Abstract
In a diverse group of organisms including plants, Caenorhabditis elegans, Drosophila, and trypanosomes, double-stranded RNA (dsRNA) is a potent trigger of gene silencing. In several model systems, this natural response has been developed into a powerful tool for the investigation of gene function. Use of RNA interference (RNAi) as a genetic tool has recently been extended to mammalian cells, being inducible by treatment with small, ≈22-nt RNAs that mimic those produced in the first step of the silencing process. Here, we show that some cultured murine cells specifically silence gene expression upon treatment with long dsRNAs (≈500 nt). This response shows hallmarks of conventional RNAi including silencing at the posttranscriptional level and the endogenous production of ≈22-nt small RNAs. Furthermore, enforced expression of long, hairpin dsRNAs induced stable gene silencing. The ability to create stable “knock-down” cell lines expands the utility of RNAi in mammalian cells by enabling examination of phenotypes that develop over long time periods and lays the groundwork for by using RNAi in phenotype-based, forward genetic selections.
The use of genetically tractable model systems has been the key to our present understanding of gene structure and function, cell and organismal biology, and, ultimately, the molecular aspects of human disease. The ability to stably knock out or knock down gene expression and, thus, function, in particular, has been paramount to the use of such models for illuminating biological function. For example, the use of conditional lethals in bacteriophage T4 allowed functional analysis of phage morphogenesis modules (1), whereas the same technique applied to yeast permitted the discovery of functional hierarchies among genes regulating cell cycle progression (2, 3). In both scenarios, cells acquire stable phenotypes through heritable genetic alterations.
Although such basic genetic approaches are virtually effortless in many model organisms, cultured mammalian cells have proven somewhat intractable, in this regard. This is largely because cultured mammalian cells are diploid and favor nonhomologous over homologous recombination. Current approaches to create stable phenotypes in mammalian cells have been often met with limited success. Dominant-negative and antisense strategies have proven inconsistent and unpredictable, thus lacking experimental rigor equivalent to a point mutation in yeast. However, one approach now used extensively in other diploid organisms has the potential to foment a revolution in mammalian somatic cell genetics. This approach is dubbed double-stranded RNA (dsRNA)–dependent posttranscriptional gene silencing, or RNA interference (RNAi).
It has become clear that dsRNA-induced silencing phenomena are present in evolutionarily diverse organisms including plants, fungi, and metazoans (reviewed in ref. 4). A combination of genetic and biochemical studies suggests that many of these phenomena share a common mechanism. The prevailing model begins with the conversion of the dsRNA silencing “trigger” into small RNAs (guide RNAs or siRNAs, ref. 5) that range in size from ≈21 to 25 nts, depending on the species of origin (6–8). These RNAs become incorporated into a multicomponent nuclease complex, which uses the sequence of the guide/siRNAs to identify and destroy homologous mRNAs (7, 8).
In several systems, dsRNA-induced silencing has been harnessed as a powerful tool for the analysis of gene function. Particularly in Caenorhabditis elegans, RNAi has emerged as the standard protocol for quickly assessing the consequences of inhibiting gene function. In fact, programs are underway to create RNAi libraries that can be used to suppress, individually, each of the ≈19,000 genes in the worm genome (9, 10). In Drosophila, the first evidence of dsRNA-induced silencing came from the study of embryos (11), and subsequently, RNAi has proven an effective tool in cultured cells and in adult insects (7, 12, 13).
Despite its utility in diverse systems, harnessing RNA to study gene function in mammals seemed potentially problematic. Indeed, mammals have evolved robust systems for responding to dsRNAs, specifically as an antiviral defense (reviewed in refs. 14 and 15). In somatic cells, dsRNA activates a variety of responses. Predominant among these is PKR, a kinase that is activated by dimerization in the presence of dsRNA (16). PKR, in turn, phosphorylates EIF2α, causing a nonspecific translational shutdown (reviewed in ref. 14). dsRNA also activates 2′-5′ oligoadenylate polymerase, the product of which is an essential cofactor for a nonspecific ribonuclease, RNase L (reviewed in ref. 17).
Recently, Tuschl and colleagues (5) have demonstrated that RNAi can be provoked in numerous mammalian cell lines through the introduction of siRNAs. These siRNAs avoid provoking the PKR response by virtue of their small size and are presumed to be incorporated into the RNAi pathway by mimicking the products of the Dicer enzyme, which catalyzes the initiation step of RNAi (18). The ability to apply RNAi in mammals will undoubtedly spark a firestorm of effort to assess the consequences of suppressing the expression of genes in cultured mammalian cells.
