Abstract
The phylogenetic relationships of amoebae are poorly resolved. To address this difficult question, we have sequenced 1,280 expressed sequence tags from Mastigamoeba balamuthi and assembled a large data set containing 123 genes for representatives of three phenotypically highly divergent major amoeboid lineages: Pelobionta, Entamoebidae, and Mycetozoa. Phylogenetic reconstruction was performed on ≈25,000 aa positions for 30 species by using maximum-likelihood approaches. All well-established eukaryotic groups were recovered with high statistical support, validating our approach. Interestingly, the three amoeboid lineages strongly clustered together in agreement with the Conosa hypothesis [as defined by T. Cavalier-Smith (1998) Biol. Rev. Cambridge Philos. Soc. 73, 203–266]. Two amitochondriate amoebae, the free-living Mastigamoeba and the human parasite Entamoeba, formed a significant sister group to the exclusion of the mycetozoan Dictyostelium. This result suggested that a part of the reductive process in the evolution of Entamoeba (e.g., loss of typical mitochondria) occurred in its free-living ancestors. Applying this inexpensive expressed sequence tag approach to many other lineages will surely improve our understanding of eukaryotic evolution.
Unicellular amoebae are possibly the simplest eukaryotic organisms in morphological terms. The locomotion of these organisms with pseudopodia provided the basis for classifying them together as Rhizopoda, one of the four classes in the classical taxonomy of protozoa. Although the old textbook description of amoebae as a “blob of cytoplasm with a nucleus” is clearly obsolete, they exhibit few morphological traits that can be used as taxonomic characters. In the past, size and shape of the body and the pseudopodia, the absence or presence of flagella or a flagellated life cycle stage, the properties of the cytoplasm and nucleus, and a few other characteristics have been used to classify amoeboid protists. This process has led to a proliferation of taxonomic schemes, none of which is fully convincing (1–3). Ultrastructural studies have disclosed a number of additional morphological features, but have helped little in putting the taxonomy of amoebae on a firm basis. The classification of the vast and diverse group of amoeboid organisms is still in constant flux, and their genuine evolutionary relationships remain uncertain. For most such organisms no molecular information is available and even rRNA-encoding DNA (rDNA) sequences have been determined for only a few species. Phylogenies based on this molecule with varying species sampling and tree reconstruction methods often suggest paraphyly of different amoeboid genera, with the following order of emergence: Physarum, Entamoeba, Dictyostelium, Mastigamoeba, and Acanthamoeba (4–8). However, a few genera, for example, Mastigamoeba and Entamoeba (9, 10), sometimes group together. Indeed, problems in tree reconstruction, such as long branch attraction artefacts (LBA) (11), affect rDNA phylogenies. Detailed studies with complex models of sequence evolution reveal that there is not enough signal in rDNA to support paraphyly of amoebae (12, 13).
Among amoeboid organisms, three extensively studied species represent some of the phenotypically most divergent groups: the cellular slime mold Dictyostelium discoideum, the pelobiont Mastigamoeba balamuthi, and the entamoebid Entamoeba histolytica. These are dramatically different in their morphology and biology. One of the most striking differences is that D. discoideum is a typical mitochondrion-containing eukaryote, whereas M. balamuthi and E. histolytica are amitochondriate (14–17). Not surprisingly, the three species are assigned to separate lineages in most taxonomic schemes (3). However, Cavalier-Smith (18) in his recent “revised six-kingdom system of life” suggested that their great phenotypic diversity notwithstanding these three organisms are closely related, and placed them in the newly erected Subphylum Conosa (Phylum Amoebozoa, Infrakingdom Sarcomastigina, Kingdom Protozoa). In the following, we refer to this grouping as the Conosa hypothesis.
Sequences of single genes or few concatenated genes did little to resolve the genuine relationship of these three organisms. Only a few genes are presently available from Mastigamoeba. In RNA polymerase II phylogenies, Acanthamoeba, Dictyostelium, and Mastigamoeba do not cluster together (19). For enolase, Mastigamoeba groups with Entamoeba but also with the flagellated protist Trypanosoma (20). Furthermore, Dictyostelium harbors two copies of this gene, rendering the interpretation of enolase tree problematic (E.B., unpublished work). A combined analysis of small and large subunit rDNAs and the two elongation factors (EF-1α and EF-2) indicates the monophyly of Conosa, but without significant statistical support, despite the use of ≈5,000 positions (21). In single gene analyses, Dictyostelium and Entamoeba do not generally group together (22–28). Yet, sometimes the same genes provide a weak support for the sister grouping of Dictyostelium and Entamoeba [e.g., cpn60 (29) or tubulin (30)]. In contrast, the monophyly of Mycetozoa (slime molds such as Dictyostelium, Physarum, and Planoprotostelium) is robustly supported by EF-1α (31) and actin (32) phylogenies. This finding is consistent with the shared presence of fused cox1 and cox2 genes on their mitochondrial genomes (33) and with the results of combined protein data analysis (34).
