Abstract
All eukaryotes contain centromere-specific histone H3 variants (CenH3s), which replace H3 in centromeric chromatin. We have previously documented the adaptive evolution of the Drosophila CenH3 (Cid) in comparisons of Drosophila melanogaster and Drosophila simulans, a divergence of ≈2.5 million years. We have proposed that rapidly changing centromeric DNA may be driving CenH3's altered DNA-binding specificity. Here, we compare Cid sequences from a phylogenetically broader group of Drosophila species to suggest that Cid has been evolving adaptively for at least 25 million years. Our analysis also reveals conserved blocks not only in the histone-fold domain but also in the N-terminal tail. In several lineages, the N-terminal tail of Cid is characterized by subgroup-specific oligopeptide expansions. These expansions resemble minor groove DNA binding motifs found in various histone tails. Remarkably, similar oligopeptides are also found in N-terminal tails of human and mouse CenH3 (Cenp-A). The recurrent evolution of these motifs in CenH3 suggests a packaging function for the N-terminal tail, which results in a unique chromatin organization at the primary constriction, the cytological marker of centromeres.
Centromeres are the chromosomal loci responsible for poleward movement at meiosis and mitosis, and are essential for the faithful segregation of genetic information. Despite their crucial role in the cell cycle, relatively little is known about how most centromeres are organized at the molecular level. In complex eukaryotes, centromeres contain large arrays of tandemly repeated satellite sequences, yet their inheritance appears to be largely sequence-independent (reviewed in refs. 1–3). Instead, the presence of specialized centromeric histones is thought to be a key determinant of centromere identity and maintenance. Centromeric histones (CenH3s) belong to the histone H3 family of proteins, based on homology in the histone fold domain, and can replace H3 in nucleosomes (4, 5). However, centromeric chromatin is distinct from the rest of the genome, as demonstrated by nuclease accessibility studies in both budding and fission yeast (6–8), as well as by topoisomerase II cleavage patterns of human centromeres (9). Instead of the regular nucleosomal arrays observed in surrounding heterochromatin, centromeric DNA appears uniformly accessible to nucleases. These findings have led to suggestions that centromeric nucleosomes are randomly arranged (6, 7). A better understanding of this atypical chromatin might be gained by a closer examination of CenH3 evolution.
The structural organization of the C-terminal histone fold domain of CenH3 is believed to be very similar to that of H3 itself, based on greater than 40% amino acid identity and analysis of reconstituted nucleosomes (2, 4). However, this domain evolves more rapidly in CenH3s than in canonical H3s. In addition, whereas H3 has a conserved 45-aa N-terminal tail, those of CenH3s vary in size from 20 to 200 aa, and cannot be aligned across different eukaryotic lineages (2).
This dichotomy in evolutionary rates between CenH3s and H3s is believed to have two causes. First, whereas the CenH3s are only required to interact with centromeric DNA, conventional H3 needs to interact with, and mediate the expression of, the rest of the genome (10). This requirement results in the latter class being one of the most evolutionarily constrained classes of eukaryotic proteins. Second, CenH3s are evolving adaptively, presumably in concert with centromeres, which consist of the most rapidly evolving DNA in the genome.
Chromosomes (and their centromeres) may compete at meiosis I for inclusion into the oocyte (11). A centromeric satellite expansion could bias centromeric strength, leading to preferred inclusion in the oocyte, but also to increased nondisjunction. A subsequent alteration of CenH3's DNA-binding preferences could restore parity among different chromosomes (2), thereby alleviating the deleterious consequences of satellite drive. Successive rounds of satellite and CenH3 drive are detected as an excess of replacement changes in CenH3. Adaptive evolution of CenH3 has been documented in the histone fold domain of Drosophila (10). The N-terminal tails of CenH3 in both Drosophila and Arabidopsis are also subject to adaptive evolution, consistent with their proposed role in contacting centromeric DNA (10, 12). In addition, deletion studies in Saccharomyces cerevisiae have revealed essential protein–protein interaction sites encoded within the N-terminal tail of CenH3 (13). Although sequence divergence precludes aligning the N-terminal tails of CenH3s from different lineages, a comprehensive study of the molecular evolution within a particular lineage of CenH3s could help elucidate function for this important feature of centromeric chromatin by revealing the architecture of evolutionary constraint.
To this end, we present a detailed analysis of the evolution of the Drosophila centromeric histone (Cid) in the melanogaster group of species. Comparison of Cid genes across this 25-million-year divergence reveal highly conserved blocks in the histone fold domain, as expected, with the DNA-binding Loop 1 region (14) evolving most rapidly. This analysis also revealed the unexpected presence of oligopeptide repeats in the N-terminal tail of at least three different lineages of Drosophila Cids. The occurrence of these expansions in some vertebrate CenH3s and their similarity to previously documented peptides that interact with the minor groove of DNA suggest an attractive model for the unique chromatin structure present at centromeres.
