Abstract
Background
Vaginal microbiota (VM) links to the risk of persistent HPV infection and the progression of cervical intraepithelial neoplasia (CIN). However, a comprehensive understanding of concurrent alterations in VM and metabolome associated with that risk remains elusive in population-based studies, particularly among Chinese women.
Methods
This study conducted an extensive analysis of VM and metabolome profiles in a cohort of 56 Chinese women, classified into HPV_C (natural clearance of HPV, n = 18) and HPV_PH(persistent HPV infection accompanied by high-grade CIN, n = 38), based on the result of 3–6 months follow-up visits.
Results
Our analysis revealed a higher prevalence of Lactobacillus-dominated samples in the HPV_C cohort. Notably, the vaginal metabolome exhibited a significant interaction with VM, with Lactobacillus emerging as a pivotal influencer. We identified 386 metabolites that significantly differentiated between HPV_C and HPV_PH groups, of which 364 were associated with VM components such as Lactobacillus, Hoylesella, Fannyhessea and Megasphaera. Further examination showed that 66 of these 364 metabolites positively correlated with Lactobacillus, including citric acid, DL-beta-Leucine, Xanoic acid and Norcholic acid. Conversely, 19 metabolites, including HPV_PH enriched maltotriose and N-Acetyl-L-aspartic acid, negatively correlated with Lactobacillus. Further analysis suggested potential bi-directional modulation between VM and persistent HPV infection accompanied by high-grade, being partially mediated by vaginal metabolites.
Conclusions
This study provides additional insights into the correlations between concurrent alterations in VM and metabolome associated with persistent HPV infection accompanied by high-grade CIN.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-025-04126-w.
Keywords: Human papillomavirus, Cervical intraepithelial neoplasia, Vaginal microbiota, Lactobacillus, Metabolome
Introduction
Human papillomavirus (HPV) infection stands as a notable precursor to cervical intraepithelial neoplasia (CIN) and cervical cancer [1], with persistent HPV infection driving carcinogenesis. While HPV vaccination and screening have reduced cervical cancer incidence, understanding modifiers of HPV persistence and CIN progression remains critical, especially in developing countries. Emerging studies implicate the vaginal microbiota (VM) as a key determinant of HPV infection outcomes, with the dysbiosis potentially exacerbating oncogenic risks through metabolic, immunological, and direct microbial interactions [2–11].
Healthy reproductive-age women typically exhibit the VM pattern dominated by Lactobacillus species [12, 13]. Furthermore, cross-sectional studies consistently reported lower Lactobacillus abundance in HPV-positive women diagnosed with high-grade CIN, in contrast to those with less severe CIN grades or no lesions [2–6]. These findings were complemented by prospective studies, which have hinted at potential causal links between vaginal Lactobacillus and both the likelihood of persistent HPV infection and the progression of CIN [7–11]. Notably, women whose VM was dominated by Lactobacillus crispatus exhibited a favorable correlation with spontaneous CIN regression. Conversely, those with VM structures lacking Lactobacillus dominance showed an increased propensity for CIN progression [7, 8].
The VM’s functional impact extends to the vaginal metabolome, a critical mediator of host-microbe-virus interactions [2, 12, 13]. Recent studies have delved into the characterization of the vaginal metabolome among women exhibiting varying statuses of HPV infection and CIN grades [2]. Notably, Ilhan ZE et al. reported a distinctive pattern of metabolite enrichment in lipids and depletion in amino acids and nucleotide metabolites, thereby linking vaginal dysbiosis to HPV infection and precancerous cervical dysplasia [2]. Specifically, the presence of anti-inflammatory nucleotides was observed to be associated with Lactobacillus dominance, regardless of CIN severity. Cervical cancer patients demonstrated an elevated abundance of amino acid metabolites compared to other HPV-positive groups, both with and without CIN. Separately, other researchers found that HPV-positive women exhibited higher concentrations of biogenic amines and phospholipids compared to HPV-negative women [12]. These studies further underscore the complex interplay between VM, vaginal metabolome, and the risk of persistent HPV infection as well as high-grade CIN.
Despite advances in understanding VM-metabolome-HPV interplay, population-specific data, particularly among Chinese women, remain scarce. To partially address this gap, we recruited a cohort of HPV-positive Chinese women and divided them into two distinct groups: those who experienced natural clearance of HPV infection (HPV_C) and those who had persistent HPV infection accompanied by high-grade CIN (HPV_PH). By integrating 16S rRNA gene sequencing with untargeted metabolomics, we aimed to: (1) delineate VM-metabolome co-alterations distinguishing HPV_C and HPV_PH groups, and (2) identify microbial taxa and metabolites potentially driving persistent HPV infection and CIN progression. Our findings will contribute invaluable population-based insights into the role of VM and associated metabolome in the risk of persistent HPV infection and the occurrence of high-grade CIN.
Materials and methods
Participant recruitment and sample collection
This study was embedded with the “Establishment and Promotion of Innovative Cervical Cancer Control Models"project conducted in Guangxi Zhuang Autonomous Region, China. Fifty-six HPV-positive women were enrolled following stringent eligibility criteria designed to minimize confounding factors: (1) age ≥ 18 years; (2) premenopausal status; (3) no sexual activity within the preceding week; (4) no vaginal douching or medication within the past week; (5) no exposure to antibiotics within the last month; (6) no hormone replacement therapy or GnRH-a administration in the past three months; (7) no uterine cavity manipulation within the prior three months; (8) absence of reproductive tract inflammation or systemic diseases (e.g., autoimmune disorders, diabetes). Following institutional review board approval, all participants provided informed consents, being classified into two groups: natural clearance of HPV infection (HPV_C, n = 18) and persistent HPV infection as well as high-grade CIN (HPV_PH, n = 38).