These contradictory, admittedly weakly supported, results of molecular phylogeny are caused by systematic and stochastic errors in tree reconstruction. Evolutionary rates are quite variable between genes as well as between species (35). For example, amoebae appear to evolve very fast for tubulin but very slowly for actin, whereas exactly the contrary is observed for ciliates (32). Variable evolutionary rates generate artificial groupings in the eukaryotic molecular tree (36), because of LBA. Taking into account among-site rate variation through a Γ law is known to alleviate the LBA problem (37), which has been successfully shown in the case of the eukaryotic rRNA tree (38). Unfortunately, the covarion-like structure of molecular markers could limit the success of this approach (13). Stochastic errors are likely also responsible for this wide range of results, because statistical supports are generally low. This finding is not unexpected because the size of the commonly used genes is ≈300 positions, which provide little information to resolve such ancient events as the diversification of eukaryotes. It is thus not surprising that much recent progress has been based on the analysis of combined protein data sets (24, 34, 39).
To overcome the lack of resolution observed in amoeba phylogenies, we have analyzed several hundred phylogenetic markers simultaneously. This analysis was achieved by the sequencing of 1,280 expressed sequence tags (EST) from M. balamuthi. Thanks to the ongoing genome projects of D. discoideum and E. histolytica, we were able to include these three amoebae in a data set of 123 genes for which orthology is undisputed. The analysis of this alignment of considerable size (25,000 aa positions and 30 species) provided very strong support for the monophyly of Conosa (M. balamuthi, D. discoideum, and E. histolytica).
Materials and Methods
Mastigamoeba ESTs.
Putative protein sequences from M. balamuthi (ATCC 30984) (15) were obtained from our EST project. Details of the procedures followed will be published elsewhere. In brief, a directionally cloned library was constructed by synthesizing cDNA from poly(A)+ RNA isolated from this organism with the use of a cDNA Synthesis Kit and cloning into the Lambda ZAP II vector with the ZAP cDNA Gigapack III Gold Cloning Kit, both from Stratagene. An aliquot of this library containing a random collection of clones was excised by superinfection with helper phage. Clones were selected randomly and sequenced on both strands. The two single-strand sequences for each clone were aligned into a contig by using the Staden assembly package. Contigs were then entered into magpie, customized for this EST project (40, 41). The sequences are available at http://niji.imb.nrc.ca/magpie/newrock/private/ and have been deposited in the GenBank database.
Construction of the Alignment.
Our aim was to find as many protein-encoding genes as possible for which an archaeal outgroup as well as a good diversity of eukaryotic phyla were available. Starting from all of the ESTs from Mastigamoeba and Porphyra yezoensis (42), tblastn searches were performed on the five complete archaeal genomes published by the end of 1999. If at least three species had a blast score lower than 10−6, a tblastn search was run against the National Center for Biotechnology Information nonredundant database. All protein sequences with a blast score lower than 10−6 were then retrieved with the program alibaba (P.L., unpublished work). Each set of sequences was aligned with clustal w (43), and the alignment was manually refined with the ed program (44). A preliminary analysis was performed by using the neighbor-joining (NJ) method (45). We retained only genes for which (i) eukaryotes were clearly monophyletic and (ii) the archaeal homologs were more similar to the eukaryotic ones than were the bacterial ones (even if a few bacterial species seem to have acquired the corresponding gene through lateral gene transfer from Archaea). We have retained 94 genes, of which 78 did not show ancient duplications, whereas 16 additional gene families contained 62 paralogs for which duplication events very likely occurred before the last common ancestor of extant eukaryotes (e.g., eight paralogs in the CCT gene).
To increase the number of represented eukaryotic phyla, for 25 species we included nucleotide sequences obtained from the web sites of ongoing EST and genome projects (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). To detect the homologs of the above-mentioned 94 genes for these 25 species, we wrote a program that launched a tblastn search by using Arabidopsis thaliana amino acid sequence as the seed, except for fungal species for which Saccharomyces cerevisiae was used (H.P., unpublished work). For genes with several paralogs, tblastn searches were performed for each paralog. All of the high-scoring segments with a blast score below 10−10 were retained and their sequences were added to the file containing the already aligned sequences, according to the alignments in the blast results. To detect obvious contaminant sequences, which can occur in large-scale sequencing projects, we constructed a maximum parsimony (MP) tree with PAUP 4b8 (46). This method is fast and not too sensitive to missing data, a frequent phenomenon because several sequences are partial. We detected exclusively mammalian contaminants for apicomplexan species and yeast ones for Dictyostelium.
To construct a consensus sequence for each species starting from the multiple sources, new options were added to the ed program (44). These allow easy handling of large number of sequences (e.g., 100 sequences for a single species). Moreover a quick removal of introns and sequence regions of poor quality (e.g., many ambiguous characters or frame shifts) was implemented. Because EST data are not free of errors, great care was taken in retaining only regions for which comparison with homologous sequences strongly suggested a high quality, sufficient for phylogenetic analysis. The quality of these consensus sequences was attested a posteriori when after their construction several sequences obtained by high-quality approaches (e.g., genomic data from rice) appeared in the data bank and showed less than 1% difference in overlapping regions.
A custom software, split-para, was written to deal with genes containing paralogs. This program allows one to create as many files containing aligned orthologous sequences and archaeal ones as there are paralogs for the gene. For each gene, only unambiguously aligned regions were retained. The alignments of the 140 orthologous genes are available from H.P. on request. We selected seven completely sequenced archaea (two crenotes and five euryotes) and all of the 23 eukaryotic groups for which most of the 140 genes are available. In several cases, we constructed chimeric sequences to represent important groups more comprehensively, primarily from the species indicated below: basidiomycetes (Cryptococcus neoformans, Coprinus cinereus, and Ustilago maydis), stramenopiles (Phytophthora infestans and Laminaria digitata), ciliates (Euplotes, Paramecium, and Tetrahymena), Sarcocystidae (Toxoplasma gondii, Neospora caninum, and Sarcocystis), chlorophytes (Chlamydomonas spp.), monocots (Oryza sativa and Zea mays), and rhodophytes (Porphyra spp.).