Methods
Drosophila species (Table 1) were obtained from the Drosophila Species Center (presently at Tucson, AZ) except for Drosophila trilutea (kind gift of Dr. M.T. Kimura, Hokkaido University, Japan). Genomic DNA was extracted from all flies and PCR was carried out to amplify the Cid gene and flanking regions as described in ref. 10. For more divergent species, additional primers were designed to internal regions of Cid that are relatively conserved, 5′-CGGCAGCTTGGGTATCAGTGT-3′ (CidmidR2), 5′-GGCCATCAATGCCATGTCNCGNACNTC-3′ (degenerate primer H3degR1), and 5′-CTGCCGTTCTCGCGTCTNGTNCGNGA-3′ (degenerate primer H3degF1). The internal Cid-specific primers were used to amplify and sequence only an internal piece of Cid, and species-specific primers were designed to amplify the entire gene. PCR products were cloned and sequenced using BigDye sequencing (Applied Biosystems). At least three independent clones were sequenced on both strands for each species. All sequences have been deposited in GenBank (accession nos. in Table 1).
Table 1.
Species used in the present study
| Drosophila species | Species group | Stock center ID | GenBank accession nos. |
|---|---|---|---|
| melanogaster | melanogaster | ref. 3 | AF259371 |
| simulans | melanogaster | ref. 3 | AF321923 |
| sechellia | melanogaster | 14021-0248.0 | AF321925 |
| mauritiana | melanogaster | 14021-0241.0 | AF321924 |
| yakuba | melanogaster | 14021-0261.0 | AF435454 |
| teissieri | melanogaster | 14021-0257.0 | AF321926 |
| orena | melanogaster | 14021-0245.0 | AF435456 |
| erecta | melanogaster | 14021-0224.0 | AF435455 |
| takahashii | takahashii | 14022-0311.0 | AF435460 |
| trilutea | takahashii | Dr. M.T. Kimura | AF435457 |
| prostipennis | takahashii | 14022-0291.0 | AF435354 |
| paralutea | takahashii | 14022-0281.0 | AF435458 |
| lutescens | takahashii | 14022-0271.0 | AF435459 |
| lucipennis | suzukii | 14023-0331.0 | AF435356 |
| mimetica | suzukii | 14023-0341.0 | AF435461 |
| rajasekhari | suzukii | 14023-0361.0 | AF435355 |
| ananassae | ananassae | 14024-0371.0 | AF435466 |
| bipectinata | ananassae | 14024-0381.0* | AF435462 |
| malerkotliana | ananassae | 14024-0391.0 | AF435463 |
| pseudoananassae | ananassae | 14024-0411.0 | AF435465 |
| parabipectinata | ananassae | 14024-0401.0 | AF435464 |
| pseudoobscura | pseudoobscura | 14011-0121.0 | AF435467 |
In addition, strains 0381.1, 0381.2, 0381.3, and 0381.4 were partially characterized for repeat expansions.
Multiple alignment of Cid sequences was carried out using CLUSTAL X (15). Phylogenetic trees were created using the neighbor-joining program (16). Bootstrap trials were conducted using the PAUP* package of programs, Version 4.0b6 (17). In case of the repeat expansions in the takahashii and ananassae subgroups, oligopeptides from all species were aligned, and a position-specific scoring matrix (PSSM) created using BLOCKMAKER (http://blocks.fhcrc.org). The PSSMs were displayed as Logos (18), a graphical representation of aligned sequences where at each position the size of each letter is proportional to the frequency of that particular residue in that position and the total height of all of the letters in the position is proportional to the conservation (information content) of the position. Letters in the logo are colored according to the physical and chemical characteristics of the amino acid residues they specify. PSSMs generated were also used to search both the nonredundant database and a collection of known centromeric histones by using the Motif Alignment Search Tool (MAST, ref. 19). The CenH3s database was created by collating information from the various sequencing projects underway. PSSMs generated were also compared with each other by using the LAMA search tool (20), which reports significance of matches by using expected number based on searching 5,000 blocks and Z-scores (number of standard deviations away from the mean match to a set of shuffled blocks).