Clinical physicians, with the assistance of dedicated research assistants, meticulously documented pertinent primary data for each participant, encompassing age, sexual history, pregnancy and abortion history, HPV genotype, pathological diagnosis, and additional relevant information. Vaginal swabs were collected at least three days post-menstruation to ensure sample uniformity and integrity. During colposcopy, skilled clinical physicians utilized vaginal swabs to obtain samples from the posterior fornix, with two swabs collected from each participant. Following collection, all swabs were stored in sterile 2 mL tubes, initially kept at −20 °C, and promptly transferred to −80 °C storage within 30 min to optimize sample preservation.
HPV testing and pathological diagnosis
HPV detection and genotyping was performed using the Roche Cobas® 4800 HPV Test System, which specifically detects HPV16/18 and 12 other high-risk HPV genotypes, namely HPV 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68.
The diagnosis of high-grade CIN was conducted by two experienced pathologists, with p16 immunohistochemical staining serving as an auxiliary diagnostic tool, in accordance with established guidelines [14, 15]. Specifically, cases with CIN1 morphology but diffuse p16 staining (> 75% epithelial layers) were upstaged to CIN2 + samples.
Microbial DNA extraction, 16S rRNA gene amplicon sequencing, and data processing
Microbial DNA was extracted from vaginal swab samples using the Qiagen DNeasy PowerSoil Pro Kit (Germany). The concentration and purity of the extracted DNA were assessed using 1% agarose gel electrophoresis on the Agilent 5400 platform (Agilent Technologies, Inc., Santa Clara, USA). For sequencing library preparation, the high-variability V4-V5 region of the 16S rRNA gene was amplified using the primers 515-FR (GTGCCAGCMGCCGCGGTAA) and 926-RR (CCGTCAATTCMTTTRAGTTT). These libraries were then sequenced on the MGISEQ-2000 platform (BGI, China) for high-throughput sequencing, generating 300 bp reads. The raw sequencing data underwent rigorous quality control and analysis using the QIIME2 pipeline, resulting in comprehensive profiles of VM.
Untargeted metabolomics analysis
To comprehensively profile the vaginal metabolome, high-resolution liquid chromatography with mass spectrometry (LC–MS) was performed on the Thermo Fisher Scientific platform (Ottawa, Canada). Raw intensity data were converted to mzXML format and ion features were extracted using Progenesis QI software (version 2.2). These features were subjected to stringent filtering criteria, excluding those missing in more than half of the quality control samples or in more than 80% of the test samples, as well as those with a relative standard deviation greater than 30%. Metabolite identification was conducted through a comprehensive search of the Human Metabolome Database (HMDB, version 5.0) and the Kyoto Encyclopedia of Genes and Genomes (KEGG, version 96.0). The resultant metabolite abundance matrix served as the basis for subsequent analyses, providing a comprehensive overview of metabolic profiles.
Bioinformatics analysis
To identify VM community state types (CSTs) at both genus and species level, unsupervised hierarchical clustering was applied based on the average Euclidean distance metric. Each CST was determined according to the dominant bacterial genera or species that constituted at least 50% of the microbial community. Permutational multivariate analysis of variance (PERMANOVA) was conducted to assess the contribution of VM CSTs to metabolic profiles using the vegan package in R software.
All continuous variables were standardized to conform to a standard normal distribution (N ~ (0, 1)) prior to the robust linear regression analysis, with grouping (HPV_C and HPV_PH) as a binomial variable and HPV16 positivity adjusted as the confounding factor. Orthogonal partial least squares discriminant analysis (OPLS-DA) was then applied to calculate variable importance in projection (VIP) for each metabolite. A total of 386 metabolites were identified as differentially abundant between those two cohorts based on the following criteria: p < 0.05 for robust linear regression analysis, VIP > 1, and fold change ≥ 1.25.
The synergistic relationships between the VM and metabolome were evaluated via O2PLS analysis, implemented in the OmicsPLS package of the R software. The spearman rho coefficient between VM components and metabolites was calculated using the R package psych, with FDR < 0.05 as statistical significant. Mediation effects were assessed via R package mediation, and then were determined based on an FDR < 0.05 for the proportion of mediation, with the lowest value of the 95% confidence interval (CI) exceeding zero. All statistical analyses and data visualization were performed using R software (version 4.0.5), with Benjamini-Hochberg (BH) correction being applied for FDR.