The power of RNAi as a genetic tool would be greatly enhanced by the ability to engineer stable silencing of gene expression. Whereas the production of small RNAs via in vivo expression is problematic, stable silencing has been induced in model organisms by directed expression of long dsRNAs (13, 19, 20). We therefore undertook an effort to identify mammalian cells in which long dsRNAs could be used as RNAi triggers in the hope that these same cell lines would provide a platform upon which to develop stable silencing strategies.
Materials and Methods
Cell Culture.
P19 mouse embryonic carcinoma cells (American Type Culture Collection, CRL-1825) were cultured in α-MEM (GIBCO/BRL) supplemented with 10% heat-inactivated FBS and 1% antibiotic/antimycotic solution (GIBCO/BRL). Mouse embryo stem cells (J1, provided by S. Kim, Cold Spring Harbor Laboratory) were cultured in DMEM containing ESgro (Chemicon) according to the manufacturer's instructions. C2C12 murine myoblast cells (gift of N. Tonks, Cold Spring Harbor Laboratory) were cultured in DMEM (GIBCO/BRL) supplemented with 10% heat-inactivated FBS and 1% antibiotic/antimycotic solution (GIBCO/BRL).
RNA Preparation.
For the production of dsRNA, transcription templates were generated by PCR; they contained T7 promoter sequences on each end of the template (see ref. 7). dsRNAs were prepared by using the RiboMax kit (Ambion, Austin, TX). Firefly and Renilla luciferase mRNA transcripts were synthesized by using the Riboprobe kit (Promega) and were gel purified before use.
Transfection and Gene Silencing Assays.
Cells were transfected with indicated amounts of dsRNA and plasmid DNA by using FuGENE6 (Roche Biochemicals) according to the manufacturer's instructions. Cells were transfected at 50–70% confluence in 12-well plates containing either 1 or 2 ml of medium per well. Dual luciferase assays (Promega) were carried out by cotransfecting cells with plasmids contain firefly luciferase under the control of SV40 promoter (pGL3-Control, Promega) and Renilla luciferase under the control of the SV40 early enhancer/promoter region (pSV40, Promega). These plasmids were cotransfected by using a 1:1 or 10:1 ratio of pGL3-control (250 ng/well) to pRL-SV40. Both ratios yielded similar results. For some experiments, cells were transfected with vectors that direct expression of enhanced green fluorescent protein (EGFP)-US9 fusion protein (21) or red fluorescent protein (pDsRed N1, CLONTECH). RNAi in S2 cells was performed as described (7).
Plasmids expressing hairpin RNAs (RNAs with a self-complimentary stem loop) were constructed by cloning the first 500 bp of the EGFP coding region (CLONTECH) into the FLIP cassette of pRIP-FLIP (E. Bernstein and G.J.H., unpublished data) as a direct repeat. The FLIP cassette contains two directional cloning sites, the second of which sports flanking LoxP sites (see Fig. 6A). The Zeocin gene (Stratagene), present between the cloning sites, maintains selection and, thus, stability of the FLIP cassette. The FLIP cassette containing EGFP direct repeats was subcloned into pcDNA3 (Invitrogen). To create an inverted repeat for hairpin production, EGFP direct repeat clones were exposed to Cre recombinase (Stratagene) in vitro and, afterward, transformed into DL759 Escherichia coli (22). These bacteria permit the replication of DNA containing cruciform structures, which tend to form from inverted repeats. DL759 transformants were screened for plasmids containing inverted repeats (≈50%).
Figure 6.
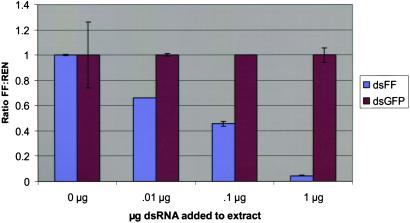
dsRNA induces silencing at the posttranscriptional level. P19 cell extracts were used for in vitro translation of firefly and Renilla luciferase mRNA (100 ng each). Translation reactions were programmed with various amounts of dsRNA 500mers, either homologous to firefly luciferase mRNA (dsLUC) or nonhomologous (dsGFP). Luciferase assays were carried out after a 1-h incubation at 30°C. Ratios of firefly to Renilla activity are normalized to no dsRNA controls. Standard deviations from the mean are shown.
Silencing of Dicer was accomplished by using a dsRNA comprising exon 25 of the mouse Dicer gene and corresponding to nucleotides 5284–5552 of the human Dicer cDNA.
In Vitro Translation and in Vitro Dicer Assays.
