Abstract
Few data exist on somatic mutation in the epithelial cell lineages that play a central role in human biology and disease. To delineate the “landscape” of somatic mutation in a human epithelial cell lineage, we determined the frequency and molecular nature of somatic mutations occurring in vivo in the X-linked HPRT gene of kidney tubular epithelial cells. Kidney epithelial mutants were frequent (range 0.5 to 4.2 × 10−4) and contained a high proportion of unreported HPRT base substitutions, −1-bp deletions and multiple mutations. This spectrum of somatic mutation differed from HPRT mutations identified in human peripheral blood T lymphocytes and from germ-line HPRT mutations identified in Lesch–Nyhan syndrome or hyperuricemia patients. Our results indicate that DNA damage and mutagenesis may have unusual or mechanistically interesting features in kidney tubular epithelium, and that somatic mutation may play a more important role in human kidney disease than has been previously appreciated.
Epithelial cells constitute 60% of differentiated human cell types and are the source of 85% of adult cancers (ref. 1 and http://www.cancer.org/eprise/main/docroot/STT/content/STT_1x_2001_Facts_and_Figures.pdf). Despite their importance in normal biology and disease pathogenesis, remarkably few data exist on either the frequency or the molecular spectrum of somatic mutation in human epithelial cell lineages. To delineate the somatic mutational “landscape” of a human epithelial cell lineage—the quantitative and molecular aspects of somatic mutation as a function of age—we used primary epithelial cell cloning and DNA sequencing to characterize somatic mutation in vivo in the X-linked HPRT gene of kidney cortical tubular epithelial cells.
In initial work we demonstrated that primary tubular epithelial cell clones could be grown directly from human kidney tissue. We also showed that mutant tubular epithelial cells, recovered by growth in the purine analogue 6-thioguanine (TG), were surprisingly frequent (range 0.5 to 24 × 10−4). Mutant frequency among 80 donors of ages 3–94 yr increased ≈1%/yr of donor age and was ≥ 10-fold higher in kidney than in peripheral blood T lymphocytes of normal, age-matched donors (2–9). Most TG-resistant kidney tubular epithelial cells from single donors contained different HPRT mutations, thus establishing a close correspondence between mutant and mutation frequencies (2, 3, 10).
To provide a more detailed picture of the quantitative and molecular features of in vivo somatic mutation in human kidney tubular epithelial cells, we determined the frequency and molecular spectrum of HPRT mutations in 96 TG-resistant mutants from 19 kidney tissue donors. We have also compared kidney mutations with somatic HPRT mutations identified in human peripheral blood T lymphocytes (5–9) or with germ-line HPRT mutations identified in Lesch–Nyhan (LN) syndrome or hyperuricemia (HRH) patients (11). The high frequency and unusual spectrum of HPRT mutations in TG-resistant tubular epithelial cells indicate that DNA damage or mutagenesis may have unusual or mechanistically interesting features in kidney, and that somatic mutation may play a more important role in human renal disease pathogenesis than had been previously appreciated.
Materials and Methods
Cell Culture.
Control and TG-resistant primary kidney tubular epithelial cell clones were grown from kidney tissue as described (2). Tissue was obtained from unused transplant kidneys (n = 8; Northwest Kidney Center, Seattle) or as normal tissue from nephrectomy kidneys (n = 11; University of Washington Hospital, Seattle, and Swedish Hospital, Seattle). None of the tissue donors had received prior radiation or chemotherapy. Mutants were recovered by cell growth for 18–21 days in the presence of 60 μM (10 μg/ml) TG. All kidney tissue was collected and used under approval of the University of Washington Human Subjects Review Board.
HPRT Mutation Analysis.
Nucleic acid isolation and HPRT cDNA amplification from TG-resistant or control cell clones were performed as described (2). The complete 9-exon, 657-bp HPRT ORF of reverse transcriptase–PCR (RT-PCR) products was directly sequenced, and mutations were verified by sequencing both DNA strands. Exon skip/exclusion mutations identified in RT-PCR products were further characterized by amplifying and sequencing skipped or excluded exons and their splice junctions from genomic DNA. The HPRT primers used for amplification and sequencing are shown in Table 5, which is published as supporting information on the PNAS web site, www.pnas.org, or have been described (12). DNA sequencing was performed by using Big Dye or Rhodamine Dye Terminator Cycle Sequencing kits (Applied Biosystems/Perkin–Elmer Cetus). Sequencing reactions were purified on Centri-sep columns (Princeton Separations, Adelphia, NJ) before analysis on an Applied Biosystems Prism model 377 sequencer. Mutations are reported here following guidelines of the Nomenclature Working Group (13) and have been referenced to the “sense” (coding or mRNA equivalent) strand of the human HPRT cDNA (14) or the HPRT gene (15).
Statistical Analyses.
The association of multiply mutant clones with donor age, gender, or donor type was examined by using the Wilcoxon test (age) or the Fisher exact test (gender and donor type, i.e., transplant vs. nephrectomy). A Monte Carlo method was used to determine whether the same base substitution, e.g., an A>G transition, occurred more often in multiply mutant clones than would be expected by chance alone, given the observed distribution of base substitutions in all mutants. Expected distributions for the number of matching substitutions out of the observed total of six multiple substitution mutation pairs were generated by repeatedly drawing six pairs of base substitutions (with replacement) from the observed distribution of kidney base substitution types (Table 1). This expected distribution was then compared with observed data to obtain the P value.