Results
Study population
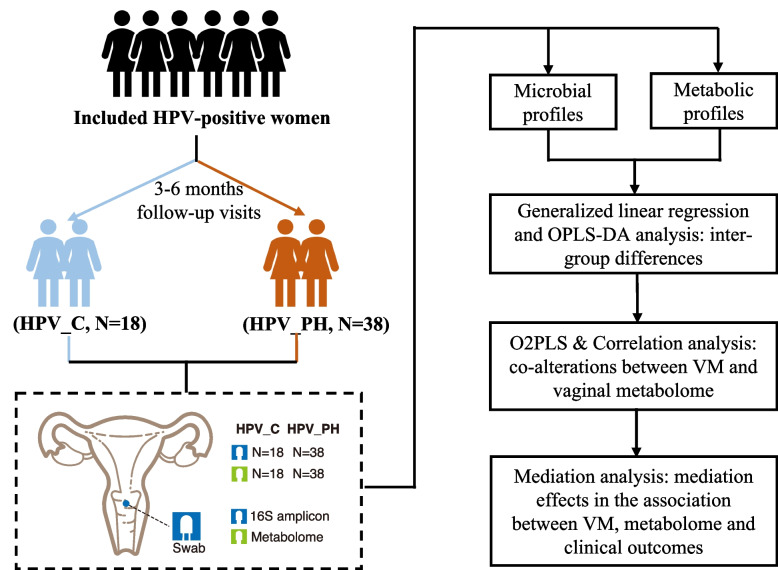
The final cohort comprised 56 HPV-positive female participants, who were stratified into two groups based on prospective investigations after 3–6 months follow-up visits: natural clearance of HPV infection (HPV_C, n = 18) and persistent HPV infection accompanied by high-grade CIN (HPV_PH, n = 38) (Fig. 1, Table 1). Each participant contributed two vaginal swabs at follow-up visits, which were utilized for VM and vaginal metabolome profiling.
Fig. 1.
Study design. All the vaginal swabs were collected at the time when natural HPV clearance or persistent HPV infection accompanied by high-grade CIN was determined. HPV_C: natural clearance of HPV infection; HPV_PH: persistent HPV infection accompanied by high-grade cervical intraepithelial neoplasia (CIN); VM: vaginal microbiome; OPLS-DA: Orthogonal Partial Least Squares-Discriminant Analysis; O2PLS: Two-Way Orthogonal Partial Least Squares
Table 1.
Information of included 56 women
| Clinical outcome | HPV_C (N = 18) | HPV_PH (N = 38) | P value |
|---|---|---|---|
|
Age (mean: min–max) |
40.8(30–57) | 41.1(30–53) | 0.73 |
| HPV infection(baseline) | |||
| HPV16 + | 0.0%(0/18) | 26.3%(10/38) | 0.021 |
| HPV18 + | 0.0%(0/18) | 5.3%(2/38) | 1 |
| Other 12 HPV genotypes + | 100.0%(18/18) | 100.0%(38/38) | 1 |
| HPV infection(follow-up visit) | |||
| HPV16 + | NA | 26.3%(10/38) | |
| HPV18 + | NA | 5.3%(2/38) | |
| Other 12 HPV genotypes + | NA | 78.9%(30/38) | |
| No. of sex partner (mean: min–max) | 1(0–2) | 0.8(0–2) | 0.18 |
|
Age with first sex activity (mean: min–max) |
22.3(18–26) | 21.2(15–32) | 0.17 |
| Gestation | 94.4%(17/18) | 92.1%(35/38) | 1 |
| Abortion | 38.9%(7/18) | 42.1%(16/38) | 1 |
| CST(genus level): LD | 77.8%(14/18) | 68.4%(26/38) | 0.54 |
| CST(species level) | |||
| Lactobacillus crispatus-dominant (CST_I) | 11.1%(2/18) | 18.4%(7/38) | 0.70 |
| Lactobacillus iners-dominant (CST_III) | 66.7%(12/18) | 50.0%(19/38) | 0.27 |
CST Community state type, LD Lactobacillus-dominant. For Age, No. of sex partner and Age with first sex activity, statistic significance was assessed by Wilcoxon rank sum test with the p value was adjusted via BH correction. For other factors, statistic significance was assessed via Fisher’s exact test
Regarding the demographic characteristics, no statistically significant differences were observed between the HPV_C and HPV_PH groups (Table 1). The proportion of HPV16-positive individuals was higher in the HPV_PH group compared to the HPV_C group (Table 1). An examination of other phenotypic factors, such as smoking status, did not yield statistically significant differences between HPV_C and HPV_PH cohorts (Table 1).
Differences of VM structures between HPV_C and HPV_PH populations
Analysis indicated disparities in VM compositions between HPV_C and HPV_PH cohorts (Fig. S1A). Then we observed an increase in the proportion of Lactobacillus-dominant (LD) VM community state types (CSTs) within the HPV_C group (77.8%) compared to the HPV_PH group (68.4%), though there was no statistic significance (Fig. 2A-B, Table 1). Further delving into the data, we found a decrease in the prevalence of Lactobacillus crispatus-dominant (CST_I) pattern (11.1% vs 18.4% in the HPV_PH group) and a corresponding increase in Lactobacillus iners-dominant (CST_III) pattern (66.7% vs 50.0% in the HPV_PH group) within HPV_C cohorts (Fig. 2C, Table 1, Fig. S1B).
Fig. 2.
Distinct VM structures between HPV_C and HPV_PH group. A Distribution of genus-level community state types (CSTs). This panel depicts the distribution of CSTs in both HPV_C and HPV_PH cohorts. The proportion of Lactobacillus-dominant (LD) CSTs (77.8% and 68.4% in HPV_C and HPV_PH group, respectively) are distinct between the two groups, though there is no statistic significance with Fisher’s exact test p value = 0.54. B VM compositions at genus level. C VM composition at species level
Specifically, microbial samples classified as non-Lactobacillus-dominant (NLD) CSTs exhibited a diverse composition, primarily comprising Fannyhessea and Megasphaera species, including Fannyhessea vaginae and Megasphaera lornae (Fig. 2B-C, Table S1−2).