To detect natural selection on individual codon positions, we used the ADAPTSITE package of programs (ref. 21, http://mep.bio.psu.edu/adaptivevol.html). Briefly, the alignment and phylogenetic tree are used to infer ancestral codons at all interior nodes of the tree. Next, the total number of synonymous and nonsynonymous substitutions are calculated for each codon and a global average is computed. Adaptive evolution is inferred when the rate of nonsynonymous substitutions per nonsynonymous site (rN) exceeds that of synonymous substitutions per synonymous site (rS) at a significant level. Conversely, if rS < rN, purifying selection is inferred. The 9-aa expansions in both the takahashii and ananassae groups were examined for evidence of natural selection. We used adaptsite-d (Poisson) to calculate rN and rS followed by adaptsite-t to evaluate statistical significance.
PDB structures of the nucleosome core particle (1AOI, ref. 14) were displayed using the SWISSPDB VIEWER (Version 3.7b2, http://www.expasy.ch/spdbv).
Results
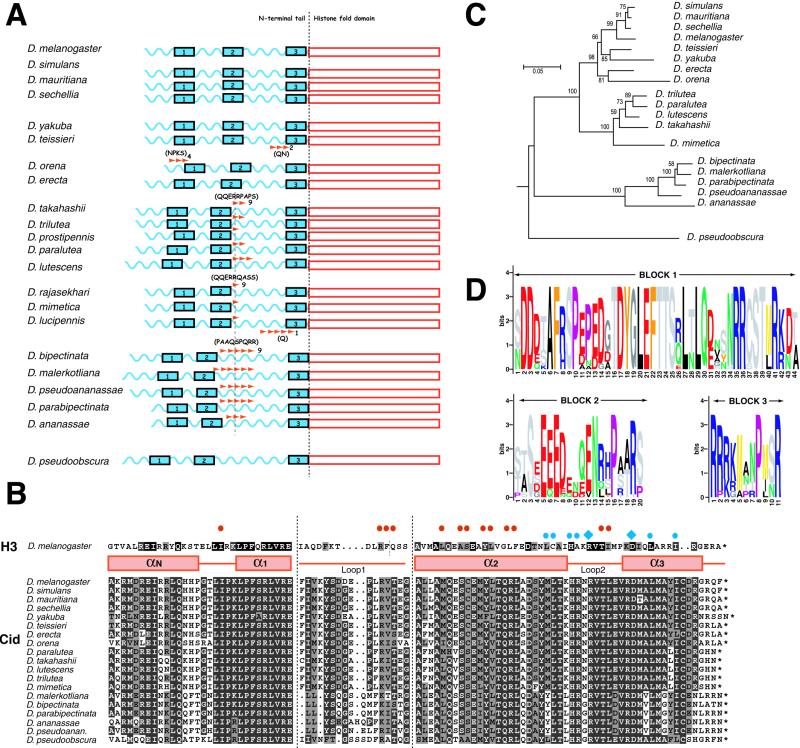
To amplify a phylogenetic range of Cid sequences (Table 1), we used a PCR strategy using internal primers in conjunction with primers to upstream and downstream genes (10). In all 22 species studied, the Cid gene lacks introns and varies from 639 to 879 nucleotides in length. Most of the length variation in Cid can be attributed to the N-terminal tail, whereas the histone fold domain is nearly constant in length (schematized in Fig. 1A).
Figure 1.
Phylogenetic conservation of Cid, the Drosophila CenH3, in the melanogaster group. (A) Schematic representation of Cid from different Drosophila species. The N-terminal tail is shown as a wavy blue line and the histone fold domain in red. The relative locations of three conserved sequence blocks in the tail are indicated as blue boxes (not drawn to scale). Each oligopeptide expansion is represented by red arrowheads; the amino acid repeat length is indicated by a number following the expansion and the number of iterations are indicated by the number of arrowheads. In addition, the consensus sequence of the repeats is shown in parentheses. (B) A multiple alignment of the histone fold domain of Drosophila Cids and D. melanogaster H3. Black dots represent gaps in the alignment. The alignment has been shaded to a 75% consensus by using MACBOXSHADE, with identical and similar residues shown in black and gray, respectively. Secondary structure assignments are from ref. 14 with the exception of Loop 1, which has been redefined to encompass the flanking variable sequence (between the dotted vertical lines). For instance, because of insertions/deletions, it is unclear exactly where the α1 helix ends and the α2 helix begins within Loop 1 of Cid. H3 contacts with itself, another H3 molecule and H4 are indicated by blue diamonds, blue dots, and red dots, respectively (14). (C) Cid gene phylogeny. Bootstrap support (percentage of 1,000 trials) is shown next to each node. (D) The three conserved blocks in Cid's N-terminal tail in Logos format (Methods).