Table 1.
Kidney epithelial HPRT mutations
| Class | Kidney, n | Epithelium*, % |
|---|---|---|
| Base substitutions | 55 | 66 |
| Transitions | 26 | 31 |
| A>G | 4 | 5 |
| G>A | 16 | 19 |
| C>T | 3 | 4 |
| T>C | 3 | 4 |
| Transversions | 29 | 35 |
| A>C | 2 | 2 |
| A>T | 1 | 1 |
| C>A | 1 | 1 |
| C>G | 2 | 2 |
| G>C | 4 | 5 |
| G>T | 6 | 7 |
| T>A | 6 | 7 |
| T>G | 7 | 8 |
| Deletions | 23 | 28 |
| − 1 bp | 13 | 16 |
| − 2–29 bp | 10 | 12 |
| Insertions | 3 | 4 |
| + 1 bp | 2 | 2 |
| + 17 bp | 1 | 1 |
| Other/complex* | 2 | 2 |
| Total | 83 | 100 |
| Exon skip, cause undetermined† | 9 | |
| None‡ | 13 |
Other/complex mutations: c.77_79delATCinsTT, observed once in each of two different donors (see Table 6).
Exon skip mutations that could not be fully characterized.
TG-resistant mutants that lacked mutations in the HPRT ORF.
The distribution of distances between mutations in multiply mutant clones was analyzed by comparing the observed distribution against the distribution expected under the assumption that multiple mutations were generated uniformly and independently in the HPRT gene. An initial (expected hypothetical) distribution was generated by determining the distance between single nucleotide mutation pairs within mutants for 50,000 simulated data sets each comprising seven multiply mutant clones (six clones with pairs of mutations, and one clone with four mutations). This distribution was generated by using mutations occurring anywhere along 40 kb of the HPRT gene under the assumptions of uniformity and independence. The HPRT gene sequence was then used to generate an expected observable distribution based on 50,000 data sets generated as described above with the exception that only observable mutations, i.e., those falling in HPRT exons, were included. This expected observable distribution and the distribution of intermutational distances observed in kidney were tested for equality by using the Kolmogorov–Smirnov (K-S) test. P values for the K-S test will be small when the assumptions of mutational uniformity and/or independence are violated. An analogous method was used for T lymphocyte data. A second method of comparing expected observable and experimentally observed intermutational distance distributions was to perform a standard χ2 test for goodness of fit for six different intermutational distance categories chosen to reflect the experimental data. The test was performed for all six categories simultaneously as well as for each individual category or bin.
The proportions of types of mutations by source were compared by using Pearson's χ2 test or Fisher's exact test, as appropriate. Proportions of mutational sites that were unique for each data set were compared by using the same methods. The spectrum of HPRT mutations in kidney epithelium, peripheral blood T lymphocytes, or the germ line were compared by using the exact hypergeometric test, an extension of the Fisher exact test that is very reliable for analyses of the sparse tables encountered in this context (16).
Significance levels for single nucleotide substitution mutation “hotspot” identification were determined under the null hypothesis that the N mutations in each data set had an equal probability of hitting any of the estimated 300 mutable and observable sites in the HPRT coding region. To calculate the P value for a potential hotspot, 106 data sets were generated under the null hypothesis for each observed data set, conditioning on N. The P value for X hits at a single position was taken to be the percent of data sets at which the maximum frequency reached or exceeded X.
Results
Kidney Epithelial Mutant Frequency and Spectrum.
TG-resistant and control primary tubular epithelial cell clones were grown from kidney cortical tissue samples from 19 adult donors (age range 31 to 77 yr; Table 2). The colony-forming efficiency of kidney cortical cell suspensions (mean 2.4 × 105 colonies/g tissue) and the frequency of TG-resistant mutants (mean 1.9 × 10−4; Table 2) were comparable to previous results (2, 3). Ninety-six TG-resistant mutants, as well as control clones from each donor, were isolated for HPRT mutation analyses: an average of five mutants/donor (mean 4.9, range 1 to 15) were analyzed.
Table 2.
Kidney tissue mutant frequency
| Source | Gender | Number | Age
|
Mutant frequency* (× 10−4)
|
||
|---|---|---|---|---|---|---|
| Mean | Range | Mean | Range | |||
| Nephrectomy | Male | 6 | 59.8 | 44–74 | 2.2 | 1.0–3.4 |
| Female | 5 | 54.0 | 31–77 | 2.2 | 0.8–4.2 | |
| Transplant | Male | 8 | 51.3 | 34–67 | 1.4 | 0.5–3.2 |
| Total | 19 | 54.7 | 31–77 | 1.9 | 0.5–4.2 | |
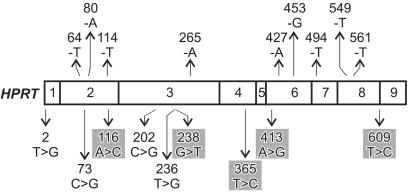
A majority of TG-resistant mutants (83/96 or 86%) contained HPRT mutations. The remaining 13 mutants, from seven different donors, produced a normal-length RT-PCR product that contained no mutation in the HPRT ORF (Table 1 and Table 6, which is published as supporting information on the PNAS web site). Base substitutions were 66% (55/83) of the HPRT mutations identified in TG-resistant mutants (Table 1). Substitutions were observed at 51 different HPRT cDNA positions and included all 12 base substitution types. Transitions and transversions (n = 26 and 29, respectively) were equally frequent, but occurred preferentially at G:C base pairs (32 substitutions at G:C vs. 23 at A:T base pairs). Among the base substitutions five (5/55, 9%) identify new human HPRT mutable sites and an additional four (4/55, 7.3%) were new substitutions at known mutable sites (refs. 17 and 18; Fig. 1).