Phylogenetic Analysis.
Phylogenetic trees were based on the analysis of amino acid sequences with maximum likelihood (ML), MP, and NJ methods with the programs PROTML 2.3 (47) and TREE-PUZZLE 5.0 (48), PAUP 4d8 (46), and MUST 3.0 (44), respectively. We constructed concatenated data sets of the 140 genes for the 30 species, allowing a variable number of missing species per gene.
Because of computing time and memory limitations, a first fusion, allowing only two missing species (56 genes only) and comprising 10,037 positions was used to select the most likely topologies. First, ML trees were obtained by the quick add search, with the JTT model of amino acid substitution and retention of the 2,000 top-ranking trees. Bootstrap values (BVs) were computed by the RELL method (49). This first ML analysis, together with MP and NJ bootstrap analyses, allowed us to define several phylogenetic constraints that were biologically reasonable and generally supported by the data (except for the position of nematodes, see ref. 50): the phylogenies of Archaea the phylogenies of Archaea (Aeropyrum, Sulfolobus), (Pyrococcus, (Methanococcus, (Thermoplasma, (Archaeoglobus, (Halobacterium))))); Opisthokonta ((Basidiomycetes, (((Candida, Saccharomyces), Neurospora), Schizosaccharomyces)) (mammals, (Caenorhabditis, Drosophila))); Plantae, (((Arabidopsis, monocots), green algae), (red algae, nucleomorph of Guillardia)); kinetoplastids (Trypanosoma, Leishmania); and alveolates ((Sarcocystidae, Plasmodium), ciliates). However, as an exhaustive search was unrealistic even with these constraints (more than 2 million possible topologies), we separately added two different constraints [the monophyly of Conosa, found in preliminary analysis, 31,185 possible topologies, and the grouping Plantae (alveolates, stramenopiles); ref. 34; 10,395 possible topologies], and we retained the 1,000 best topologies for both exhaustive protml searches. This resulted in a set of 1,961 topologies (39 being common to the two searches), which were used for further analysis. Because half of the studied topologies did not support the monophyly of Conosa, our approach should not introduce a bias favoring its recovery.
The detailed phylogenetic analysis was performed on the 123 genes with a maximum of seven missing sequences. Departing from the standard use of concatenated sequences, we computed the likelihood of the 1,961 topologies for each gene and selected the best topology as the one that had the minimal sum of likelihood values of all genes. This allowed the branch lengths and the α parameter (when used) to be different for each gene, an important consideration given the variability of evolutionary rates between species and between genes (35). Because the model used for the concatenated sequences is nested within this model, one can perform a log-likelihood ratio test to test which model is the best (51). Twice the difference between the likelihood of concatenated sequences and the sum of the likelihood values of the 123 genes has to be compared with χ2 statistics with a number of degrees of freedom equal to the number of additional free parameters. In this case, the number of free parameters was the number of branches, 57 (2 × 30 − 3), plus the α parameter (when used) multiplied by the number of genes minus one (122), that is 6,954 degrees of freedom (7,076 with a Γ law).
To handle rate variation among sites, we computed likelihood values by using a Γ law model (eight discrete classes). In this case, because of computing time, we retained only 200 topologies among the 1,961 previously studied. The topologies were ranked in decreasing order of likelihood obtained in the analysis performed without Γ law. We kept the first 100 topologies, expected to also be the best ones with a Γ correction, and an additional 100 evenly spaced topologies to verify that the Γ correction does not completely modify the ranking with these constraints. Furthermore, to verify that missing sequences did not affect our results, we partitioned the complete data set between genes displaying fewer than two missing species (56 genes) and the remaining ones. For each of the 1,961 topologies, likelihoods were summed for each of the two subsets, leading to 1,961 pairs of values. If missing species significantly affected the results, then the correlation should be weaker than the one obtained by random partitioning. The significance of the correlation coefficient is thus assessed by computing the distribution produced by 10,000 random partitions of the data set (pools of 56 and 67 genes).
The reliability of the nodes was evaluated with a bootstrap analysis on the genes with 2,000 replicates, by using the same principle as the RELL method (49). To assess the influence of the number of genes on the phylogenetic inference, we applied a modified version of the PRN method of Lecointre et al. (52). For 11 different numbers of genes (n = 10, 20 … 110), we randomly drew n genes without replacement (jackknife), on which the bootstrap (drawing with replacement) was performed. The jackknife steps were repeated 1,000 times, and the mean and variance of BVs were computed for some selected nodes.
Results and Discussion
Data Set Construction.
By retrieving data from GenBank and current genome and EST sequencing projects, we were able to identify 140 orthologous genes for which an archaeal outgroup was available. To more specifically address the question of the phylogeny of amoebae, we sequenced 1,280 ESTs from the amitochondriate pelobiont, M. balamuthi. With a selection of 30 species representing most of the major eukaryotic phyla (animals, fungi, plants, red algae, stramenopiles, and alveolates) and three lineages of amoebae (Dictyostelium, Entamoeba, and Mastigamoeba), we retained 123 genes showing at most seven missing species, which provided 25,000 unambiguously aligned amino acid positions. Because of the computing time required to deal with such a huge data set, a first analysis was performed by assuming that all of the sites evolve at the same rate. We verified that missing species had little effect on the significance of our results, because the correlation coefficient found for the initial partition (i.e., genes with fewer than two missing species versus the remaining ones) was higher than the one found for 8% of the random partitions.