We present a multiple alignment of the histone fold domain from various Drosophila Cids along with the Drosophila H3 sequence for comparison (Fig. 1B). Most residues involved in mediating the histone–histone interactions of H3 (14, 22) are well conserved in Cid, consistent with their structural role. In contrast, the Loop 1 region of Cid, which is predicted to form an extensive DNA-interaction domain with Loop 2 of H4, is more variable (Fig. 1B). All CenH3s have a longer Loop 1 region, consistent with its predicted role in conferring some degree of DNA-binding specificity (23). We have previously shown that the two replacements fixed between Drosophila melanogaster and Drosophila simulans in Loop 1 have been subject to adaptive evolution (10). In the present analysis, comparison of pairs of closely related species indicate additional positions in Loop 1 that have undergone frequent episodes of amino acid replacements. For instance, two replacements separate Drosophila yakuba from Drosophila teissieri [GAT GAA GCA (amino acids DEA) versus GAT GGA GAA (DGE), respectively], whereas four replacements separate Drosophila erecta from Drosophila orena [TTG AGG GTC TCC GAG GGC (LRVSEG) versus TTG AAG ATC AGC GTG GCA (LKISVA), respectively] in their Loop 1 regions (Fig. 1B). The adaptive evolution that characterizes Loop 1 may be indicative of changing DNA specificity (2, 10). In addition to regions subject to adaptive evolution, our analysis also highlights residues in Loop 1 that are well conserved and likely play a structural role in Cid function instead of conferring DNA specificity.
Based on the structure of the nucleosome (14, 22), there are additional DNA-interaction sites of structural significance on the H3 molecule that are well conserved in Cid. From the multiple alignment, we find that both the N-terminal helix αN and the carboxy-terminus of the protein are also evolving rapidly. α N is predicted to make specific contacts with the DNA gyres as the N-terminal tail of H3 exits the nucleosome (14, 22) and like Loop 1, this region may also confer some discrimination in DNA binding.
Based on a multiple alignment of the nucleotide sequences of the histone fold domain (data not shown), we can reconstruct the phylogeny of the Cid gene in the melanogaster group of species (Fig. 1C). As expected for an essential single-copy gene, the phylogeny of Cid is congruent with previous phylogenies of this group of species (ref. 24, for example). The divergence between the melanogaster species group and the ananassae species groups is estimated as at least 12.7 million years (based on paraphyly of ananassae, montium, and melanogaster species groups), whereas that between the obscura and melanogaster groups is estimated as 24 million years (25). Despite a total alignment length of only 300 nucleotides, we observe a high resolution in the phylogeny. Because eukaryotes are presumed to have one characteristic CenH3, it might serve as a universal phylogenetic marker in groups that have been traditionally hard to resolve.
In contrast to the histone fold domain, only few segments of the N-terminal tail are conserved. We identified three stretches of conserved protein sequences which we have termed Blocks 1–3 respectively (Figs. 1 A and D). Block 1 is 44 aa long and occurs close to the N-terminal end of the protein. We have shown the conservation of Block 1 in Logos format in Fig. 1D, where the “tallest” residues are invariant across all species sampled by our study. Block 1 is unique in that we were unable to find any other significant hits in the nr-database when using a MAST search. Block 2 is 20 aa long and occurs in the middle of the N-terminal tail. It primarily consists of a stretch of acidic residues flanked by a pair of well conserved serines. We could find no homologous sequences to Block 2 in the database other than those in very acidic proteins. Block 3 is 11 aa long and immediately abuts the histone fold domain. It is characterized by a basic region as well as invariant proline and arginine residues. The position of the third block is significant because the N-terminal tail of histone H3 is predicted to exit the nucleosome through a minor groove channel precisely at this junction (14). Block 3 resembles a putative nuclear localization signal encoded by four consecutive basic residues (26). There is precedent for some histones relying on an NLS-dependent pathway of nuclear import (27), but this has yet to be demonstrated for any CenH3.
Most of the length polymorphism in different Cid genes can be directly traced to oligopeptide repeat expansions in the N-terminal tail (Fig. 1A). These oligopeptides vary both in repeat lengths as well as the number of iterations. Thus, Drosophila lucipennis has a pentameric glutamine (Q) expansion, whereas D. teissieri and D. orena have three repeats of (QN) and (NPKS), respectively (Fig. 1A). The diverse location of these expansions and their absence in closely related species attest to their independent evolutionary origins. In the lineages leading to the takahashii, suzukii and ananassae species groups (Table 1), the expansions occur in the same location (dashed vertical line, Fig. 1A) and are each 9 aa residues long. In the takahashii and suzukii sister groups, copy number varies from 1 to 3, whereas in the ananassae group, it varies from 3 to 6 copies (Fig. 1A).