Figure 1.
Newly identified human somatic HPRT −1-bp deletions and base substitutions in kidney tubular epithelial cells. The central rectangle represents the HPRT ORF with previously unreported single base deletions shown above and base substitutions below the rectangle. Exon boundaries are indicated by vertical lines in the cDNA; individual exons are numbered. Deleted or substituted bases are indicated together with the mutation position in the human HPRT cDNA, where the A of the cDNA ATG start codon is position 1. Gray shading indicates substitutions that identify mutable sites. Changes are given by using the sense (coding) strand of human HPRT cDNA as a reference.
Deletions were the second most frequent mutation type identified in TG-resistant mutants (23/83, 28%; Table 1). The most frequent deletion, of −1 bp, occurred preferentially at A:T base pair positions (11/13 or 85%; Table 6). A majority of these −1-bp deletions (nine of 11, 82%) had not been reported before (Fig. 1; ref. 18). Ten additional deletions of 2–29 bp are detailed in Table 6. Several of these larger deletions occurred in or adjacent to short palindromic or direct repeat sequences (Table 6). Large HPRT deletions, of ≥50 bp, have been documented in kidney and appear to represent approximately the same fraction of mutations in kidney as is T lymphocytes (≈15%; refs. 2–4). These have not been fully characterized at the nucleotide sequence level and thus have not been included here.
Additional mutations included two +1-bp insertions and a +17-bp insertion that was an exact, direct duplication of HPRT nucleotides c.580_596; and c.77_79ATC>TT, observed once in each of two different donors. In contrast, two instances of apparent clonality, in which different mutants from the same donor contained identical HPRT mutations, were observed (mutants 55–012 and 64–079). Eight “exon skip/exclusion” mutations that involved one or more HPRT exons were identified, but could not be fully characterized because of the lack of adequate genomic DNA samples.
Thirteen mutants that had a normal-length RT–PCR product but no mutation in the HPRT ORF were identified. Similar TG-resistant mutants have been identified in human peripheral blood T lymphocyte populations (see, e.g., ref. 5) and may arise in at least some instances as a result of HPRT regulatory mutations or gene silencing. Methylation can modulate HPRT gene expression (see, e.g., ref. 19), and methylation-induced gene silencing of, e.g., the VHL (von Hipple–Lindau) tumor suppressor gene, has been documented in tubular epithelial cells (20). It was not possible to rule out clonality in donors with multiple TG-resistant mutants that lacked HPRT mutations (mutants 58–011 and 68–004).
Multiple HPRT Mutations in Kidney Mutants.
An unexpectedly high fraction of TG-resistant kidney epithelial mutants contained multiple HPRT mutations. Multiple mutant clones were 7% (7/96) of all mutants, contained 19% (16/83) of HPRT mutations, and were found in six of 19 donors (Tables 3 and 6). In contrast, multiple HPRT mutations separated by >1 bp are rare (≤0.3%) in human T lymphocyte and germ-line HPRT mutational spectra (5–9, 21). Kidney multiply mutant clones were not associated with donor age (P = 0.96), gender (P = 1.0), or donor type (transplant vs. nephrectomy; P = 0.44). Six multiply mutant clones contained two different HPRT mutations: one pair of adjacent base substitutions flanking the exon 8/splice donor junction, and five additional mutation pairs separated by from 2 bp to >25 kb in the HPRT gene. One mutant contained four different HPRT mutations, separated by from 14 bp to 4,864 bp, in four different HPRT exons (58–010; Table 3).
Table 3.
Multiple HPRT mutations in tubular epithelial cells
| Mutant* | Mutation 1† | Mutation 2 | Distance, bp‡ |
|---|---|---|---|
| 54-026 | c.609T>C§ | IVS8+1G>A§ | 1 |
| 58-010a | c.413A>G | c.427delA | 14 |
| 58-010b | c.427delA | c.496A>G | 4,864 |
| 58-010c | c.496A>G | c.540A>G | 215 |
| 61-001¶ | Δe2-6‖ | IVS7-1G>C | 5,013 |
| 61-006 | c.71T>A | c.73C>G | 2 |
| 62-027 | c.365T>C | c.427delA | 7,024 |
| 65-014 | c.478G>A | c.485G>A | 7 |
| 68-010 | c.77_79ATC>TT** | c.526C>T** | 25,025 |
Mutant designations uniquely identify both the donor and mutant (see Table 6). Mutations are designated following Mutation Nomenclature Working Group guidelines (44).
Mutation positions are referenced to the HPRT cDNA sense strand sequence in which the A of the ATG start codon is position 1.