Likelihood Summation Versus Gene Concatenation.
Instead of concatenating genes as is generally done (24, 34), we followed the method proposed by Yang (51), by computing the likelihood for each gene and selecting the topology that minimizes the sum of the likelihood. This means that a different set of parameters was allowed for each gene, i.e., branch lengths and, when used, the α parameter of the Γ law were different for each gene. The model corresponding to the analysis of the concatenated sequences is nested within this model, because its constraint is that branch lengths and the α parameter are the same for all genes. We thus compared the fit of the two models with a log likelihood ratio test. Although our model had 6,954 additional parameters, it gave a significantly better fit to the data than the simplest one: 2ΔlnL = 2 × (771, 803–757, 078) = 29,450 (for P = 0.01, the χ2 limit is 6,954), indicating that the evolutionary rates on the branches of the phylogeny were significantly not proportional among the genes studied. The correlation between the likelihoods of the two models was good (r2 = 0.91), although the best tree was not the same. This finding indicated that the analysis of concatenated sequences was a good approximation for searching in the tree space, and we indeed used it to select the 2,000 most likely topologies on which we applied the most complex analysis (i.e., summing the likelihood of all of the genes).
Monophyly of Conosa.
The most likely phylogeny without a Γ law correction is shown in Fig. 1. All of the nodes indicated by an asterisk were constrained, as they were recovered with high BVs by the analysis of the concatenated sequences with ML, MP, and NJ methods. Yet, in a few cases, the cryptophyte nucleomorph (MP) and Caenorhabditis (NJ) emerged earlier than expected in the tree (50, 53), probably because of a LBA artifact. However, our large data set strongly recovered the monophyly of all of the eukaryotic phyla and three superphyla (Alveolata, Opisthokonta, and Plantae). The most interesting feature of this tree was the sister-group relationship of Entamoeba and Mastigamoeba, this group being clustered with Dictyostelium, thus confirming the clade Conosa (54). These two nodes were strongly supported by BVs (greater than 95%). These values may be inflated because computation imposes the use of RELL bootstrap instead of the standard one (49, 55). This is a robust molecular demonstration of the monophyly of an amoeba clade, consisting of Mycetozoa (represented by Dictyostelium), Entamoebidae, and Pelobionta (represented by Mastigamoeba). This finding is in sharp contrast with many analyses based on rRNA that suggested paraphyly of these organisms. The use of ≈25,000 characters instead of ≈1,000 is a likely explanation for this difference.
Figure 1.
ML tree based on 25,032 aa positions. * indicates a constrained node. We used the JTT model, without taking into account among-sites rate variation. The branch lengths have been computed on the concatenated sequences. BVs were obtained by bootstrapping the 123 genes.
Yet, in our phylogeny (Fig. 1), three nodes, such as the position of Conosa as sister group of opisthokonts, were not resolved (BVs between 55% and 68%) despite the very large data set. Finally, the early emergence of diplomonads and kinetoplastids received a high BV. Given the fact that their sequences display many autapomorphies (especially insertions/deletions for diplomonads) and the very long branch of the archaeal outgroup, these positions are likely caused by a LBA artifact (25, 56). This artifact has been shown to be important for short sequences (36), and it is all the more expected when long sequences are used [as the LBA artifact is a case where phylogenetic reconstruction methods are inconsistent (11), i.e., converge toward the wrong answer when more data are taken into account].
Impact of Rate-Across-Sites Correction.
The use of a more adequate model of sequence evolution is known to reduce the impact of LBA (37). We therefore took into account the variation of the evolutionary rate across sites by applying a Γ law, but with the number of topologies reduced to 200 because of prohibitive computation time (several months on a Sun Ultra 10 computer). A log-likelihood test clearly demonstrated that this model provided a much better fit to the data, despite its 122 additional free parameters: 2ΔlnL = 2 × (757, 078–727, 183) = 59,790 (for P = 0.01, the χ2 limit is 161). Yet, the phylogeny with a Γ model (Fig. 2) was quite similar to the one that did not take it into account (Fig. 1). The monophyly of Conosa was again recovered, with slightly higher BVs (97% and 98%). This finding strongly suggested that the recovery of this clade was not caused by a tree reconstruction artifact, a potential problem because the long branch of Entamoeba could have been attracted toward the base of the tree by the Archaea through LBA artifact and would indeed disrupt the monophyly of Conosa. The support for the sister group between Conosa and animals/fungi clade increased to 93%, in agreement with previous results showing a sister group between Mycetozoa and Opisthokonta (24, 34).
Figure 2.
ML tree based on 25,032 aa positions, taking into account among-sites rate variation (JTT + Γ model). See Fig. 1 for details.