Logarithmically growing cells were harvested in PBS containing 5 mM EGTA washed twice in PBS and once in hypotonic buffer (10 mM Hepes, pH 7.3/6 mM β-mercaptoethanol). Cells were suspended in 0.7 packed-cell volumes of hypotonic buffer containing Complete protease inhibitors (Roche Molecular Biochemicals) and 0.5 units/ml of RNasin (Promega). Cells were disrupted in a Dounce homogenizer with a type B pestle, and lysates were centrifuged at 30,000 × g for 20 min. Supernatants were used in an in vitro translation assay containing capped m7G(5′)pppG firefly and Renilla luciferase mRNA or in in vitro Dicer assays containing 32P-labeled dsRNA. For in vitro translation assays, 5 μl of extract were mixed with 100 ng of firefly and Renilla mRNA along with 1 μg of dsRNA (or buffer)/10 mM DTT/0.5 mM spermidine/200 mM Hepes, 3.3 mM MgOAc/800 mM KOAc/1 mM ATP/1 mM GTP/4 units of Rnasin/215 μg of creatine phosphate/1 μg of creatine phosphate kinase/1 mM amino acids (Promega). Reactions were carried out for 1 h at 30°C and quenched by adding 1× passive lysis buffer (Promega). Extracts were then assayed for luciferase activity. In vitro assays for Dicer activity were performed as described (18).
Construction of Stable Silencing Lines.
Ten-centimeter plates of P19 cells were transfected with 5 μg of GFP hairpin expression plasmid and selected for stable integrants by using G-418 (300 ng/ml) for 14 days. Clones were selected and screened for silencing of GFP.
Results
RNAi in Pluripotent Murine P19 Cells.
It has long been clear that the nonspecific responses to dsRNA are attenuated during early development. In fact, injection of dsRNA into early-stage mouse embryos can induce sequence-specific silencing of both exogenous and endogenous genes (23, 24). Consistent with the possibility that RNAi might extend to mammals, homologs of the proteins that participate in this response can be easily identified in the mouse and human genomes (reviewed in ref. 4).
We sought to determine whether long dsRNA triggers could induce sequence-specific silencing in cultured murine cells, both to develop this approach as a tool for probing gene function and to allow mechanistic studies of dsRNA-induced silencing to be propagated to mammalian systems. We, therefore, attempted to extend previous studies in mouse embryos (23, 24) by searching for RNAi-like mechanisms in pluripotent, embryonic cell types.
We surveyed a number of cell lines of embryonic origin for the degree to which generalized suppression of gene expression occurred upon introduction of dsRNA. As an assay, we tested the effects of dsRNA on the expression of GFP as measured in situ by counting fluorescent cells. As expected, in both human embryonic kidney cells (293) and mouse embryo fibroblasts, GFP expression was virtually eliminated irrespective of the sequence of the cotransfected dsRNA (not shown). In some pluripotent teratocarcinoma and teratoma cell lines (e.g., N-Tera1, F9), the PKR response was attenuated but still evident (not shown); however, in contrast, transfection of nonhomologous dsRNAs had no effect on the expression of reporter genes (e.g., GFP, luciferase) either in mouse embryonic stem cells (not shown) or in p19 embryonal carcinoma cells (Fig. 1).
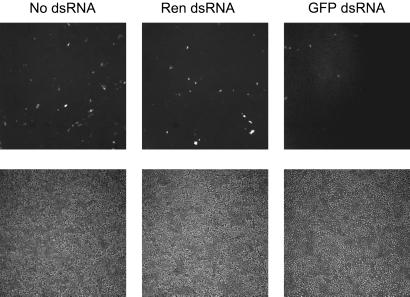
Figure 1.
RNAi in P19 embryonal carcinoma cells. Ten-centimeter plates of P19 cells were transfected by using 5 μg of GFP plasmid and 40 μg of the indicated dsRNA (or no RNA). Cells were photographed by fluorescent and phase-contrast microscopy at 72 h after transfection; silencing was also clearly evident at 48 h posttransfection.
Transfection of P19 embryonal carcinoma cells with GFP in the presence of cognate dsRNA corresponding to the first ≈500 nts of the GFP coding sequence had a strikingly different effect. GFP expression was eliminated in the vast majority of cotransfected cells (Fig. 1), suggesting that these cultured murine cells might respond to dsRNA in a manner similar to that which we had previously demonstrated in cultured, Drosophila S2 cells (7).