Distinct vaginal metabolic profiles between HPV_C and HPV_PH populations
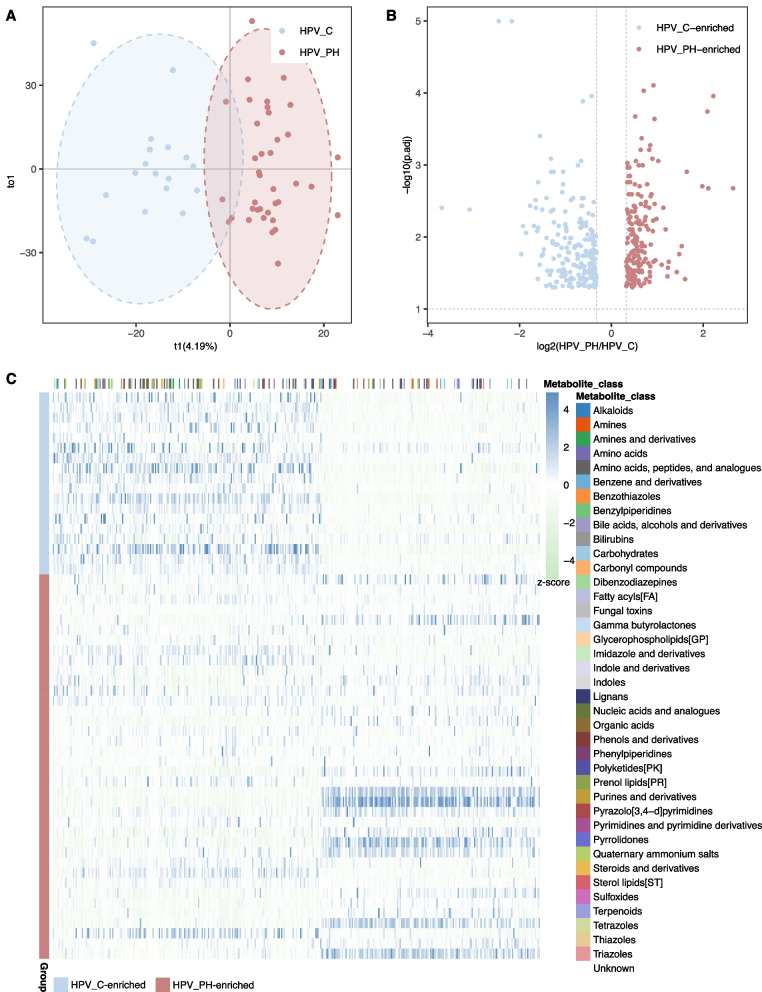
Utilizing untargeted metabolomics, we identified a total of 2,279 metabolites. OPLS-DA analysis indicated notable separation of microbial samples between HPV_C and HPV_PH group, based on metabolic profiles (Fig. 3A). Among these metabolites, the robust linear regression analysis pinpointed 386 metabolites that were differentially produced between the two cohorts following standards: p < 0.05, VIP(Variance Inflation Factor) > 1 and fold change ≥ 1.25 (Fig. 3B, Table S3).
Fig. 3.
Vaginal metabolic profiles differed between HPV_C and HPV_PH cohort. A OPLS-DA indicated separation of microbial samples between HPV_C and HPV_PH group, based on metabolic profiles. B Robust linear regression analysis indicated 386 metabolites with significant variations between two cohorts (p < 0.05, VIP(Variance Inflation Factor) > 1 and fold change ≥ 1.25). FDR was conducted via BH correction. C The heatmap depicts the normalized intensity (z-score) of 386 differentially produced metabolites, alongside functional classifications associated with these metabolites
Of these 386 metabolites, 213 demonstrated an enrichment while 173 exhibited a depletion among HPV_C populations (Fig. 3C, Table S3). Among those 213 HPV_C-enriched metabolites, 75 were annotated with known classifications, mainly constituting pathways related to amino acids, peptides and analogues, amino acid as well as indoles and their derivatives (Fig. 3C, Table S3). As for 173 HPV_C-depleted metabolites, 31 had known functional classifications such as amino acids, peptides and analogues as well as fatty acyls (Fig. 3C, Table S3).
Co-alterations of vaginal microbial and metabolic profiles across HPV_C and HPV_PH cohorts
Permutational Multivariate Analysis of Variance (PERMANOVA) showed notable contribution of genus-level CSTs (LD and NLD CSTs) to variations of metabolic profiles (FDR < 0.0001) (Table S4). There were also significant variations of metabolic profiles between NLD and CST_I as well as CST_III (FDR < 0.001) (Table S4). However, differences of metabolic profiles between CST_I and CST_III were not statistically notable (FDR > 0.05) (Table S4). This suggested a low degree of metabolic profile heterogeneity within the LD CST cohort.
The two-way orthogonal partial least squares (O2PLS) analysis, bolstered by loading value assessments, further uncovered the most significant role of Lactobacillus in determining metabolic profiles, being followed by Dialister, Hoylesella and Prevotella (Table S5).