Distance between mutation positions in the HPRT gene in DNA base pairs.
Mutations at successive nucleotides in the HPRT gene (g.40109–40110).
Two different mutants from donor 61 contained the same mutation pair.
The endpoints of this deletion have not been defined.
Amplified cDNA contained two subpopulations, one each showing the designated mutations.
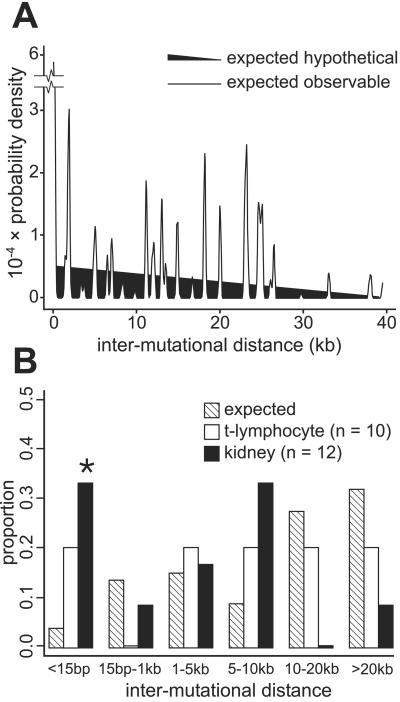
The distance between pairs of mutations in multiple mutant clones differed from the expected observable distribution for pairs of independent HPRT mutations (Fig. 2A; P = 0.0018). A subset of these mutations were closely spaced: four of 12 mutation pairs were separated by an average of only 6 bp (range 1 to 14 bp), a significantly higher fraction than would be expected by chance alone (Fig. 2B; P = 0.003). A second interesting feature of multiple mutations was molecular identity: the same base substitution (e.g., A>G three times in mutant 58–010, or G>A twice in mutant 65–014) was observed more often than would be expected by chance alone (P = 0.0015), despite there being no difference in the overall distribution of base substitution types in mutants containing single or multiple mutations (P = 0.18).
Figure 2.
Distribution of multiple mutations in the human HPRT gene. (A) The probability distribution for distances between pairs of mutations occurring uniformly and independently across the portion of the human HPRT gene containing the nine HPRT exons (expected hypothetical distribution) is shown as a filled triangle falling monotonically from shortest and longest distances. In contrast, the distribution of pairs of mutations that fall in or immediately adjacent to one or more HPRT exon (expected observable distribution) is highly nonuniform, as indicated by the jagged line. (B) Proportion of multiple mutation pairs from kidney and peripheral blood T lymphocytes (23) as a function of distance between mutations versus expectation from A. The distribution of distances between mutations differed from expectation for kidney (P < 4 × 10 −4) although not for T lymphocytes (P = 0.15; results not shown), as did the proportion of closely spaced kidney mutations separated by <15 bp (*, P = 0.004, versus 0.07–0.96 for other length classes).
These unusual features of multiple mutations suggest that at least a portion may have a mechanistically linked origin, a conclusion reinforced by our failure to find individual mutations in multiple mutant clones in other mutants from the same donor. A recent analysis of multiple mutations in the lacI transgene of Big Blue mice also has identified what may be mechanistically linked mutations by using a statistical approach similar to ours (22). In contrast, multiple HPRT mutations in normal peripheral blood T lymphocytes from patients treated for acute lymphocytic leukemia appear to have arisen sequentially and had an intermutation distance distribution that closely approximated the expectation for independent mutations (ref. 23; Fig. 2B; P = 0.79).
Human Somatic and Germ-Line HPRT Mutations Compared.
The recent publication of large series of in vivo HPRT mutations from peripheral blood T lymphocytes (502 mutations; refs. 5–9) and from LN syndrome or HRH patients (270 mutations; ref. 21) allowed us to compare in vivo mutations arising in the HPRT gene in two different somatic cell lineages or the human germ line. These somatic and germ-line mutations are summarized in Table 4, with the exception of incompletely characterized exon skip mutations. Germ-line LN or HRH mutations were further divided for analyses because of differences in the stringency of selection: LN mutations resemble kidney and T lymphocyte mutations in severity, as they reduce HPRT protein production or enzymatic activity to ≤2% of wild-type levels, whereas HRH mutations are almost exclusively missense mutations that lead to retention of ≥10% of wild-type HPRT protein and activity levels (11, 21). The use of a common RT-PCR/DNA sequencing strategy to characterize most of the mutations summarized in Table 4 minimized mutation detection bias and provided comparable sets of mutations for subsequent spectrum and hotspot analyses (see below).
Table 4.