To quantify the extent to which dsRNA could induce sequence-specific gene silencing, we used a dual luciferase reporter assay similar to that which had first been used to demonstrate RNAi in Drosophila embryo extracts (25). P19 EC cells were transfected with a mixture of two plasmids that individually direct the expression of firefly luciferase and Renilla luciferase. These were cotransfected with no dsRNA, with dsRNA that corresponds to the first ≈500 nts of the firefly luciferase, or with dsRNA corresponding to the first ≈500 nts of GFP as a control. Cotransfection with GFP dsRNA gave luciferase activities that were similar to the no-dsRNA control, both in the firefly/Renilla activity ratio and in the absolute values of both activities. In contrast, in cells that received the firefly luciferase dsRNA, the ratio of firefly to Renilla luciferase activity was reduced by up to 30-fold (250 ng, Fig. 2B). For comparison, we carried out an identical set of experiments in Drosophila S2 cells. Although qualitatively similar results were obtained, the silencing response was more potent. At equivalent levels of dsRNA, S2 cells suppressed firefly luciferase activity to virtually background levels (not shown).
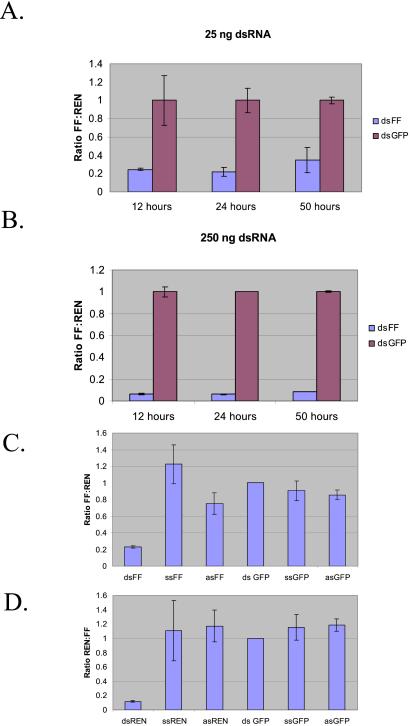
Figure 2.
RNAi of firefly and Renilla luciferase in P19 cells. (A) P19 cells were transfected with plasmids that direct the expression of firefly and Renilla luciferase and dsRNA 500 mers (25 or 250 ng, as indicated), that were either homologous to the firefly luciferase mRNA (dsFF) or nonhomologous (dsGFP). Luciferase activities were assayed at various times after transfection, as indicated. Ratios of firefly to Renilla activity are normalized to dsGFP controls. (B and C) P19 cells in 12-well culture dishes (2 ml of media) were transfected with 0.25 μg of a 9:1 mix of pGL3-Control and pRL-SV40 as well as 2 μg of the indicated RNA. Extracts were prepared 9 h after transfection. (B) Ratio of firefly to Renilla luciferase is shown. (C) Ratio of Renilla to firefly luciferase is shown. Values are normalized to dsGFP. The average of three independent experiments is shown; error bars indicate standard deviation.
The complementary experiment, in which dsRNA was homologous to Renilla luciferase, was also performed. Again, in this case, suppression of the expression of the Renilla enzyme was ≈10-fold (Fig. 2D). Thus, the dsRNA response in P19 cells was flexible, and the silencing machinery was able to adapt to dsRNAs directed against any of the reporters that were tested.
We took two approaches to test whether this response was specific for dsRNA. Pretreatment of the trigger with purified RNase III, a dsRNA-specific ribonuclease, before transfection greatly reduced its ability to provoke silencing (not shown). Furthermore, transfection of cells with single-stranded antisense RNAs directed against either firefly or Renilla luciferase had little or no effect on expression of the reporters (Fig. 2 C and D). Considered together, these results provided a strong indication that double-stranded RNAs provoke a potent and specific silencing response in P19 embryonal carcinoma cells.
Efficient silencing could be provoked with relatively low concentrations of dsRNA (25 ng/ml culture media; see Fig. 2A). The response was concentration-dependent, with maximal suppression of ≈20-fold being achieved at a dose of 1.5 μg/ml culture media.
Silencing was established rapidly and was evident by 9 h posttransfection (the earliest time point examined). Furthermore, the response persisted without significant changes in the degree of suppression for up to 72 h following a single dose of dsRNA.
RNAi in Embryonic Stem Cells.
To assess whether the presence of a sequence-specific response to dsRNA was a peculiarity of P19 cells or whether it also extended to normal murine embryonic cells, we performed similar silencing assays in mouse embryonic stem cells. Cotransfection of embryonic stem cells with noncognate dsRNAs (e.g., GFP), again, had no dramatic effect on either the absolute values or the ratios of Renilla and firefly luciferase activity (Fig. 3). However, transfection with either firefly or Renilla luciferase dsRNA dramatically and specifically reduced the activity of the targeted enzyme (Fig. 3).