Stramenopiles were sister group of alveolates instead of Plantae, albeit with a low support (62%). This grouping, called chromalveolates, was proposed first by Cavalier-Smith based on morphological criteria (18) and has received some support from the analysis performed on the combination of four genes (≈60%) (34). The most convincing evidence in favor of the monophyly of this clade is the presence of a duplicated copy of glyceraldehyde-3-phosphate dehydrogenase that is targeted to the chloroplast specifically in chromalveolates (57). The lack of monophyly as shown in Fig. 1 was probably caused by a LBA artifact generated by the long branch of alveolates, which was canceled out when among-site rate variation was taken into account. Yet, diplomonads and kinetoplastids still robustly emerged early (Fig. 2). This could be correct, but because the ML method is known to be inconsistent because of the covarion-like substitution pattern (58), we believed that these positions were caused by LBA.
Nevertheless, the improvement provided by the Γ law is very significant and much larger than the improvement provided by the separate analysis of the 123 genes. The best tree (Fig. 2) had a lnL of −727,183 (sum of likelihood with a Γ model), −740,344 (likelihood of concatenated sequences with a Γ model), −757,078 (sum of likelihood without a Γ model), and −771,803 (likelihood of concatenated sequences without a Γ model). The comparison between the likelihood of concatenated sequences with a Γ model and the sum of likelihood without a Γ model is difficult because the two models are not nested. However, the less parameter-rich model (one additional parameter, the α parameter) shows a lnL greater by 16,734 than the most parameter-rich model (6,954 additional parameters), which is completely the opposite of the expected. This finding strongly suggests that analysis of concatenated sequences with a Γ law provides better results than a separate analysis without Γ law does. Thus, the variation of evolutionary rate between positions was more important for inferring phylogeny than the variation of evolutionary rate between genes (i.e., the fact that branch lengths are not proportional).
How Many Genes To Be Considered?
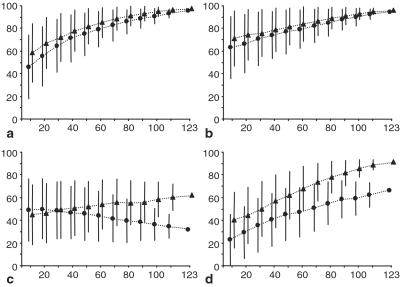
We have shown that the use of a huge data set (30 species, 123 genes, ≈25,000 positions) provided a robust answer for many nodes in the eukaryotic phylogeny, especially to the difficult question of the monophyly of Conosa. Yet, one can ask whether such a large-scale and time-consuming approach is necessary instead of the analysis of few genes [e.g., four (34) or 13 (24)]. A jackknifing of the genes was performed and the evolution of BVs for four nodes was studied, using the same approach as Lecointre et al. (52). The support for the monophyly of Entamoeba/Mastigamoeba (Fig. 3a) and Conosa (Fig. 3b) steadily increased with the number of genes used, with or without a Γ law model. Even if the mean BVs were rather high for 20 genes (between 60% and 70%), the standard deviation was quite large, indicating that even a large data set of 20 genes could provide a strong support for the paraphyly of Conosa. It is nevertheless possible that a single “lucky” gene could give a correct answer to a difficult problem. Actually, if it has undergone a considerable acceleration of its evolutionary rate on an internal branch (short in terms of time), this branch will be long and easy to recover. The branches at the base of chromalveolates for glyceraldehyde-3-phosphate dehydrogenase (57) and at the base of triploblastic animals for rDNA (35) are two clear examples. However, it is difficult to ascertain whether a single gene really provides the correct answer, especially because a lateral gene transfer or an unknown bias are possible alternative explanations. The use of a large number of genes allowed us to reduce the impact of stochastic errors and lateral gene transfers and to be very confident in the monophyly of this large group of amoebas.
Figure 3.
Effect of the number of genes studied on the evolution of BVs for the grouping of Entamoeba/Mastigamoeba (a), for the monophyly of Conosa (b), for the monophyly of chromalveolates (c), and for the sister group of Conosa and Opisthokonta (d). ● indicate the mean BVs without Γ law correction, and ▴ indicate the mean BVs with Γ law correction. Vertical bars correspond to 1 SD computed from 1,000 jackknife replicates.
However, Fig. 3 illustrates one possible pitfall of such a massive approach. Without Γ correction, the support in favor of the monophyly of chromalveolates (Fig. 3c) decreased when adding data and probably will converge toward a value of 0, because the ML tree reconstruction method used is inconsistent. Conversely, the use of a Γ correction drastically changed the pattern, because BVs increased and probably will converge toward 100% as more genes are added. An important effect of the Γ correction was also evident for the sister grouping of Conosa and Opisthokonta, the support increasing faster with the correction than without. These two cases demonstrated that when large data sets are used (e.g., 25,000 positions) it is of utmost importance that the tree reconstruction method be consistent. Unfortunately, this was probably not the case here because our method did not handle covarion structure, in particular the fact that the number of variable positions was different among lineages. Because diplomonads (especially Giardia) are known to display more variable positions than other eukaryotes (25), their well-supported early branching is probably caused by the inconsistency of the ML method (55, 58).
The Origin of the Amitochondriate Phenotype in Conosa.
Pelobionts and entamoebids for a long time have been regarded as ancestrally primitive and premitochondriate (e.g., refs. 14–16 and 59). The best-known pelobiont, Pelomyxa palustris, is a large multinucleate amoeba without mitochondria but that harbors three different prokaryotic endosymbionts (60), which was proposed to correspond to an intermediate stage in the emergence of the mitochondrion-containing eukaryotic cell (61). These notions, however, have been dramatically challenged in recent years (62).