Distribution of HPRT mutations in kidney epithelial cells, T lymphocytes, and germ line
| Class | Kidney epithelium*
|
T-lymphocyte set 1†
|
T-lymphocyte set 2‡
|
LN germ line§
|
HRH germ line§
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | |
| Base substitutions | 55 | 66 | 288 | 83 | 95 | 62 | 149 | 73 | 28 | 97 |
| Transitions | 26 | 31 | 152 | 44 | 42 | 27 | 88 | 43 | 19 | 66 |
| Transversions | 29 | 35 | 136 | 39 | 53 | 35 | 61 | 30 | 9 | 31 |
| Tandem | (2) | (4) | (0) | (0) | (0) | |||||
| Deletions | 23 | 28 | 54 | 15 | 53 | 35 | 28 | 14 | 1 | 3 |
| − 1 bp | 13 | 16 | 21 | 6 | 21 | 14 | 8 | 4 | 0 | 0 |
| − >1 bp | 10 | 12 | 33 | 9 | 32 | 21 | 20 | 10 | 1 | 3 |
| Insertions | 3 | 4 | 7 | 2 | 5 | 3 | 26 | 13 | 0 | 0 |
| + 1 bp | 2 | 2 | 6 | 2 | 2 | 1 | 15 | 7 | 0 | 0 |
| + >1 bp | 1 | 1 | 1 | 0 | 3 | 2 | 11 | 5 | 0 | 0 |
| Other/complex | 2 | 2 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 |
| Total | 83 | 100 | 349 | 100 | 153 | 100 | 205 | 100 | 29 | 100 |
There were significant differences in the proportion of HPRT base substitution mutations in kidney as compared with T lymphocyte set 1 (66% vs. 83%; P = 0.002) or with HRH germ-line mutations (66% vs. 97%; P = 0.0031). The difference in proportion of base substitutions in the two T lymphocyte data sets (83% vs. 62%) was significant (P = 2.3 × 10−6), as was the difference between LN and HRH germ-line data sets (73% vs. 97%; P = 0.01). There was a striking bias in deletions over insertions among somatic (kidney and T lymphocyte) as compared with germ-line HPRT mutations: 7 to 10.6:1 vs. 1.1:1, respectively (P = 2.45 × 10−8).
The use of a hypergeometric test (16) to compare Table 4 base substitution mutational spectra indicated that the spectrum of kidney base substitutions was substantially different from that observed in T lymphocytes or LN germ-line data (P = ≤10−6 to 0.0541). Despite the significant difference noted above in the proportion of base substitutions in the two T lymphocyte data sets (83% and 62%), the spectrum of base substitutions in these data sets were similar (P = 0.3159). Kidney and HRH germ-line mutation spectra were not significantly different (P = 0.305); however, the small size of the data sets may have limited the power of this analysis.
A Monte Carlo analysis of Table 4 data was used to identify HPRT base pair positions at which there was a statistically significant excess (P < 0.05) of base substitutions in either T lymphocytes and/or LN germ line. Ten such hotspot positions were identified in T lymphocyte (TL) or LN germ line: c.143G (TL), c.146T (TL), c.151C (LN), c.197G (TL), c.207_212G6 (TL, LN), c.222C (LN), c.508C (TL, LN), c.606G (TL), c.611A (TL), and c.617G (TL). Several of these 10 positions have been identified in other human HPRT mutation hotspot analyses (8, 17, 18, 24). Kidney base substitutions including the two most frequent were found at hotspot positions c.197, c.207_212, c.508, and c.606. We could not formally identify hotspot positions in kidney as the total number of kidney base substitutions (n = 55) was small, as were the maximum number of independent mutations at suspected hotspots (e.g., c.197, n = 3). However, there was a significant difference in the proportion of kidney base substitutions at unique positions (51/55, or 93%), as compared with either T lymphocyte (118/288 and 57/95, 41% and 60%) or LN germ-line mutations (89/149 or 60%; P = 6 × 10−12 to 4 × 10−5). In contrast, HRH germ-line mutations had a comparable fraction of base substitutions at unique positions (24/28 or 86%, vs. 93% in kidney; P = 0.43).
Mutagenesis might have interesting mechanistic features at several HPRT base pair positions in addition to those identified as hotspots. For example, all three types of base substitution and 1-bp deletion or insertion mutations were identified at c.125T and c.568G, in addition to the c.207–212G6 hotspot, in both T lymphocyte and germ-line data. A second example was the occurrence of different molecular types of mutations in short sequence blocks of 3–7 nt; e.g., c.535_539GTTGG (−1, −3, −4, and −24 dels and four different base substitutions among 18 mutations in four data sets) and c.610_616CATGTTT (−1, −2, and −18 dels and eight base substitutions among 38 mutations in four data sets).
Discussion
We used a combination of primary epithelial cell cloning and DNA sequencing to determine the frequency and molecular spectrum of somatic mutations occurring in vivo in human kidney tubular epithelial cells. The impetus for this work came from the large body of data implicating somatic mutation in human cancer pathogenesis (reviewed in refs. 25 and 26) and the comparative lack of quantitative or molecular data on in vivo somatic mutation in normal human epithelial cells. Several features of the human HPRT gene facilitated this analysis. HPRT is X-linked, and thus a single active allele is transcribed in somatic cells. Mutations in this single active allele ranging from single base substitutions to megabase DNA rearrangements have been documented to convert the cellular phenotype to TG resistance, and the resulting mutant cells can be quantitatively recovered by simple purine analogue selections (27). The HPRT gene and protein have been thoroughly characterized (11, 14, 15), and a wide range of sequence information, probes, primers, and extensive databases of human HPRT mutation are available (17, 18). This extensive background has facilitated mutational analysis using HPRT as a target, and the results of these analyses strongly suggest that HPRT reflects features of mutagenesis at many other chromosomal target genes in human somatic cells.