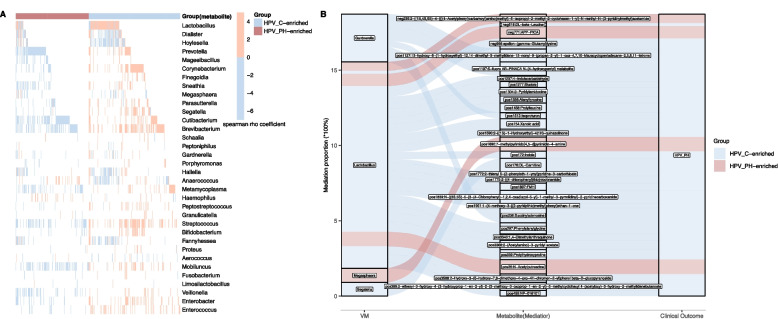
To investigate co-alterations between VM and vaginal metabolome, we applied spearman rho coefficient to quantify the correlations between VM genera and 386 differentially abundant metabolites. Under the stringent threshold of FDR < 0.05, we identified the correlations between VM genera with 364 of 386 differentially produced metabolites, among which 85 correlated notably with Lactobacillus (Fig. 4A, Table S6). For those 85 metabolites, 17 were enriched in HPV_PH samples, among which N-Acetylputrescine, N-Acetyl-L-aspartic acid and maltotriose correlated negatively with Lactobacillus (Fig. 4A, Table S6). Sixty-four of 68 HPV_C-enriched metabolites had positive correlations with Lactobacillus level, including 4-Methylcatechol, succinyladenosine, DL-Carnitine, indole, gamma-Glutamyltryptophan, citric acid, DL-beta-Leucine, tryptophan, asparagine and gamma-Glutamyltyrosine (Fig. 4A, Table S6). The four HPV_C-enriched metabolites, which were negatively correlated with Lactobacillus level, were xylonic acid, isoproturon, fumaric acid and beta-D-Fructose 6-phosphate (Fig. 4A, Table S6). Compared to Lactobacillus, most of above-mentioned metabolites had oppositive correlations with Fannyhessea, Hoylesella, Prevotella, Megasphaera and Gardnerella (Table S6).
Fig. 4.
The correlation between VM, metabolome and persistent HPV infection accompanied by high-grade CIN. A Being assessed by spearman rho coefficient, this heatmap illustrates the correlation between VM components and metabolites that significantly differentiate HPV_C and HPV_PH group. Red and blue lines signify positive and negative correlations (FDR < 0.05), respectively. B This analysis explores the mediation effects of differentially produced metabolites (mediator) in potential modulation of VM on the risk of persistent HPV infection accompanied by high-grade CIN. Mediation effects were determined based on an FDR < 0.05 for the proportion of mediation, with the lowest value of the 95% confidence interval (CI) exceeding zero. FDR was conducted via BH correction
Inferred mediation effects of vaginal metabolites in correlations between VM and persistent HPV infection accompanied by high-grade CIN
Applying mediation analysis, we found that persistent HPV infection accompanied by high-grade CIN seemed to impact the growth of Lactobacillus via the mediation of Fannyhessea and Megasphaera (Fig. S2A). Then 9 of 386 differentially produced metabolites mediated the inhibition of Fannyhessea on the growth of Lactobacillus, among which 8 were enriched in HPV_C group and negatively correlated with Fannyhessea such as prolylleucine as well as alpha-fluoromethylhistidine(FMH), and 1 HPV_PH-enriched N-Acetyl-L-aspartic acid (Fig. S2B).
Further analysis also suggested the potential modulation of Lactobacillus on persistent HPV infection mediated by 25 differentially produced metabolites that notably correlated with Lactobacillus (Fig. 4B). Those metabolites included prolylleucine, 4-indolecarbaldehyde, sktole, alanyltyrosine, alpha-fluoromethylhistidine(FMH), phenylalanylglycine, prolylhydroxyproline and DL-beta-Leucine (Fig. 4B).
Among those differentially produced metabolites that correlated with non-Lactobacillus genera, we found the mediation effects of six metabolites on the impact of Megasphaera, Gardnerella or Segatella on persistent HPV infection accompanied by high-grade CIN (Fig. 4B). Those metabolites encompassed isoproturon, succinyladenosine and 7-methylpyrimido[4,5-d]pyrimidin-4-amine (Fig. 4B).
Discussion
Our findings corroborate prior evidence demonstrating reduced prevalence of LD VM profiles in women with persistent HPV infection and high-grade CIN, compared to those achieving natural viral clearance [2–6]. Nevertheless, the functional viewpoint remain to be fully explored in those differences. Several studies indicated variations of VM functional communities and metabolic profiles among women with distinct states of HPV infection and CIN grade [2, 12, 13, 16–20]. Expanding on this, our analysis revealed 386 metabolites with differed levels between HPV_C and HPV_PH cohorts. This suggested distinct VM-host interactions that differed with the risk of persistent HPV infection and CIN progression, partially aligning with previous reports [2, 13, 16, 18, 21, 22].
Among 213 HPV_C-enriched metabolites, succinyladenosine had the most significant fold change and correlated positively with vaginal Lactobacillus. Though its role remains to be explored, previous reports indicated the co-alteration of succinyladenosine with lactic acid in response to environmental changes [23], with latter modulated epithelial integrity and immunity [24, 25]. However, several studies indicated higher levels of succinyladenosin in tumors [26, 27]. Thus, further researches should be conducted to unravel the specific role of succinyladenosine in the risk of persistent HPV infection and CIN progression. We also found the positive association of vaginal Lactobacillus with 4-indolecarbaldehyde, which had broad-spectrum antibacterial effects and related to nutrient absorption as well as metabolisms [28, 29]. Moreover, Lactobacillus-correlated 3'-Adenosine monophosphate (3'-AMP) inhibited viral infection [30]. Despite more evidence is needed, those findings provide hints for the association of vaginal Lactobacillus with metabolic profiles in persistent HPV infection with high-grade CIN.
Among 173 HPV_PH-enriched metabolites, we found the notably increased level of maltose and maltotriose, which were the common products of vaginal glycogen degraded by non-Lactobacillus components [31–33]. Studies demonstrated that maltotriose inhibited vaginal colonization by L. crispatus due to dissipated alpha-glucan degrading activity, in agreement with carbon catabolite repression [34]. Those findings partially explained the negative correlation between maltotriose and vaginal Lactobacillus.