The origin of Dictyostelium, Mastigamoeba, and Entamoeba from a common ancestor provides strong arguments against the ancestral nature of pelobionts and entamoebids. The results show that mitochondriate and amitochondriate phenotypes do not define major lineages and indicate that organisms with highly different metabolic properties can evolve within a single lineage. They also suggest that the ancestral metabolic phenotype in this lineage was mitochondriate and that the amitochondriate condition developed by a drastic reduction of the mitochondrial compartment. To assume development in the opposite direction would imply that the typical mitochondrion of slime molds is the result of convergent evolution, a highly untenable position. The outgroup position of Dictyostelium to the Entamoeba/Mastigamoeba clade probably reflects a genuine relationship and is not caused by LBA (see branch lengths in Figs. 1 and 2). This conclusion is concordant with the increasingly accepted notion that extant amitochondriate protists arose by regressive evolution from mitochondriate ancestors and do not represent primitive, premitochondrial organisms.
The small double membrane-bounded mitosome (crypton) of Entamoeba is probably the product of the reduction of the mitochondrial machinery (63, 64). Organelles of similar morphology are also present in Mastigamoeba and other pelobionts (15, 17), but their functional properties have not yet been investigated. Mastigamoeba is a free-living organism, whereas Entamoeba is an intestinal parasite. The common ancestor of the two organisms, in which the regression most likely occurred, was probably free-living, thus the regression probably preceded the establishment of a parasitic lifestyle by the ancestor of Entamoeba. Hence, much of the highly reduced cellular makeup of this organism could not be attributed to its parasitic nature.
The coexistence of mitochondriate and amitochondriate, free-living and parasitic organisms in a single clade, opens up interesting possibilities to dissect the steps leading from the ancestral free-living forms to the simplified forms as they emerged on the same lineage. In comparative studies of these forms, special attention will have to be paid to those differences of the metabolic machinery that are related to the lack of classical mitochondrial ATP-producing functions (16, 65–67). Although information on the metabolism of Mastigamoeba and the pelobionts is close to nil, the EST project has detected the presence of a number of genes coding for enzymes known to be present in Entamoeba but absent from multicellular eukaryotes. Our views on Dictyostelium metabolism are also incomplete, but the available data indicate no striking differences from multicellular eukaryotes. The tentative implication of these considerations is that the transition from the mitochondriate to amitochondriate condition was accompanied by significant changes of the enzymatic composition in the metabolic machinery.
Sequencing ESTs of Protists, a Powerful Method to Resolve Eukaryotic Phylogeny.
The sequencing of about 1,280 ESTs of Mastigamoeba allowed us to locate confidently this species within the eukaryotic tree, something that has not been possible previously, based on the analysis of morphology (17) and a few genes (4, 19–28). Because this sequencing approach is not very expensive, it could be readily applied to many eukaryotic phyla for which pure culture is possible and a genome project is inconceivable (e.g., Euglena gracilis, genome size ≈3 × 109 or Amoeba proteus, ≈3 × 1011). This approach, if applied to many protists, will greatly advance the resolution of difficult questions on eukaryotic phylogeny through the possible discovery of a few lucky genes and even more certainly through a combined analysis, above all if tree reconstruction methods that correctly handle covarion structure are used.
Supplementary Material
Acknowledgments
We thank Dr. F. Schuster (Brooklyn College, Brooklyn, NY) for providing the organism used and giving patient advice on its cultivation and handling, R. K. Singh and his coworkers (National Research Council, Halifax Institute of Marine Biosciences) for the sequencing of the cDNA clones, and Drs. C. Brochier, S. Gribaldo, D. Horner, and D. Moreira for critical reading of the manuscript. This research was supported by National Science Foundation Grant MCB 9615659 and National Institutes of Health Grant AI11942 to M.M. We thank the C. neoformans Genome Project, the Stanford Genome Technology Center (http://www-sequence.stanford.edu), funded by the National Institute of Allergy and Infectious Diseases/National Institutes of Health under Cooperative Agreement AI47087, the Neurospora Sequencing Project, and the Whitehead Institute/Massachusetts Institute of Technology Center for Genome Research (www-genome.wi.mit.edu).