The high frequency and unusual spectrum of kidney HPRT mutations suggest that DNA damage or mutagenesis may be substantially different in kidney epithelium than in peripheral blood T lymphocytes or the germ line. This point is reinforced by the finding that 26% (18/70) of the single position mutations identified in kidney (nine base substitutions and nine −1-bp deletions) had not been reported among the >2,500 HPRT base substitution or single base deletion mutations already identified in the human HPRT gene (Fig. 1; refs. 17 and 18). Kidney epithelial cells, especially those in the nephron convoluted tubule segments, contain abundant mitochondria and consume large amounts of oxygen to generate the energy required for glomerular filtrate processing (28). Oxidant exposure has been shown to induce HPRT mutations in primary human peripheral blood lymphocytes in culture (see, e.g., ref. 29 and references therein). However, kidney tubular epithelial cells do not accumulate disproportionately high numbers of oxidative damage-induced HPRT mutations such as G>A transitions, G>T transversions, or CC>TT tandem transitions (reviewed in ref. 30). It was surprising as well that virtually all kidney base substitutions were at unique positions, as opposed to T lymphocytes or the germ line where sizeable fractions of mutations occurred at hotspot positions (see above). Mutagenesis at three of the mutation hotspots we identified—c.143G, c.151C, and c.508C—appears to be largely the result of deamination of 5-methylcytosine in 5mCpG dinucleotides, leading to C>T transitions (24). Despite the clear importance of this mechanism for mutagenesis in both germ-line and T lymphocyte data (n = 67), only one such mutation was identified in kidney (c.508C>T). The absence of oxidative damage- or deamination-induced mutations in kidney suggests that other types of DNA damage, e.g., from endogenous reactive nitrogen species or circulating mutagens that are concentrated or metabolized in kidney, may be the most important sources of kidney epithelium somatic mutation (31).
Several other differences in the spectrum of mutations among kidney, T lymphocyte, and germ-line mutations provide additional insight into lineage-specific differences in mutagenesis. For example, CpG hotspots such as c.508C are prominent in germ-line and somatic lineages, whereas others such as c.151C are prominent only in germ line or, in the case of c.143G, only in T lymphocytes. Other hotspots such as c.197G are prominent in somatic lineages, but absent from germ line. A third example was the striking asymmetry in the ratio of deletions to insertions: deletions are strongly favored in somatic lineages, as compared with the germ line where equal proportions of deletions and insertions were observed (Table 4). Moreover, −1-bp deletions were favored over larger deletions in kidney although not in T lymphocytes (57% versus 40%).
The mutational data in Table 4 also helped us to identify sequence contexts that may be important for promoting specific types of in vivo mutations in the HPRT gene. For example, all three types of base substitution as well as insertion or deletion mutations were identified at single base pairs (e.g., c.125T or c.566G) that reside in sequence blocks that might be prone to strand slippage or misalignment during DNA replication or repair synthesis (e.g., c.122_128TAATTAT and c.564_569TAGGAT). Other hotspots whose sequence context might promote misalignment-mediated mutagenesis include c.197G, the first G in a (TG)3 dinucleotide run, and the c.207_212G6 mutational hotspot. A third example, from kidney, was the −A deletion from c.426_427AA found in the sequence block CAAAACAATG. This single base deletion was observed in three different kidney tissue donors (53–008, 58–010, and 62–027) although not in T lymphocytes or the germ line. Complex mutational events such as these might be promoted by high levels or specific types of DNA base damage in conjunction with promutagenic DNA sequence contexts (32, 33).
The spectrum of kidney epithelial HPRT mutations included a surprisingly high fraction of multiple mutations. A better understanding of mechanistic pathways that generate multiple mutations are of particular interest in the context of mutation-driven disease processes such as neoplasia. One mechanism that could generate closely spaced multiple mutations is the replication of unusual types of DNA damage or clustered sites of damage (e.g., oxidative damage). Recently identified error-prone, lesion-replicating DNA polymerases might play an important role in this process (33–36), although nothing is known as yet about the role of these polymerases in generating mutations in human kidney epithelial cells.
Repetitive strand slippage or misalignment during DNA synthesis, as noted above, could also generate multiple, closely spaced mutations (33). However, among the somatic and germ-line HPRT mutations summarized in Table 4, multiple mutations have thus far been observed almost exclusively in kidney epithelium and appear to arise in kidney by a mechanism other than strand slippage or templating. The nonuniform distribution of expected distances between pairs of independent HPRT mutations emphasizes the need to test experimental data against expectation to identify mutations that may have arisen by any concerted mutagenesis mechanism (Fig. 2).
Cell division potential and somatic selection, in addition to DNA damage, are likely to be important determinants of mutant frequency and mutational spectrum in many human cell lineages. Mammalian kidney tubular epithelial cells retain cell division potential and appear to divide throughout life. This largely latent cell division potential can be readily revealed by tubular injury (37, 38). Slow mitotic cycling may allow large numbers of tubular epithelial cells to capture DNA damage as somatic mutations during life, while retaining the potential for further clonal expansion. The retention of cell division potential by a majority of tubular epithelial cells differs markedly from many other epithelia, e.g., skin or gut (38), but makes good “design sense:” it provides an efficient way to continually replace small numbers of injured or dead epithelial cells throughout the nephron or kidney functional unit. Somatic mutations that arise in the HPRT gene as part of this renewal program are likely to have long persistence times, as there appears to be little or no somatic selection against HPRT-deficient cells in mammalian kidney cortex (39). These features of the normal biology of kidney tubular epithelial cells may thus conspire to facilitate mutagenesis, and to promote somatic mutation accumulation, over time.