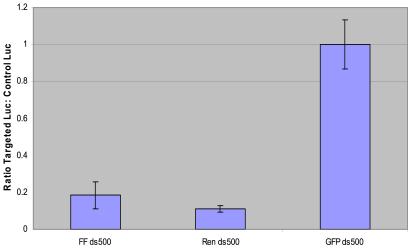
Figure 3.
Specific silencing of luciferase expression by dsRNA in murine embryonic stem cells. Mouse embryonic stem cells in 12-well culture dishes (1 ml of media) were transfected with 1.5 μg of dsRNA along with 0.25 μg of a 10:1 mixture of the reporter plasmids pGL3-Control and pRL-SV40. Extracts were prepared and assayed 20 h after transfection. The ratio of firefly to Renilla luciferase expression is shown for FF ds500; the ratio of Renilla to firefly is shown for Ren ds500. Both are normalized to ratios from the dsGFP transfection. The average of three independent experiments is shown; error bars indicate standard deviation.
This result suggests that RNAi can operate in multiple murine cell types of embryonic origin, including normal embryonic stem cells. The ability to provoke silencing in a cell type that is normally used for the generation of genetic, mosaic animals suggests the possibility of eventually testing the biological effects of silencing both in culture and in reconstituted animal models.
RNAi in Murine Somatic Cells.
RNAi effector pathways are likely to be present in mammalian somatic cells, based on the ability of siRNAs to induce transient silencing (5). Furthermore, we have shown that RNAi initiator and effector pathways clearly exist in embryonic cells that can enforce silencing in response to long dsRNA triggers. We therefore sought to test whether the RNAi machinery might exist intact in some somatic cell lines.
Transfection of HeLa cells with luciferase reporters in combination with long dsRNA triggers caused a nearly complete suppression of activity, irrespective of the RNA sequence. In a murine myoblast cell line, C2C12, we noted a mixture of two responses. dsRNAs homologous to firefly luciferase provoked a sequence-specific effect, producing a degree of suppression that was slightly more potent than was observed upon transfection with cognate ≈21-nt siRNA (ref. 5; Fig. 4A). However, with long dsRNA triggers, the specific effect was superimposed upon a generalized suppression of reporter gene expression that was presumably because of PKR activation (Fig. 4B).
Figure 4.
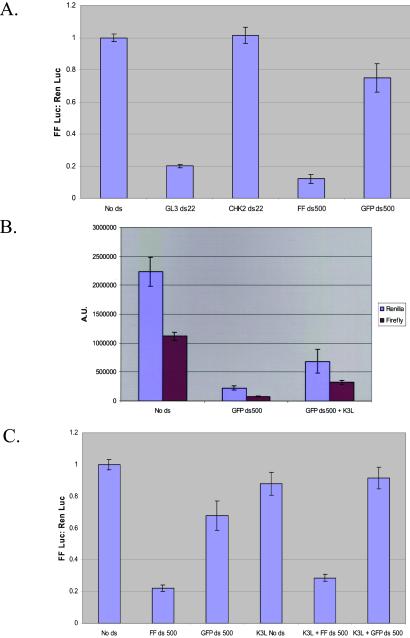
RNAi in C2C12 murine myoblast cells. (A) Mouse C2C12 cells in 12-well culture dishes (1 ml of media) were transfected with 1 μg of the indicated dsRNA along with 0.250 μg of the reporter plasmids pGL3-Control and pRL-SV40. Extracts were prepared and assayed 24 h after transfection. The ratio of firefly to Renilla luciferase expression is shown; values are normalized to ratios from the no dsRNA control. The average of three independent experiments is shown; error bars indicate standard deviation. (B) C2C12 cells cotransfected with 1 μg of either plasmid alone or a plasmid containing a hyperactive mutant of vaccinia virus K3L (26). The absolute counts of Renilla and firefly luciferase activity are shown. (C) The ratios of firefly/Renilla activity from B, normalized to no dsRNA controls.
Numerous mammalian viruses have evolved the ability to block PKR as an aid to efficient infection. For example, adenoviruses express VA RNAs, which mimic dsRNA with respect to binding but not to activation of PKR (16). Vaccinia virus uses two strategies to evade PKR. First is expression of E3L, which binds and masks dsRNAs (26). The second is expression of K3L, which binds and inhibits PKR via its ability to mimic the natural substrate of this enzyme, eIF2α (26).
Transfection of C2C12 cells with a vector that directs K3L expression attenuates the generalized repression of reporter genes in response to dsRNA. However, this protein had no effect on the magnitude of specific inhibition by RNAi (Fig. 4C).
