Abstract
HemK, a universally conserved protein of unknown function, has high amino acid similarity with DNA-(adenine-N6) methyl transferases (MTases). A certain mutation in hemK gene rescues the photosensitive phenotype of a ferrochelatase-deficient (hemH) mutant in Escherichia coli. A hemK knockout strain of E. coli not only suffered severe growth defects, but also showed a global shift in gene expression to anaerobic respiration, as determined by microarray analysis, and this shift may lead to the abrogation of photosensitivity by reducing the oxidative stress. Suppressor mutations that abrogated the growth defects of the hemK knockout strain were isolated and shown to be caused by a threonine to alanine change at codon 246 of polypeptide chain release factor (RF) 2, indicating that hemK plays a role in translational termination. Consistent with such a role, the hemK knockout strain showed an enhanced rate of read-through of nonsense codons and induction of transfer-mRNA-mediated tagging of proteins within the cell. By analysis of the methylation of RF1 and RF2 in vivo and in vitro, we showed that HemK methylates RF1 and RF2 in vitro within the tryptic fragment containing the conserved GGQ motif, and that hemK is required for the methylation within the same fragment of, at least, RF1 in vivo. This is an example of a protein MTase containing the DNA MTase motif and also a protein-(glutamine-N5) MTase.
HemK was first identified in Escherichia coli as a suppressor of the light-sensitive phenotype of a protoporphyrin IX-accumulating mutant (hemH) (1), and genes that code for highly homologous proteins have been found not only in eubacteria but also in eukaryotes, such as yeast (GenPept accession no. AAA99648), Arabidopsis thaliana (GenPept accession no. AAD26417), and humans (GenPept accession no. AAD26417). Also, similar proteins but without the N′-terminal portion (a region of about 50 aa in the bacterial homologs) are present in both types of archaea, Euryarchaeota, such as Pyrococcus abyssi (GenPept accession no. CAB49354), and Crenarchaeota, such as Aeropyrum pernix (GenPept accession no. BAA79522).
In E. coli, hemK is a member of a multifunctional operon consisting of at least hemA, the product of which catalyses the conversion from glutamyl-tRNA to glutamate-1-semialdehyde at the first step of porphyrin biosynthesis, prfA, encoding polypeptide chain release factor (RF) 1, and hemK in that order. Based on the phenotype of the initial mutant, we proposed the hypothesis that HemK functions in some step of porphyrin biosynthesis. However, no direct evidence for such activity has been reported to date. In addition, whereas the tripartite operon structure is conserved only among some members of the γ subclass of proteobacteria (2, 3), the link between the last two genes, prfA and hemK, is more widely conserved in eubacteria (4, 5), implying a possible relationship of HemK to translation.
On the other hand, significant similarity is found between the primary structure of HemK homologs and those of S-adenosyl-l-methionine (SAM)-dependent methyl transferases (MTases). Especially, the DNA-(adenine-N6) MTase family, which methylates the exocyclic amino nitrogen attached to the C-6 position of adenine, show the highest similarity with the “HemK family” (6). Indeed all of the HemK homologs share a common signature with the “N6 adenine-specific DNA methylases” ([LIVMAC]-[LIVFYWA]-x-[DN]-P-P-[FYW], PROSITE accession no. PS00092). However, no DNA MTase activity of the HemK homologs has yet been reported.
In this study, we constructed a series of hemK knockouts of E. coli K12, found that they grew very slowly under all of the conditions examined, and used them to elucidate the cellular function and activity of hemK. By examining the knockouts and the suppressors that partially rescue their growth, we found that HemK catalyses the methylation of RF1 and RF2, and that hemK knockout causes defects in translational termination.
Materials and Methods
Bacterial Strains.
All bacterial strains used are derivatives of E. coli K-12 and are listed in Table 1.
Table 1.
Characteristics of bacterial strains
| Bacterial strain | Relevant feature(s) | Reference(s)* |
|---|---|---|
| LE392 | supE44 supF58 hsdR514 galK2 | 7 |
| galT22 metB1 trpR55 lacY1 | ||
| LK783 | LE392 ΔhemK∷Cm | |
| LLR201, HLR201 | Faster growing suppressor strains from LK783 | |
| CA293 | HfrC lacZoc659trpam8relA1 spoT1 | 8 and 9 |
| CK783 | CA293 ΔhemK∷Cm | |
| W3110 | IN(rrnD-rrnE)1 rph-1 | 9 |
| WK103 | W3110 ΔhemK∷Cm | |
| CA274 | HfrC lacZam56trpam8relA1 spoT1 | 9 |
| C600 | thr-1 leuB6 lacY1 supE44 rfbD1 | 9 |
| thi-1 mcrA1 fhuA21 cyn-1 | ||
Unless otherwise noted, the source was this study.
Plasmid and Phage Clones.
