.0014-3820.2000.tb01231.x. [DOI] [PubMed] [Google Scholar]
- 32.Carson H L, Clague D A. In: Hawaiian Biogeography: Evolution on a Hotspot Archipelago. Wagner W L, Funk VA, editors. Washington, DC: Smithsonian Institution Press; 1995. [Google Scholar]
- 33.Campbell K E, Marcus L. Natural History of Los Angeles County, Science Series. 1992;36:395–412. [Google Scholar]
- 34.James H F, Burney D A. Biol J Linn Soc. 1997;62:279–297. [Google Scholar]
- 35.Aldrich J W. Wilson Bull. 1946;58:94–103. [Google Scholar]