These results raise the possibility that, at least in some cell lines and/or cell types, blocking nonspecific responses to dsRNA will enable the use of long dsRNAs for the study of gene function. This might be accomplished through the use of viral inhibitors, as described here, or through the use of cells isolated from animals that are genetically modified to lack undesirable responses.
Stable Suppression of Gene Expression Using RNAi.
To date, dsRNAs have been used to induce sequence-specific gene silencing in either cultured mammalian cells or in embryos only in a transient fashion. However, the most powerful applications of genetic manipulation are realized only with the creation of stable mutants. The ability to induce silencing by using long dsRNAs offers the opportunity to translate into mammalian cells work from model systems such as Drosophila, plants, and C. elegans wherein stable silencing has been achieved by enforced expression of hairpin RNAs (13, 19, 20).
P19 EC cells were transfected with a control vector or with an expression vector that directs expression of a ≈500-nt GFP hairpin RNA from an RNA polymerase II promoter (cytomegalovirus). Colonies arising from cells that had stably integrated either construct were selected and expanded into clonal cell lines. Each cell line was assayed for persistent RNAi by transient cotransfection with a mixture of two reporter genes, dsRED to mark transfected cells and GFP to test for stable silencing.
Transfection of clonal P19 EC cells that had stably integrated the control vector produced equal numbers of red and green cells, as would be expected in the absence of any specific silencing response (Fig. 5B), whereas cells that express the GFP hairpin RNA gave a very different result. These cells expressed the dsRED protein with an efficiency comparable to that observed in cells containing the control vector. However, the cells failed to express the cotransfected GFP reporter (Fig. 5B). These data provide a strong indication that continuous expression of a hairpin dsRNA can provoke stable, sequence-specific silencing of a target gene.
Figure 5.
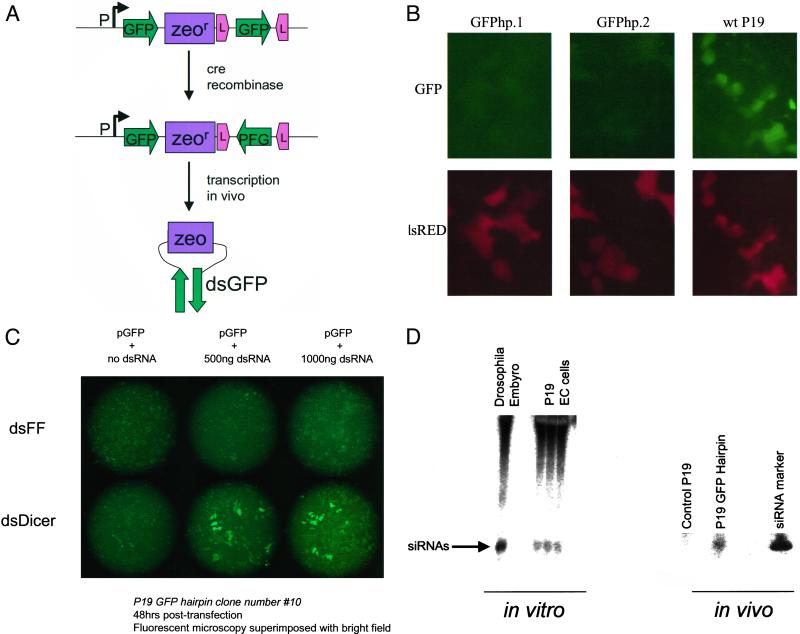
Expression of a hairpin RNA produces P19 EC cell lines that stably silence GFP. (A) A cartoon of the FLIP cassette used to construct the GFP hairpin. GFP represents the first 500 coding base pairs of EGFP. Zeo, zeocin resistance gene; L, Lox; P, the cytomegalovirus promoter in the expression plasmid pcDNA3. Homologous GFP fragments are first cloned as direct repeats into the FLIP cassette. To create inverted repeats for hairpin production, the second repeat is flipped by using Cre recombinase. When transcribed, the inverted repeat forms a GFP dsRNA with a hairpin loop. (B) P19 cell lines stably expressing the GFP hairpin plasmid, GFPhp.1 (clone 10) and GFPhp.2 (clone 12), along with wt P19 were transfected with 0.25 μg each of GFP and RFP reporter genes. Fluorescence micrographs were taken by using filters appropriate for GFP and RFP. Magnification is 200×. (C) P19 GFPhp.1 cells were transfected with pEGFP and 0, 0.5, or 1 μg of Dicer or firefly dsRNA. Fluorescence micrographs were taken at 48 h posttransfection and are superimposed with bright field images to reveal non-GFP expressing cells. Magnification is 100×. (D) In vitro and in vivo processing of dsRNA in P19 cells. In vitro Dicer assays were performed on S2 cells and three independently prepared P19 extracts by using 32P-labeled dsRNA (30°C for 30 min). A Northern blot of RNA extracted from control and GFPhp.1 P19 cells shows the production of ≈22mer RNA species in hairpin-expressing cells but not in control cells. Blots were probed with a 32P-labeled “sense” GFP transcript.