The high-frequency, age-dependent increase and unusual spectrum of somatic mutations in tubular epithelial cells suggest that somatic mutation may play a role in human kidney disease pathogenesis. For example, somatic mutation accumulation could promote decrements in tubular function or nephron number that accompany aging (reviewed in ref. 40). Somatic mutations could also promote tubular epithelial proliferative diseases such as polycystic kidney disease (41) or neoplasia (20, 42). The renal cortical cysts of inherited polycystic kidney disease, the common benign adenomas that increase in frequency and size with age (43), and renal cell carcinoma, the most common kidney epithelial malignancy, all arise from tubular epithelial cells (20, 42). It will be important to determine whether kidney tubular epithelium reflects the landscape of somatic mutation in other human epithelial cell lineages. Human epithelial mutagenesis mechanisms may be more interesting than previously thought and the role of somatic mutation in epithelial disease pathogenesis more important than currently appreciated.
Supplementary Material
Acknowledgments
We thank George Martin and Chuck Ogburn for help in the early phases of this project and Bryn Bridges, Jan Drake, John Heddle, Larry Loeb, Darryl Shibata, and Bernard Strauss for helpful discussion or for reading drafts of this manuscript. We thank the University of Washington Center for Ecogenetics and Environmental Health (National Institute on Environmental Health Sciences Grant P30ES07033) Molecular Biomarker Laboratory for help with DNA sequencing. This work was supported by National Institute on Aging and National Cancer Institute grants (to R.J.M.), National Cancer Institute Grant R29 CA77607 (to M.J.E.), and National Institute on Aging Training Grant AG00057 (to L.M.C.).
Abbreviations
- TG
6-thioguanine
- LN
Lesch–Nyhan
- HRH
hyperuricemia
- RT-PCR
reverse transcriptase–PCR
References
- 1.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D, editors. Molecular Biology of the Cell. New York: Garland; 1994. pp. 1138–1193. [Google Scholar]
- 2.Martin G M, Ogburn C E, Colgin L M, Gown A M, Edland S D, Monnat R J., Jr Hum Mol Genet. 1996;5:215–221. doi: 10.1093/hmg/5.2.215. [DOI] [PubMed] [Google Scholar]
- 3.Colgin L M. Ph.D. thesis. Seattle: Univ. of Washington; 1997. [Google Scholar]
- 4.Cole J, Skopek T R. Mutat Res. 1994;304:33–105. doi: 10.1016/0027-5107(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 5.Burkhart-Schultz K J, Thompson C L, Jones I M. Carcinogenesis. 1996;17:1871–1883. doi: 10.1093/carcin/17.9.1871. [DOI] [PubMed] [Google Scholar]
- 6.Burkhart-Schultz K J, Jones I M. Environ Mol Mutagen. 1997;30:371–384. [PubMed] [Google Scholar]
- 7.Podlutsky A, Osterholm A M, Hou S M, Lambert B. Carcinogenesis. 1998;19:557–566. doi: 10.1093/carcin/19.4.557. [DOI] [PubMed] [Google Scholar]
- 8.Podlutsky A, Hou S-M, Nyberg F, Pershagen G, Lambert B. Mutat Res. 1999;431:325–339. doi: 10.1016/s0027-5107(99)00176-1. [DOI] [PubMed] [Google Scholar]
- 9.Hackman P, Hou S-M, Nyberg F, Pershagen G, Lambert B. Mutat Res. 2000;468:45–61. doi: 10.1016/s1383-5718(00)00039-5. [DOI] [PubMed] [Google Scholar]
- 10.Colgin L M, Hackmann A F M, Monnat R J., Jr Hum Mutat. 1999;13:504–505. doi: 10.1002/(SICI)1098-1004(1999)13:6<504::AID-HUMU15>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 11.Jinnah H A, Friedmann T. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 2001. pp. 2537–2570. [Google Scholar]
- 12.Gibbs R A, Nguyen P-N, Edwards A, Civitello A B, Caskey C T. Genomics. 1990;7:235–244. doi: 10.1016/0888-7543(90)90545-6. [DOI] [PubMed] [Google Scholar]
- 13.den Dunnen J T, Antonarakis S E. Hum Mutat. 2000;15:2–7. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 14.Jolly D J, Okayama H, Berg P, Esty A C, Filpula D, Bohlen P, Johnson G G, Shively J, Hunkapillar T, Friedmann T. Proc Natl Acad Sci USA. 1983;80:477–481. doi: 10.1073/pnas.80.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards A, Voss H, Rice P, Civitello A, Stegemann J, Schwager C, Zimmermann J, Erfle H, Caskey C T, Ansorge W. Genomics. 1990;6:593–608. doi: 10.1016/0888-7543(90)90493-e. [DOI] [PubMed] [Google Scholar]