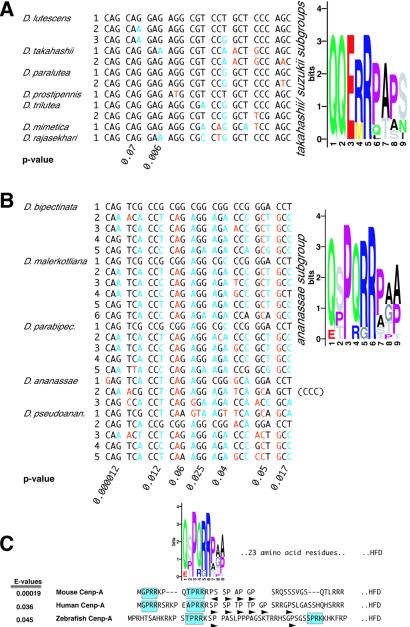
To investigate the evolutionary significance of the oligopeptide repeat expansions in Cid's N-terminal tail, we aligned all instances of the repeat expansion in the takahashii/suzukii and ananassae species groups (Fig. 2 A and B). From the multiple synonymous (blue) and replacement (red) substitutions, it is evident that the repeats are ancient and have not been subject to concerted evolution (which would lead to low nucleotide diversity). Despite their age, the amino acid residues encoded at many of the nine positions are subject to purifying selection (P values in Fig. 2 A and B). Indeed, in comparisons of these repeats within strains of Drosophila bipectinata, we find six synonymous and no replacement polymorphisms. The number of iterations of the repeat is not fixed in a species. For example, three strains of D. bipectinata have five copies of each repeat, whereas two strains have six. The Logo of the nine-residue stretch clearly highlights conserved and rapidly evolving residues. The two consensus oligopeptides from takahashii and ananassae groups are similar to each other in the last four positions (Z score: 4.2; 5.7 expected hits in a LAMA search of 5,000 blocks).
Figure 2.
Molecular evolution of oligopeptide expansions in CenH3s. (A) Multiple alignment of the oligopeptide expansions in the pair of takahashii/suzukii species groups. All synonymous and replacement substitutions are shown in blue and orange relative to the first line of the alignment. Eight codon positions had substitutions that could be evaluated for natural selection by using Adaptsite; of these, seven positions had rn < rS (2 significant, P values shown), whereas codon 4 has rn > rS, although only marginally so. Also presented is a Logo of the repeat consensus, which clearly highlights the conservation of the first five codons relative to the last four. (B) Expansions in the ananassae species group. All positions have rn < rS (7 significant, P values shown), indicating a stringent purifying selection. This conservation is confirmed by the Logo of the ananassae consensus shown in C. (C) Match of ananassae consensus to vertebrate Cenp-As. E-values reported are from a search of about 15 putative CenH3s. The entire N-terminal tails of the human, mouse, and zebrafish Cenp-As (Danio rerio Cenp-A, accession nos. BF156113, BI877040, and BF158223; Washington Univ. St. Louis Zebrafish EST Project) are shown upstream of the histone fold domain. In addition to the ananassae consensus, we could identify matches to the SPKK motifs (blue boxes) as well as short dipeptide motifs that include a proline at every alternate site (dark arrowheads).
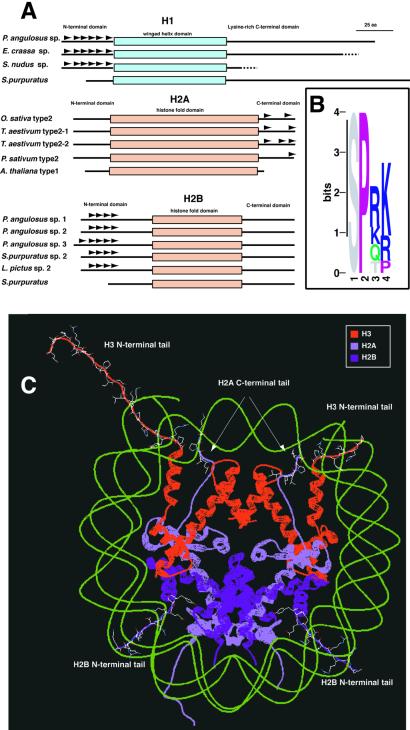
The first five positions of the ananassae consensus are similar to previously defined four-residue-long SPKK motifs (Z score: 3.4; 20 expected hits in a LAMA search of 5,000 blocks), known to mediate histone interactions with linker DNA in the minor groove. SPKK motifs have been found in the N-terminal tails of sea urchin sperm histones H1 and H2B, as well as the C-terminal tails of a class of angiosperm histone H2As (Fig. 3A, refs. 28–30). A Logo consensus of these motifs is shown in Fig. 3B. SPKK motifs are also found embedded in larger repeats within the carboxy-terminal tails of linker histone H1s (not shown; ref. 31).