In Drosophila S2 cells and C. elegans (18, 27–30), RNAi is initiated by the Dicer enzyme, which processes dsRNA into ≈22-nt siRNAs (18). In both, S2 cells and C. elegans experiments by using dsRNA to target Dicer suppress the RNAi response (18, 27, 29). Whether Dicer plays a central role in hairpin-induced gene silencing in P19 cells was tested by transfecting P19 cells stably transfected with GFP hairpin constructs with mouse Dicer dsRNA (see Materials and Methods). Treatment with Dicer dsRNA, but not control dsRNA, resulted in derepression of GFP (Fig. 5C).
dsRNA Induces Posttranscriptional Silencing.
A key feature of RNAi is that it exerts its effect at the posttranscriptional level by destruction of targeted mRNAs (reviewed in ref. 4). To test whether dsRNAs induced silencing in mouse cells via posttranscriptional mechanisms, we used an assay identical to that, used initially to characterize RNAi responses in Drosophila embryo extracts (25). We prepared lysates from P19 EC cells that were competent for in vitro translation of capped mRNAs corresponding to Renilla and firefly luciferase. Addition of nonspecific dsRNAs to these extracts had no substantial effect on either the absolute amount of luciferase expression or on the ratio of firefly to Renilla luciferase (Fig. 6). In contrast, addition of dsRNA homologous to the firefly luciferase induced a dramatic and dose-dependent suppression of activity. Addition of RNA corresponding to only the antisense strand of the dsRNA had little effect, comparable to a nonspecific dsRNA control, and pretreatment of the dsRNA silencing trigger with RNase III greatly reduced its potential to induce silencing in vitro. A second hallmark of RNAi is the production of small, ≈22-nt siRNAs, which determine the specificity of silencing. We found that such RNA species were generated from dsRNA in P19 cell extracts (Fig. 5D, in vitro), indicative of the presence of a mouse Dicer activity. These species were also produced in cells that stably express GFP hairpin RNAs (Fig. 5D, in vivo). Considered together, the posttranscriptional nature of dsRNA-induced silencing, the association of silencing with the production of ≈22-nt siRNAs, and the dependence of this response on Dicer, a key player in the RNAi pathway, strongly suggests that dsRNA suppresses gene expression in murine cells via a conventional RNAi mechanism.
Discussion
The discovery that dsRNA could induce gene silencing in organisms as diverse as plants and parasitic protozoans has raised the possibility that RNAi might be a nearly universal mechanism of gene silencing. This notion has been supported by the identification of homologs of proteins that participate in the silencing process in virtually all genomes examined to date, with the exception of Saccharomyces cerevisiae (reviewed in ref. 4). The first indications that this response might also extend to mammals came from the observation that injection of dsRNAs into early mouse embryos induced sequence-specific silencing (23, 24). Recent work by Tuschl and colleagues (5) had shown that siRNAs can induce silencing in numerous mammalian cell lines, presumably by entering the RNAi pathway. However, both in mouse embryos and previous mammalian cell culture studies, silencing was transient.
As an extension of these pioneering studies, we have demonstrated that dsRNA can induce potent and specific gene silencing in mouse embryonic cell lines. Specifically, we have shown that silencing can be induced by long dsRNAs in mouse embryonal carcinoma cell lines, in normal mouse embryonic stem cells, and in some mouse somatic cells. There are several indications that this phenomenon might be mechanistically related to RNA interference pathways that have been characterized in plants, C. elegans, and Drosophila. First, induction of silencing requires dsRNA. Second, in vitro studies suggest that silencing occurs at the posttranscriptional level. Third, silencing is correlated with the appearance of ≈22-nt siRNAs homologous to the gene that is being suppressed. However, final placement of the phenomenon reported here within the pantheon of dsRNA-induced silencing mechanisms will require a characterization of the protein and/or ribonucleoprotein machinery, which enforces suppression. A significant step toward this goal has been taken by the demonstration that Dicer is required for dsRNA-induced silencing in P19 cells.