Consistent with previous reports [7, 8, 35], we found the potential bi-directional modulations between VM and HPV infection in this study. Lebeau et al. demonstrated the inhibition of HPV infection on the growth of vaginal Lactobacillus, while our prior findings indicated elevated levels of vaginal Lactobacillus after surgically removing high-grade cervical lesions thus eliminating HPV infection [35, 36]. In consistence, we found the potential modulation of persistent HPV infection accompanied high-grade CIN on vaginal Lactobacillus, being mediated by Fannyhessea and associated metabolites. This is partially aligned with the positive association of Fannyhessea with HPV infection and negative association of Fannyhessea with Lactobacillus [2–6]. We also found the mediation of several metabolites in the association between Lactobacillus and persistent HPV infection accompanied by high-grade CIN. Though the role of those metabolites need to be further explored, our findings suggested the potential modulation of vaginal Lactobacillus on the risk of persistent HPV infection.
The limitations of the current study necessitate acknowledgment. First, the modest sample size present constraints in drawing definitive conclusions, such as overfitting and reduced power in association analysis. However, these limitations were partially addressed by implementing stringent inclusion criteria that excluded participants with prevalent vaginal infections and menopause, factors known to profoundly impact the VM [37]. Second, while 16S rDNA amplicon sequencing provided valuable insights, it limited our capacity to comprehensively understand the functional communities within the VM [38, 39]. Consequently, we were unable to delve deeply into intra-group heterogeneity and inter-group variations among Lactobacillus strains and their functional landscapes, highlighting the need for future studies employing more exhaustive sequencing methodologies, such as shotgun metagenomics, to bridge these gaps. Lastly, although our analysis unveiled VM-metabolite associations and predicted mediation effects, further studies should be needed to validate inferred causal relationships in those associations.
Supplementary Information
Acknowledgements
Not applicable.
Clinical trial number
Not applicable.
Authors’ contributions
D.H., and W.R. conceived the study. W.R., D.H. and L.C. recruited and selected attenders. W.D., Y.Q. and X.R. performed the sample storage and associated experimental analysis. D.W. and Y.Q. performed data analysis and drafted the manuscript. W.R. and D.H. polished the manuscript. All authors reviewed the manuscript.
Funding
This study was supported by National Key R&D Program of China(2024YFC2707503), Shenzhen Public Platform for Preservation of Fertility and Reproduction (XMHT20220104049), Shenzhen Key Medical Discipline Construction Fund (SZXK027) and Shenzhen Science and Technology Program (JCYJ20240813120012017).
Data availability
The raw data of VM have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences that are accessible at https://ngdc.cncb.ac.cn/gsa/search?searchTerm = CRA024190. Metabolome data are accessible at https://ngdc.cncb.ac.cn/search/specific?db = omix&q = OMIX009604.
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Peking University Shenzhen Hospital (registration number: 2023–016). We obtained signed consents from all participants who were fully informed. We declare that our study adhered to the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qin Yang, Wenkui Dai and Di Wu contributed equally to this work.
Contributor Information
Ruifang Wu, Email: wurfpush@126.com.
Hui Du, Email: duhui_107108@163.com.
References
- 1.Lei J, Ploner A, Elfström KM, Wang J, Roth A, Fang F, Sundström K, Dillner J, Sparén P. HPV vaccination and the risk of invasive cervical cancer. N Engl J Med. 2020;383(14):1340–8. 10.1056/NEJMoa1917338. [DOI] [PubMed] [Google Scholar]
- 2.Ilhan ZE, Łaniewski P, Thomas N, Roe DJ, Chase DM, Herbst-Kralovetz MM. Deciphering the complex interplay between microbiota, HPV, inflammation and cancer through cervicovaginal metabolic profiling. EBioMedicine. 2019;44:675–90. 10.1016/j.ebiom.2019.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo C, Dai W, Zhou Q, Gui L, Cai H, Wu D, Hou J, Li C, Li S, Du H, Wu R. Cervicovaginal microbiota significantly changed for HPV-positive women with high-grade squamous intraepithelial lesion. Front Cell Infect Microbiol. 2022;12:973875. 10.3389/fcimb.2022.973875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Qiu X, Wang W, Li D, Wu A, Hong Z, Di W, Qiu L. Human papillomavirus infection and cervical intraepithelial neoplasia progression are associated with increased vaginal microbiome diversity in a Chinese cohort. BMC Infect Dis. 2020;20(1):629. 10.1186/s12879-020-05324-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitra A, MacIntyre DA, Lee YS, Smith A, Marchesi JR, Lehne B, Bhatia R, Lyons D, Paraskevaidis E, Li JV, Holmes E, Nicholson JK, Bennett PR, Kyrgiou M. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep. 2015;5:16865. 10.1038/srep16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng JJ, Song JH, Yu CX, Wang F, Wang PC, Meng JW. Difference in vaginal microecology, local immunity and HPV infection among childbearing-age women with different degrees of cervical lesions in Inner Mongolia. BMC Womens Health. 