Abbreviations
- rDNA
rRNA-encoding DNA
- BV
bootstrap value
- EST
expressed sequence tag
- LBA
long branch attraction
- ML
maximum likelihood
- NJ
neighbor joining
- MP
maximum parsimony
Footnotes
References
- 1.Lee J J, Hutner S H, Bovee E G. An Illustrated Guide to the Protozoa. Lawrence, KS: Society of Protozoologists; 1985. [Google Scholar]
- 2.Page F C. Arch Protistenkd. 1987;133:199–217. [Google Scholar]
- 3.Corliss J O. Acta Protozool. 1994;33:1–51. [Google Scholar]
- 4.Hinkle G, Leipe D D, Nerad T A, Sogin M L. Nucleic Acids Res. 1994;22:465–469. doi: 10.1093/nar/22.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pawlowski J, Bolivar I, Fahrni J F, Cavalier-Smith T, Gouy M. Mol Biol Evol. 1996;13:445–450. doi: 10.1093/oxfordjournals.molbev.a025605. [DOI] [PubMed] [Google Scholar]
- 6.Stiller J, Hall B. Mol Biol Evol. 1999;16:1270–1279. doi: 10.1093/oxfordjournals.molbev.a026217. [DOI] [PubMed] [Google Scholar]
- 7.Cavalier-Smith T, Chao E E. Arch Protistenkd. 1996;147:227–236. [Google Scholar]
- 8.Milyutina I A, Aleshin V V, Mikrjukov K A, Kedrova O S, Petrov N B. Gene. 2001;272:131–139. doi: 10.1016/s0378-1119(01)00556-x. [DOI] [PubMed] [Google Scholar]
- 9.Silberman J D, Clark C G, Diamond L S, Sogin M L. Mol Biol Evol. 1999;16:1740–1751. doi: 10.1093/oxfordjournals.molbev.a026086. [DOI] [PubMed] [Google Scholar]
- 10.Pawlowski J, Bolivar I, Fahrni J F, De Vargas C, Bowser S S. J Eukaryotic Microbiol. 1999;46:612–617. doi: 10.1111/j.1550-7408.1999.tb05137.x. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein J. Syst Zool. 1978;27:401–410. [Google Scholar]
- 12.Kumar S, Rzhetsky A. J Mol Evol. 1996;42:183–193. doi: 10.1007/BF02198844. [DOI] [PubMed] [Google Scholar]
- 13.Philippe H, Germot A. Mol Biol Evol. 2000;17:830–834. doi: 10.1093/oxfordjournals.molbev.a026362. [DOI] [PubMed] [Google Scholar]
- 14.Bakker-Grunwald T, Wostmann C. Parasitol Today. 1993;9:27–31. doi: 10.1016/0169-4758(93)90161-8. [DOI] [PubMed] [Google Scholar]
- 15.Chavez L A, Balamuth W, Gong T. J Protozool. 1986;33:397–404. doi: 10.1111/j.1550-7408.1986.tb05630.x. [DOI] [PubMed] [Google Scholar]
- 16.Reeves R E. Adv Parasitol. 1984;23:105–142. doi: 10.1016/s0065-308x(08)60286-9. [DOI] [PubMed] [Google Scholar]
- 17.Walker G, Simpson A G B, Edgcomb V, Sogin M L, Patterson D J. Eur J Protistol. 2001;37:25–49. [Google Scholar]
- 18.Cavalier-Smith T. Biol Rev Cambridge Philos Soc. 1998;73:203–266. doi: 10.1017/s0006323198005167. [DOI] [PubMed] [Google Scholar]
- 19.Stiller J W, Riley J, Hall B D. J Mol Evol. 2001;52:527–539. doi: 10.1007/s002390010183. [DOI] [PubMed] [Google Scholar]
- 20.Hannaert V, Brinkmann H, Nowitzki U, Lee J A, Albert M-A, Sensen C W, Gaasterland T, Müller M, Michels P, Martin W. Mol Biol Evol. 2000;17:989–1000. doi: 10.1093/oxfordjournals.molbev.a026395. [DOI] [PubMed] [Google Scholar]
- 21.Arisue, N., Hashimoto, T., Lee, J. A., Moore, D. V., Gordon, P., Sensen, C. W., Gaasterland, T., Hasegawa, M. & Müller, M. (2002) J. Eukaryotic Microbiol., in press. [DOI] [PubMed]
- 22.Fast N M, Logsdon J M, Jr, Doolittle W F. Mol Biol Evol. 1999;16:1415–1419. doi: 10.1093/oxfordjournals.molbev.a026052. [DOI] [PubMed] [Google Scholar]
- 23.Moreira D, Le Guyader H, Philippe H. Mol Biol Evol. 1999;16:234–245. doi: 10.1093/oxfordjournals.molbev.a026105. [DOI] [PubMed] [Google Scholar]
- 24.Moreira D, Le Guyader H, Philippe H. Nature (London) 2000;405:69–72. doi: 10.1038/35011054. [DOI] [PubMed] [Google Scholar]
- 25.Germot A, Philippe H. J Eukaryotic Microbiol. 1999;46:116–124. doi: 10.1111/j.1550-7408.1999.tb04594.x. [DOI] [PubMed] [Google Scholar]
- 26.Roger A J, Smith M W, Doolittle R F, Doolittle W F. J Eukaryotic Microbiol. 1996;43:475–485. doi: 10.1111/j.1550-7408.1996.tb04507.x. [DOI] [PubMed] [Google Scholar]
- 27.Roger A J, Svärd S G, Tovar J, Clark C G, Smith M W, Gillin F D, Sogin M L. Proc Natl Acad Sci USA. 1998;95:229–234. doi: 10.1073/pnas.95.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edlind T D, Li J, Visvesvara G S, Vodkin M H, McLaughlin G L, Katiyar S K. Mol Phylogenet Evol. 1996;5:359–367. doi: 10.1006/mpev.1996.0031. [DOI] [PubMed] [Google Scholar]
- 29.Horner D S, Embley T M. Mol Biol Evol. 2001;18:1970–1975. doi: 10.1093/oxfordjournals.molbev.a003737. [DOI] [PubMed] [Google Scholar]
- 30.Keeling P J, Doolittle W F. Mol Biol Evol. 1996;13:1297–1305. doi: 10.1093/oxfordjournals.molbev.a025576. [DOI] [PubMed] [Google Scholar]
- 31.Baldauf S L, Doolittle W F. Proc Natl Acad Sci USA. 1997;94:12007–12012. doi: 10.1073/pnas.94.22.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Philippe H, Adoutte A. In: Evolutionary Relationships Among Protozoa. Coombs G, Vickerman K, Sleigh M, Warren A, editors. Dordrecht, the Netherlands: Kluwer; 1998. pp. 25–56. [Google Scholar]