Figure 3.
SPKK motifs in histone tails. (A) Schematics of sea urchin sperm H1 and H2B, and angiosperm H2A histones. Representative histones (28–30) are shown from sea urchin (sperm): Parechinus angulosus, Echinolampas crassa, Strongylocentrotus nudus, Strongylocentrotus purpuratus, and Lytechinus pictus; and angiosperms: Oryza sativa, Triticum aestivum, Pisum sativum, and Arabidopsis thaliana. Each instance of an SPKK motif is shown with a dark arrowhead. For comparison, a histone of each type lacking these motifs is also presented. (B) Logo of a multiple alignment of all SPKK motifs shown in A. (C) A schematic representation of the nucleosome core structure (14) highlighting the tails of H2A, H2B, and (Cen)H3 where SPKK expansions have been found and their proximity to the minor groove of the DNA wrapped around the histone octamer. Note that the two molecules of H3 have different lengths of N-terminal tails in the structure. For clarity, the two molecules of histone H4 are omitted.
The SPKK class of minor groove binding motifs includes substantial variation at the primary sequence level. The structure of SPKK bound to DNA has not been solved, but NMR and structural modeling suggest that SPKK forms a turn stabilized by one or two hydrogen bonds (29, 32). The structure is called an Asx turn if the hydrogen bond is between the OH or CO side chain of the ith amino acid (S, T, D, or N) and the main chain NH of the i+2 amino acid (A or a hydrophilic amino acid; e.g., K,R,E,T). In a beta turn there is a hydrogen bond between the main chain CO of the ith amino acid and the main chain NH of i+3. The only strict requirement is a P in the i+1 position. A variety of SPKK motif-containing peptides have been shown to bind in the minor groove of DNA (29, 33, 34), with each turn binding 2 bp. Given the inherent limitations in comparing four residue motifs and the variety of SPKK motifs that can bind in the minor groove (only a subset of which is represented in Fig. 3B), the similarity of the ananassae oligonucleotide consensus to the SPKK motif is compelling.
Using the consensus created from the ananassae group, we used the MAST program (20) to search the nonredundant database of proteins. Surprisingly, the top hit found was the mouse CenH3, the Cenp-A protein (E-value = 1). Given that our search query was only nine amino acids long and that the match to Cenp-A was in the N-terminal tail, we found this match especially noteworthy. In addition to mouse, we could find matches to the ananassae group consensus in zebrafish and human Cenp-A N-terminal tails at approximately similar locations upstream of the histone fold domain. We confirmed these matches by a MAST search of a database consisting of 15 putative CenH3s obtained from various nucleotide databases (E-values in Fig. 2C). The ananassae consensus motifs were not found in CenH3s from fungi, nematodes, plants, or even D. melanogaster and Drosophila pseudoobscura. Therefore, the ananassae-vertebrate sequence similarity likely represents an example of evolutionary convergence. In addition to the ananassae consensus repeats, the three Cenp-A sequences have an additional SPKK motif (GPRR in human and mouse Cenp-A) as well as recurring dipeptide motifs that include a proline at every other position (GP, SP, etc.).
Discussion
Centromeric chromatin is distinct from bulk chromatin, most conspicuously by the presence of CenH3. CenH3 is the only protein that is constitutively present at the centromere of every eukaryote examined so far (2). A specialized chromatin structure at the centromere is suggested by nuclease digestion experiments (6–8) and may be important for the epigenetic maintenance of the centromere through multiple cell divisions. In addition, this chromatin probably plays a key role in assembling the kinetochore at meiosis and mitosis. Thus, CenH3 could be expected to both interact with the underlying centromeric DNA, as well as interact with other proteins (including other CenH3 molecules) to provide the foundation for the kinetochore (35).
In S. cerevisiae, deletion analysis has shown that the N-terminal tail of CenH3 (Cse4) includes a 33-amino acid stretch (END domain) that is essential for function (13). By two-hybrid and dosage suppression of temperature-sensitive alleles, the END domain has been shown to interact with kinetochore components (13). Using an evolutionary approach, we have identified three conserved blocks in the N-terminal tail of Drosophila Cid. By analogy with END in Cse4, we suggest that at least two of these three Cid blocks may also mediate interactions with other centromere–kinetochore determinants (Block 3 in Fig. 1D may interact with DNA). Unlike the single nucleosome centromere of S. cerevisiae, metazoan centromeres are hundreds of kilobases in length. Thus, some of the interactions involving Cid's N-terminal tail may occur between two successive Cid molecules in a centromeric nucleosome array. These blocks can now be used in protein interaction screens to elucidate the next level of centromere organization.