2019;19(1):109. 10.1186/s12905-019-0806-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitra A, MacIntyre DA, Ntritsos G, Smith A, Tsilidis KK, Marchesi JR, Bennett PR, Moscicki AB, Kyrgiou M. The vaginal microbiota associates with the regression of untreated cervical intraepithelial neoplasia 2 lesions. Nat Commun. 2020;11(1):1999. 10.1038/s41467-020-15856-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Usyk M, Zolnik CP, Castle PE, Porras C, Herrero R, Gradissimo A, Gonzalez P, Safaeian M, Schiffman M, Burk RD, Costa Rica HPV Vaccine Trial (CVT) Group. Cervicovaginal microbiome and natural history of HPV in a longitudinal study. PLoS Pathog. 2020;16(3):e1008376. 10.1371/journal.ppat.1008376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dareng EO, Ma B, Adebamowo SN, Famooto A, Ravel J, Pharoah PP, Adebamowo CA. Vaginal microbiota diversity and paucity of Lactobacillus species are associated with persistent hrHPV infection in HIV negative but not in HIV positive women. Sci Rep. 2020;10(1):19095. 10.1038/s41598-020-76003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Paola M, Sani C, Clemente AM, Iossa A, Perissi E, Castronovo G, Tanturli M, Rivero D, Cozzolino F, Cavalieri D, Carozzi F, De Filippo C, Torcia MG. Characterization of cervico-vaginal microbiota in women developing persistent high-risk Human Papillomavirus infection. Sci Rep. 2017;7(1):10200. 10.1038/s41598-017-09842-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berggrund M, Gustavsson I, Aarnio R, Lindberg JH, Sanner K, Wikström I, Enroth S, Bunikis I, Olovsson M, Gyllensten U. Temporal changes in the vaginal microbiota in self-samples and its association with persistent HPV16 infection and CIN2. Virol J. 2020;17(1):147. 10.1186/s12985-020-01420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borgogna JC, Shardell MD, Santori EK, Nelson TM, Rath JM, Glover ED, Ravel J, Gravitt PE, Yeoman CJ, Brotman RM. The vaginal metabolome and microbiota of cervical HPV-positive and HPV-negative women: a cross-sectional analysis. BJOG. 2020;127(2):182–92. 10.1111/1471-0528.15981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu H, Liu M, Song Y, Liu L, Xu F, Chen J, Zhan H, Zhang Y, Chen Y, Lu M, Chen D. Metabolomics variation profiling of vaginal discharge identifies potential targets for cervical cancer early warning. Acta Biochim Biophys Sin (Shanghai). 2022;54(10):1561–5. 10.3724/abbs.2022133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurman RJ, Carcangiu ML, Herrington CS. World Health Organisation Classification of Tumours of the Female Reproductive Organs. 4th Revised ed. Int Agency Res Cancer. 2014;6:172–82.
- 15.Carozzi F, Gillio-Tos A, Confortini M, Del Mistro A, Sani C, De Marco L, Girlando S, Rosso S, Naldoni C, Dalla Palma P, Zorzi M, Giorgi-Rossi P, Segnan N, Cuzick J, Ronco G, NTCC working group. Risk of high-grade cervical intraepithelial neoplasia during follow-up in HPV-positive women according to baseline p16-INK4A results: a prospective analysis of a nested substudy of the NTCC randomised controlled trial. Lancet Oncol. 2013;14(2):168–76. 10.1016/S1470-2045(12)70529-6. [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Shui Y, Qian Y. A Crosstalk Analysis of high-risk human papillomavirus, microbiota and vaginal metabolome in cervicovaginal microenvironment. Microb Pathog. 2024;194:106826. 10.1016/j.micpath.2024.106826. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Wu X, Li D, Huang R, Deng X, Li M, Du F, Zhao Y, Shen J, Chen Y, Zhang P, Hu C, Xiao Z, Wen Q. HPV-associated cervicovaginal microbiome and host metabolome characteristics. BMC Microbiol. 2024;24(1):94. 10.1186/s12866-024-03244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norenhag J, Edfeldt G, Stålberg K, Garcia F, Hugerth LW, Engstrand L, Fransson E, Du J, Schuppe-Koistinen I, Olovsson M. Compositional and functional differences of the vaginal microbiota of women with and without cervical dysplasia. Sci Rep. 2024;14(1):11183. 10.1038/s41598-024-61942-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brotman RM, Shardell MD, Gajer P, Tracy JK, Zenilman JM, Ravel J, et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis. 2014;210(11):1723–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belleti R, Marcolino LD, Novak J, Ferreira CST, do Nascimento Bolpetti A, da Silva Pinto GV, et al. Cervicovaginal loads of Gardnerella spp. are increased in immunocompetent women with persistent high-risk human papillomavirus infection. J Med Microbiol. 2022;71(5):001527. [DOI] [PubMed]