- 16.Piegorsch W W, Bailer A J. Genetics. 1994;136:403–416. doi: 10.1093/genetics/136.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cariello N F, Skopek T R. J Mol Biol. 1993;231:41–57. doi: 10.1006/jmbi.1993.1255. [DOI] [PubMed] [Google Scholar]
- 18.Cariello N F, Douglas G R, Gorelick N J, Hart D W, Wilson J D, Soussi T. Nucleic Acids Res. 1998;26:198–199. doi: 10.1093/nar/26.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C, Yang M C K, Ang T P. J Biol Chem. 2001;276:320–328. doi: 10.1074/jbc.M007096200. [DOI] [PubMed] [Google Scholar]
- 20.Reuter V E, Presti J C., Jr Semin Oncol. 2000;27:124–137. [PubMed] [Google Scholar]
- 21.Jinnah H A, De Gregorio L, Harris J C, Nyhan W L, O'Neill J P. Mutat Res. 2000;463:309–326. doi: 10.1016/s1383-5742(00)00052-1. [DOI] [PubMed] [Google Scholar]
- 22.Buettner V L, Hill K A, Scaringe W A, Sommer S S. Mutat Res. 2000;452:219–229. doi: 10.1016/s0027-5107(00)00090-7. [DOI] [PubMed] [Google Scholar]
- 23.Finette B A, Homans A C, Albertini R J. Science. 2000;288:514–517. doi: 10.1126/science.288.5465.514. [DOI] [PubMed] [Google Scholar]
- 24.O'Neill J P, Finette B A. Environ Mol Mutagen. 1998;32:188–191. doi: 10.1002/(sici)1098-2280(1998)32:2<188::aid-em16>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 25.Vogelstein B, Kinzler K W. The Genetic Basis of Human Cancer. New York: McGraw–Hill; 1998. [Google Scholar]
- 26.Ehrlich M. DNA Alterations in Cancer: Genetic and Epigenetic Changes. Natick, MA: Eaton; 1999. [Google Scholar]
- 27.Fenwick R G. In: Molecular Cell Genetics. Gottesman M, editor. New York: Wiley; 1985. pp. 333–373. [Google Scholar]
- 28.Gullans S R. In: Brenner and Rector's The Kidney. Brenner B M, editor. Philadelphia: Saunders; 2000. pp. 215–246. [Google Scholar]
- 29.Díaz-Llera S, Podlutsky A, Österholm A M, Hou S-M, Lambert B. Mutat Res. 2000;469:51–61. doi: 10.1016/s1383-5718(00)00058-9. [DOI] [PubMed] [Google Scholar]
- 30.Wang D, Kreutzer D A, Essigmann J M. Mutat Res. 1998;400:99–115. doi: 10.1016/s0027-5107(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 31.Marnett L J, Plastaras J P. Trends Genet. 2001;17:214–221. doi: 10.1016/s0168-9525(01)02239-9. [DOI] [PubMed] [Google Scholar]
- 32.Seo K-Y, Jelinsky S A, Loechler E L. Mutat Res. 2000;463:215–246. doi: 10.1016/s1383-5742(00)00047-8. [DOI] [PubMed] [Google Scholar]
- 33.Kunkel T A, Bebenek K. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 34.Goodman M F, Tippin B. Nat Rev Mol Cell Biol. 2000;1:101–109. doi: 10.1038/35040051. [DOI] [PubMed] [Google Scholar]
- 35.Harfe B D, Jinks-Robertson S. Mol Cell. 2000;6:1491–1499. doi: 10.1016/s1097-2765(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 36.Friedberg E C, Fischhaber P L, Kisker C. Cell. 2001;107:9–12. doi: 10.1016/s0092-8674(01)00509-8. [DOI] [PubMed] [Google Scholar]
- 37.Cuppage F E, Chiga M, Tate A. Lab Invest. 1972;26:122–126. [PubMed] [Google Scholar]
- 38.Wright N, Alison M, editors. The Biology of Epithelial Cell Populations. Oxford: Clarendon; 1984. pp. 981–1003. [Google Scholar]
- 39.Ansell J D, Samuel K, Whittingham D G, Patek C E, Hardy K, Handyside A H, Jones K W, Muggleton-Harris A L, Taylor A H, Hooper M L. Development (Cambridge, UK) 1991;112:489–498. doi: 10.1242/dev.112.2.489. [DOI] [PubMed] [Google Scholar]
- 40.Choudhury D, Palmer B, Levi M. In: The Kidney: Physiology and Pathophysiology. Seldin D W, Giebisch G, editors. Philadelphia: Lippincott Williams & Wilkins; 2000. pp. 2571–2595. [Google Scholar]
- 41.Qian F, Watnick T J. Mol Genet Metabol. 1999;68:237–242. doi: 10.1006/mgme.1999.2896. [DOI] [PubMed] [Google Scholar]
- 42.Cohen H T. Curr Opin Nephrol Hypertens. 1999;8:325–331. doi: 10.1097/00041552-199905000-00008. [DOI] [PubMed] [Google Scholar]
- 43.Grignon D J, Eble J N. Semin Diagnostic Pathol. 1998;15:41–53. [PubMed] [Google Scholar]
- 44.Antonarakis S E Nomenclature Working Group. Hum Mutat. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.