In two genera with complex centromeres, CenH3s appear to be undergoing adaptive evolution (10, 12). Because female meiosis in plants and most metazoans is asymmetric, centromeres have the opportunity to compete for inclusion into the egg (11). Left unchecked, this competition could lead to runaway satellite expansions and increased nondisjunction as centromeres of differing strengths undergo meiosis (2). By altering its DNA-binding specificity, CenH3 could suppress such a drive process. This model predicts that the youngest satellites found in centromeric regions would represent the extant centromere, a prediction that has recently been confirmed by high-resolution mapping of the human X centromere (36). Under this model, fungal CenH3s are not expected to evolve adaptively both because they are not subject to meiotic asymmetries and because they rely on other factors for their centromeric localization (8, 37).
Adaptive evolution of Cid has been mapped to both the Loop 1 region of the histone fold domain as well as to multiple locations of the N-terminal tail in comparisons of D. melanogaster and D. simulans (10). In addition, adaptive evolution was detected for the N-terminal tail in Arabidopsis CenH3 (12). We have proposed that altered DNA-binding specificity drives CenH3's adaptive evolution. Because this adaptation has occurred in multiple segments of Cid's N-terminal tail, the N-terminal tail must make extensive contacts with the linker DNA in centromeric chromatin.
One special instance of a likely CenH3-DNA interaction is evident in the case of SPKK-containing oligopeptides we find in the ananassae species' Cid and vertebrate Cenp-A. Even in the absence of SPKK motifs, conventional histone tails interact extensively with the linker DNA (38). When found in H2A C-terminal or H2B N-terminal histone tails, SPKK-containing motifs confer an additional protection of 16 bp of linker DNA in reconstitution experiments (30, 39). The N-terminal tails of H2B and H3 are perfectly positioned to interact with the minor groove of DNA as they exit the nucleosome (Fig. 3C) in minor groove channels (14). Similarly, while the winged helix domain of linker histones is located at the nucleosome dyad, the positively charged N- and C-terminal tails interact extensively with the linker DNA (40). This interaction is weaker in rat testis H1 (H1t), which lacks SPKK motifs, compared with somatic H1s, which have them (41). Thus, the presence of minor groove interactions mediated by CenH3's N-terminal tail might result in uniform nuclease accessibility of centromeric chromatin. A comparison of H3 and H1 tails is particularly relevant as the H1 binding site on the nucleosome is very close to where the N-terminal tails of H3s exit. We propose that the minor groove binding repeats in CenH3 N-terminal tails play a similar role to H1 tails by neutralizing phosphates in the linker DNA—i.e., they shield charges to allow superhelical bending and collapse into a higher order chromatin structure. This structure would be seen as the primary constriction, the cytological marker for centromere position. It will be interesting to determine whether centromeric chromatin is especially lacking in linker histones.
Two of the best characterized minor-groove binding motifs, SPKK and AT-hooks (42), show no sequence similarity to each other, suggesting that we have explored only a subset of potential minor-groove binding motifs. In addition to the SPKK-bearing motifs, we have found additional types of repeats both in Cenp-A as well as in the different lineages of Drosophila, such as the takahashii consensus (Fig. 2A) and NPKS expansions in D. orena (Fig. 1A), that may all carry out the same function—i.e., bind in the minor groove of centromeric DNA. These peptides, like SPKK, are predicted to have a strong preference for AT-rich DNA, and may only recognize architectural DNA features such as minor groove shape rather than specific bases.
In conclusion, our examination of evolutionary constraint in the Drosophila CenH3 lineage has revealed that the N-terminal tail comprises a combination of interaction sites. We have inferred putative protein interaction blocks that might lay the foundation for kinetochore formation, DNA binding regions subject to adaptive evolution, and nonspecific minor groove binding motifs. The resulting complex with DNA may form the universal cytological feature of centromeres, the primary constriction.
Acknowledgments
We thank the Drosophila Species Center for the various fly strains used, Jorja Henikoff for help with the Blocks, LAMA, MAST, and Adaptsite searches, and Judith O'Brien for technical assistance. We also thank Kami Ahmad, Jennifer Cooper, and Paul Talbert for stimulating discussions and for comments on the manuscript. We are supported by the Helen Hay Whitney Foundation (H.S.M.), the Damon Runyon Cancer Research Foundation (D.V.), and the Howard Hughes Medical Institute (S.H.).