- 21.Novak J, Belleti R, da Silva Pinto GV, do Nascimento Bolpetti A, da Silva MG, Marconi C. Cervicovaginal Gardnerella sialidase-encoding gene in persistent human papillomavirus infection. Sci Rep. 2023;13(1):14266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Usyk M, Schlecht NF, Pickering S, Williams L, Sollecito CC, Gradissimo A, et al. molBV reveals immune landscape of bacterial vaginosis and predicts human papillomavirus infection natural history. Nat Commun. 2022;13(1):233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mu Y, Qi W, Zhang T, Zhang J, Mao S. Coordinated response of milk bacterial and metabolic profiles to subacute ruminal acidosis in lactating dairy cows. J Anim Sci Biotechnol. 2023;14(1):60. 10.1186/s40104-023-00859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delgado-Diaz DJ, Jesaveluk B, Hayward JA, Tyssen D, Alisoltani A, Potgieter M, et al. Lactic acid from vaginal microbiota enhances cervicovaginal epithelial barrier integrity by promoting tight junction protein expression. Microbiome. 2022;10(1):141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anton L, Sierra LJ, DeVine A, Barila G, Heiser L, Brown AG, et al. Common Cervicovaginal Microbial Supernatants Alter Cervical Epithelial Function: Mechanisms by Which Lactobacillus crispatus Contributes to Cervical Health. Front Microbiol. 2018;9:2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han J, Qin WX, Li ZL, Xu AJ, Xing H, Wu H, Zhang H, Wang MD, Li C, Liang L, Quan B, Yan WT, Shen F, Wu MC, Yang T. Tissue and serum metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin Chim Acta. 2019;488:68–75. 10.1016/j.cca.2018.10.039. [DOI] [PubMed] [Google Scholar]
- 27.Tsai CK, Yeh TS, Wu RC, Lai YC, Chiang MH, Lu KY, Hung CY, Ho HY, Cheng ML, Lin G. Metabolomic alterations and chromosomal instability status in gastric cancer. World J Gastroenterol. 2018;24(33):3760–9. 10.3748/wjg.v24.i33.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Y, Huang S, Zhu S, Gao B. Antibacterial Activity and Mechanism of Taxillμs chinensis (DC.) Danser and Its Active Ingredients. Int J Mol Sci. 2024;25(19):10246. 10.3390/ijms251910246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uengwetwanit T, Uawisetwathana U, Arayamethakorn S, Khudet J, Chaiyapechara S, Karoonuthaisiri N, Rungrassamee W. Multi-omics analysis to examine microbiota, host gene expression and metabolites in the intestine of black tiger shrimp (Penaeus monodon) with different growth performance. PeerJ. 2020;8:e9646. 10.7717/peerj.9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salmin AF, Pesant MJ, Burgher Y, Provost C, Labrie J, Jacques M, Gagnon CA, Beaudry F. Untargeted and targeted metabolomics reveal that adenosine nucleotides released in Actinobacillus pleuropneumoniae supernatant inhibit porcine reproductive and respiratory syndrome virus replication. Talanta. 2022;242:123315. 10.1016/j.talanta.2022.123315. [DOI] [PubMed] [Google Scholar]
- 31.Bhandari P, Tingley J, Abbott DW, Hill JE. glycogen-degrading activities of catalytic domains of α-Amylase and α-Amylase-pullulanase enzymes conserved in gardnerella spp. from the vaginal microbiome. J Bacteriol. 2023;205(2):e0039322. 10.1128/jb.00393-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhandari P, Tingley JP, Palmer DRJ, Abbott DW, Hill JE. Characterization of an α-glucosidase enzyme conserved in gardnerella spp. isolated from the human vaginal microbiome. J Bacteriol. 2021;203(17):e0021321. 10.1128/JB.00213-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhandari P, Hill JE. Transport and Utilization of Glycogen Breakdown Products by Gardnerella spp. from the Human Vaginal Microbiome. Microbiol Spectr. 2023;11(2):e0443522. 10.1128/spectrum.04435-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hertzberger R, May A, Kramer G, van Vondelen I, Molenaar D, Kort R. Genetic elements orchestrating lactobacillus crispatus Glycogen metabolism in the Vagina. Int J Mol Sci. 2022;23(10):5590. 10.3390/ijms23105590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lebeau A, Bruyere D, Roncarati P, Peixoto P, Hervouet E, Cobraiville G, Taminiau B, Masson M, Gallego C, Mazzucchelli G, Smargiasso N, Fleron M, Baiwir D, Hendrick E, Pilard C, Lerho T, Reynders C, Ancion M, Greimers R, Twizere JC, Daube G, Schlecht-Louf G, Bachelerie F, Combes JD, Melin P, Fillet M, Delvenne P, Hubert P, Herfs M. HPV infection alters vaginal microbiome through down-regulating host mucosal innate peptides used by Lactobacilli as amino acid sources. Nat Commun. 2022;13(1):1076. 10.1038/s41467-022-28724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai W, Du H, Zhou Q, Li S, Wang Y, Hou J, Guo C, Yang Q, Li C, Xie S, Li SC, Wu R. Metabolic profiles outperform the microbiota in assessing the response of vaginal microenvironments to the changed state of HPV infection. NPJ Biofilms Microbiomes. 2024;10(1):26. 10.1038/s41522-024-00500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brotman RM, Shardell MD, Gajer P, Fadrosh D, Chang K, Silver MI, Viscidi RP, Burke AE, Ravel J, Gravitt PE. Association between the vaginal microbiota, menopause status, and signs of vulvovaginal atrophy. Menopause. 2018;25(11):1321–30. 10.1097/GME.0000000000001236. [DOI] [PubMed] [Google Scholar]
- 38.Holm JB, France MT, Gajer P, Ma B, Brotman RM, Shardell M, Forney L, Ravel J. Integrating compositional and functional content to describe vaginal microbiomes in health and disease. Microbiome. 2023;11(1):259. 10.1186/s40168-023-01692-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.France MT, Fu L, Rutt L, Yang H, Humphrys MS, Narina S, Gajer PM, Ma B, Forney LJ, Ravel J. Insight into the ecology of vaginal bacteria through integrative analyses of metagenomic and metatranscriptomic data. Genome Biol. 2022;23(1):66. 10.1186/s13059-022-02635-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of VM have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences that are accessible at https://ngdc.cncb.ac.cn/gsa/search?searchTerm = CRA024190. Metabolome data are accessible at https://ngdc.cncb.ac.cn/search/specific?db = omix&q = OMIX009604.