We have demonstrated that stable, sequence-specific silencing can be induced by enforcing endogenous expression of RNA hairpins. The ability to create permanent cell lines with a desired loss-of-function phenotype extends the utility of RNAi as method for probing gene function in mammalian cells. This capability enables the production of large numbers of silenced cells for biochemical analysis and permits the evaluation of phenotypes over long time spans. However, perhaps the two most important ramifications of stable RNAi are the ability to harness this technology for unbiased, phenotype-based genetic selections and the possibility that stably silenced, embryonic cell lines might ultimately be used to reconstitute animals containing a specifically silenced locus.
Acknowledgments
We thank Scott Hammond and Emily Bernstein for critical reading of the manuscript. Large quantities of purified RNase III were a kind gift of Al Nicholson (Wayne State University). P.J.P. is an Arnold and Mabel Beckman Anderson Fellow of the Watson School of Biological Sciences and thanks Richard M. Paddison for academic support. A.A.C. is a George A. and Marjorie H. Anderson Fellow of the Watson School of Biological Sciences and is a predoctoral fellow of the Howard Hughes Medical Institute. G.J.H. is a Rita Allen Foundation scholar. This work was supported in part by National Institutes of Health Grant RO1-GM62534 and R21-CA91069 (to G.J.H.) and by a grant from Genetica, Inc. (Cambridge, MA).
Abbreviations
- dsRNA
double-stranded RNA
- RNAi
RNA interference
- siRNA
small interfering RNA
- EGFP
enhanced green fluorescent protein
References
- 1.Edgar R S, Leilausis I. Genetics. 1964;49:649–662. doi: 10.1093/genetics/49.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartwell L H, Culotti J, Pringle J R, Reid B J. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 3.Hartwell L H, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 4.Hammond S M, Caudy A A, Hannon G J. Nat Rev Genet. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton A J, Baulcombe D C. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 7.Hammond S M, Bernstein E, Beach D, Hannon G J. Nature (London) 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 8.Zamore P D, Tuschl T, Sharp P A, Bartel D P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 9.Fraser A G, Kamath R S, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Nature (London) 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 10.Gonczy P, Echeverri G, Oegema K, Coulson A, Jones S J, Copley R R, Duperon J, Oegema J, Brehm M, Cassin E, et al. Nature (London) 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 11.Kennerdell J R, Carthew R W. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 12.Clemens J C, Worby C A, Simonson-Leff N, Muda M, Maehama T, Hemmings B A, Dixon J E. Proc Natl Acad Sci USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennerdell J R, Carthew R W. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 14.Williams B R. Biochem Soc Trans. 1997;25:509–513. doi: 10.1042/bst0250509. [DOI] [PubMed] [Google Scholar]
- 15.Gil J, Esteban M. Apoptosis. 2000;5:107–114. doi: 10.1023/a:1009664109241. [DOI] [PubMed] [Google Scholar]
- 16.Clarke P A, Mathews M B. RNA. 1995;1:7–20. [PMC free article] [PubMed] [Google Scholar]
- 17.Baglioni C, Nilsen T W. Interferon. 1983;5:23–42. [PubMed] [Google Scholar]
- 18.Bernstein E, Caudy A A, Hammond S M, Hannon G J. Nature (London) 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 19.Smith N A, Singh S P, Wang M B, Stoutjesdijk P A, Green A G, Waterhouse P M. Nature (London) 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- 20.Tavernarakis N, Wang S L, Dorovkov M, Ryazanov A, Driscoll M. Nat Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- 21.Kalejta R F, Brideau A D, Banfield B W, Beavis A J. Exp Cell Res. 1999;248:322–328. doi: 10.1006/excr.1999.4427. [DOI] [PubMed] [Google Scholar]
- 22.Connelly J C, Leach D R. Genes Cells. 1996;1:285–291. doi: 10.1046/j.1365-2443.1996.23024.x. [DOI] [PubMed] [Google Scholar]
- 23.Wianny F, Zernicka-Goetz M. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- 24.Svoboda P, Stein P, Hayashi H, Schultz R M. Development (Cambridge, UK) 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- 25.Tuschl T, Zamore P D, Lehmann R, Bartel D P, Sharp P A. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawagishi-Kobayashi M, Cao C, Lu J, Ozato K, Dever T E. Virology. 2000;276:424–434. doi: 10.1006/viro.2000.0561. [DOI] [PubMed] [Google Scholar]
- 27.Grishok A, Pasquinelli A E, Conte D, Li N, Parrish S, Ha I, Baillie D L, Fire A, Ruvkun G, Mello C C. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 28.Hutvagner G, McLachlan J, Pasquinelli A E, Balint E, Tuschl T, Zamore P D. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 29.Ketting R F, Fischer S E, Bernstein E, Sijen T, Hannon G J, Plasterk R H. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knight S W, Bass B L. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]