Abbreviation
- CenH3
centromeric histone H3
Footnotes
References
- 1.Karpen G H, Allshire R C. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 2.Henikoff S, Ahmad K, Malik H S. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 3.Choo K H. Dev Cell. 2001;1:165–177. doi: 10.1016/s1534-5807(01)00028-4. [DOI] [PubMed] [Google Scholar]
- 4.Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Proc Natl Acad Sci USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. . (First Published June 6, 2000; 10.1073/pnas.130189697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer D K, O'Day K, Wener M H, Andrews B S, Margolis R L. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloom K S, Carbon J. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- 7.Polizzi C, Clarke L. J Cell Biol. 1991;12:191–201. doi: 10.1083/jcb.112.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi K, Chen E S, Yanagida M. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 9.Floridia G, Zatterale A, Zuffardi O, Tyler-Smith C. EMBO Rep. 2000;1:489–493. doi: 10.1093/embo-reports/kvd110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik H S, Henikoff S. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zwick M E, Salstrom J L, Langley C H. Genetics. 1999;152:1605–1614. doi: 10.1093/genetics/152.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talbert, P. B., Masuelli, R., Tyagi, A. P., Comai, L. & Henikoff, S. (2001) Plant Cell, in press. [DOI] [PMC free article] [PubMed]
- 13.Chen Y, Baker R E, Keith K C, Harris K, Stoler S, Fitzgerald-Hayes M. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 15.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 17.Swofford D L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2000. , Version 4. [Google Scholar]
- 18.Schneider T D, Stephens R M. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey T L, Gribskov M. J Comput Biol. 1998;5:211–221. doi: 10.1089/cmb.1998.5.211. [DOI] [PubMed] [Google Scholar]
- 20.Pietrokovski S. Nucleic Acids Res. 1996;24:3836–3845. doi: 10.1093/nar/24.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki Y, Gojobori T, Nei M. Bioinformatics. 2001;17:660–661. doi: 10.1093/bioinformatics/17.7.660. [DOI] [PubMed] [Google Scholar]
- 22.Harp J M, Hanson B L, Timm D E, Bunick G J. Acta Crystallogr D. 2000;56:1513–1534. doi: 10.1107/s0907444900011847. [DOI] [PubMed] [Google Scholar]
- 23.Shelby R D, Vafa O, Sullivan K F. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goto S G, Kimura M T. Mol Phylogenet Evol. 2001;18:404–422. doi: 10.1006/mpev.2000.0893. [DOI] [PubMed] [Google Scholar]
- 25.Russo C A, Takezaki N, Nei M. Mol Biol Evol. 1995;12:391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- 26.Gorlich D. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 27.Mosammaparast N, Jackson K R, Guo Y, Brame C J, Shabanowitz J, Hunt D F, Pemberton L F. J Cell Biol. 2001;153:251–262. doi: 10.1083/jcb.153.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Holt C, de Groot P, Schwager S, Brandt W F. In: Histone Genes. Stein G S, Stein J L, Marzluff W F, editors. New York: Wiley Interscience; 1984. pp. 65–105. [Google Scholar]
- 29.Suzuki M. EMBO J. 1989;8:797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindsey G G, Orgeig S, Thompson P, Davies N, Maeder D L. J Mol Biol. 1991;218:805–813. doi: 10.1016/0022-2836(91)90268-b. [DOI] [PubMed] [Google Scholar]
- 31.Churchill M E, Travers A A. Trends Biochem Sci. 1991;16:92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki M, Gerstein M, Johnson T. Protein Eng. 1993;6:565–574. doi: 10.1093/protein/6.6.565. [DOI] [PubMed] [Google Scholar]
- 33.Churchill M E, Suzuki M. EMBO J. 1989;8:4189–4195. doi: 10.1002/j.1460-2075.1989.tb08604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khadake J R, Rao M R. Biochemistry. 1997;36:1041–1051. doi: 10.1021/bi961617p. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan K F. Curr Opin Genet Dev. 2001;11:182–188. doi: 10.1016/s0959-437x(00)00177-5. [DOI] [PubMed] [Google Scholar]
- 36.Schueler M G, Higgins A W, Rudd M K, Gustashaw K, Willard H F. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- 37.Ortiz J, Stemmann O, Rank S, Lechner J. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angelov D, Vitolo J M, Mutskov V, Dimitrov S, Hayes J J. Proc Natl Acad Sci USA. 2001;98:6599–6604. doi: 10.1073/pnas.121171498. . (First Published May 29, 2001; 10.1073/pnas.121171498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindsey G G, Thompson P. J Biol Chem. 1992;267:14622–14628. [PubMed] [Google Scholar]
- 40.Zhou Y B, Gerchman S E, Ramakrishnan V, Travers A, Muyldermans S. Nature (London) 1998;395:402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- 41.Khadake J R, Markose E R, Rao M R. Ind J Biochem Biophys. 1994;31:335–338. [PubMed] [Google Scholar]
- 42.Aravind L, Landsman D. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]