- 33.Lang B F, Gray M W, Burger G. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- 34.Baldauf S L, Roger A J, Wenk-Siefert I, Doolittle W F. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 35.Philippe H, Lopez P, Brinkmann H, Budin K, Germot A, Laurent J, Moreira D, Müller M, Le Guyader H. Philos Trans R Soc London B. 2000;267:1213–1221. doi: 10.1098/rspb.2000.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philippe H, Germot A, Moreira D. Curr Opin Genet Dev. 2000;10:596–601. doi: 10.1016/s0959-437x(00)00137-4. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z. Trends Ecol Evol. 1996;11:367–370. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- 38.Van de Peer Y, Rensing S A, Maier U G, De Wachter R. Proc Natl Acad Sci USA. 1996;93:7732–7736. doi: 10.1073/pnas.93.15.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burger G, Saint-Louis D, Gray M W, Lang B F. Plant Cell. 1999;11:1675–1694. doi: 10.1105/tpc.11.9.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon P, Gaasterland T, Sensen C W. In: Genomics. Sensen C W, editor. New York: Wiley; 2001. pp. 379–397. [Google Scholar]
- 41.Gaasterland T, Sensen C W. Biochimie. 1996;78:302–310. doi: 10.1016/0300-9084(96)84761-4. [DOI] [PubMed] [Google Scholar]
- 42.Nikaido I, Asamizu E, Nakajima M, Nakamura Y, Saga N, Tabata S. DNA Res. 2000;7:223–227. doi: 10.1093/dnares/7.3.223. [DOI] [PubMed] [Google Scholar]
- 43.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Philippe H. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 46.Swofford D L. paup* Sunderland, MA: Sinauer; 2000. [Google Scholar]
- 47.Adachi J, Hasegawa M. Comput Sci Monogr. 1996;28:1–150. [Google Scholar]
- 48.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 49.Kishino H, Miyata T, Hasegawa M. J Mol Evol. 1990;31:151–160. doi: 10.1007/BF02109497. [DOI] [PubMed] [Google Scholar]
- 50.Aguinaldo A M, Turbeville J M, Linford L S, Rivera M C, Garey J R, Raff R A, Lake J A. Nature (London) 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 51.Yang Z. J Mol Evol. 1996;42:587–596. doi: 10.1007/BF02352289. [DOI] [PubMed] [Google Scholar]
- 52.Lecointre G, Philippe H, Le H L V, Le Guyader H. Mol Phylogenet Evol. 1994;3:292–309. doi: 10.1006/mpev.1994.1037. [DOI] [PubMed] [Google Scholar]
- 53.Douglas S, Zauner S, Fraunholz M, Beaton M, Penny S, Deng L T, Wu X, Reith M, Cavalier-Smith T, Maier U G. Nature (London) 2001;410:1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- 54.Cavalier-Smith T. J Eukaryotic Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- 55.Hirt R P, Logsdon J M, Jr, Healy B, Dorey M W, Doolittle W F, Embley T M. Proc Natl Acad Sci USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stiller J W, Duffield E C, Hall B D. Proc Natl Acad Sci USA. 1998;95:11769–11774. doi: 10.1073/pnas.95.20.11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fast N M, Kissinger J C, Roos D S, Keeling P J. Mol Biol Evol. 2001;18:418–426. doi: 10.1093/oxfordjournals.molbev.a003818. [DOI] [PubMed] [Google Scholar]
- 58.Lockhart P J, Larkum A W, Steel M, Waddell P J, Penny D. Proc Natl Acad Sci USA. 1996;93:1930–1934. doi: 10.1073/pnas.93.5.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cavalier-Smith T. Biosystems. 1991;25:25–38. doi: 10.1016/0303-2647(91)90010-i. [DOI] [PubMed] [Google Scholar]
- 60.Griffin J L. J Protozool. 1988;35:300–315. doi: 10.1111/j.1550-7408.1988.tb04348.x. [DOI] [PubMed] [Google Scholar]
- 61.Whatley J M, John P, Whatley F R. Proc R Soc London Ser B. 1979;204:165–187. doi: 10.1098/rspb.1979.0020. [DOI] [PubMed] [Google Scholar]
- 62.Roger A J. Am Nat. 1999;154:S146–S163. doi: 10.1086/303290. [DOI] [PubMed] [Google Scholar]
- 63.Tovar J, Fischer A, Clark C G. Mol Microbiol. 1999;32:1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- 64.Mai Z, Ghosh S, Frisardi M, Rosenthal B, Rogers R, Samuelson J. Mol Cell Biol. 1999;19:2198–2205. doi: 10.1128/mcb.19.3.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Field J, Rosenthal B, Samuelson J. Mol Microbiol. 2000;38:446–465. doi: 10.1046/j.1365-2958.2000.02143.x. [DOI] [PubMed] [Google Scholar]
- 66.Horner D S, Foster P G, Embley T M. Mol Biol Evol. 2000;17:1695–1709. doi: 10.1093/oxfordjournals.molbev.a026268. [DOI] [PubMed] [Google Scholar]
- 67.Müller M. In: Evolutionary Relationships Among Protozoa. Coombs G, Vickerman K, Sleigh M, Warren A, editors. Dordrecht, the Netherlands: Kluwer; 1998. pp. 109–131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.