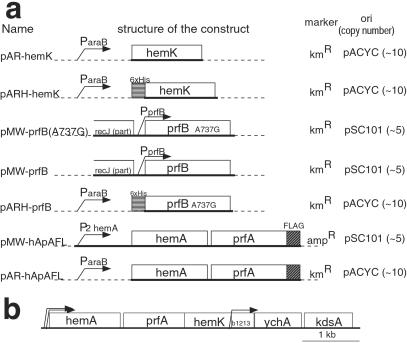
The structure of the key constructs used in this study and the original gene map around hemK are illustrated in Fig. 1. pMW118 and pMW218 (Nippon Gene, Tokyo) were used as low-copy-number vectors. To construct pMW-prfB or pMW-prfB(A737G), the 1.6-kb PvuII–EcoRI fragment containing the prfB gene from LK783 or LLR201 was inserted into the HincII and EcoRI sites of pMW218. For the controlled expression of genes, pAR3K vector (10), which harbors an arabinose-inducible araB promoter, was used. To construct pAR-hApAFL, a fragment from the initiation codon of hemA to the stop codon of prfA was amplified by PCR, and the FLAG tag coding sequence (GACTACAAGGACGATGACGATAAGTAA) was fused to its 3′ terminal. The resulting fragment was inserted into the NcoI and NarI sites of pAR3K. pMW-hApAFL was constructed by recloning the 3′ half of the hemA–prfAflag fragment from pAR-hApAFL into pMW118 in the opposite direction to the lacZα gene, and then inserting a 308-bp NheI–SalI fragment, harboring the hemA major promoter and the 5′ portion of hemA, to recreate the original structure of the hemA operon. To construct pAR3KH, the NcoI cloning site (CCATGG) of pAR3K was altered to produce the coding sequence of a 6×His tag followed by an AflIII site (CCATGAGAGGATCGCATCACCATCACCATCACATGT). pAR-hemK and pARH-hemK were constructed by amplifying the hemK coding sequence by PCR and inserting it into the NcoI or AflIII site of pAR3K or pAR3KH, respectively. The two proline codons (CCG) at amino acid positions 184 and 185 of hemK in pAR-hemK or pARH-hemK were changed to alanine codons (GCG) to produce pAR-hemKm or pARH-hemKm, respectively. pARH-prfB was constructed by amplifying the prfB coding sequence of LLR201 and inserting it into the AflIII site of pAR3KH. For the assay of translational read-through, derivatives of promoter probe vector pKK232-Z (11) were constructed. pKK232-Z(GGA) was constructed by inserting the trpA terminator and the lac promoter without an operator (11) in the SmaI site of pKK232-Z. The seventh codon of the lacZ (GGA) in pKK232-Z(GGA) was altered to each of the three nonsense codons to make pKK232-Z(UAA, UAG, or UGA). The region from the BamHI site following the lac promoter to the seventh codon of the lacZ in pKK232-Z(GGA) was deleted to make pKK232-Z(Δ). The accuracy of all of the constructions was confirmed by DNA sequencing.
Figure 1.
(a) Structures of the key constructs used in this study. Name, schematic structure of the construct, selection marker, and type of replicon (copy number per cell) is shown for each plasmid. Sequences derived from chromosomes are shown by thick lines and those from vectors by dashed lines. (b) The gene map around hemK in E. coli. The coding region of each gene (open boxes) and the locations of promoters (ref. 36, bent arrows) are shown. All genes are transcribed from left to right.
Media and Growth Conditions.
Bacteria were grown with constant aeration in LB medium (7) or on LB agar plates at 37°C unless otherwise indicated. Casamino acid broth (12), consisting of 5 g of vitamin assay casamino acid (Difco) and 5 g of NaCl per liter of water, supplemented with vitamin B1 (25 mg/liter) and tryptophan (40 mg/liter), was used for determining cell growth. Antibiotics and other compounds were supplemented at the following concentrations when needed: ampicillin, 50 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 25 μg/ml; and hemin (Sigma), 20 μg/ml.
Construction of ΔhemK Strains.
The SacII–KpnI fragment within the hemK coding region (codons 20–120) was replaced by the chloramphenicol resistance (Cmr) gene [1.1-kb AccII fragment from pHSG399 (Takara Shuzo, Kyoto)], retaining the internal promoter for downstream genes (see Fig. 1b), and the interrupted gene (ΔhemK∷Cmr) was introduced into JC7623 (recBC sbcB) by electroporation to disrupt the chromosomal hemK. The ΔhemK∷Cmr allele was introduced into other strains by P1-phage-mediated trunsduction (13), and PCR was performed to confirm the transduction.
Isolation of Suppressor Mutants from ΔhemK.
LK783 cells were mutagenized with 100 μg/ml N-methyl-N′-nitro-N-nitrosoguanidine as described (13) and plated onto LB agar plates. Colonies that grew at a faster rate were selected and purified.
Immunoblotting.
Proteins were separated by SDS/PAGE, blotted onto Hybond-ECL membranes (Amersham Pharmacia), and detected with mAbs against FLAG peptide (Anti-FLAG M2, Sigma-Aldrich) by using chemiluminescence techniques.
Isolation of 6×His- or FLAG-Tagged Proteins.
Unless otherwise noted, RF1-FLAG or 6×His-RF2 was purified from CK783 transformed with pAR-hApAFL or pARH-prfB, respectively. 6×His-HemK and its mutant were purified from JM109 transformed with pARH-hemK or pARH-hemKm, respectively. Cells were grown in 50 ml of LB broth at 37°C to about 0.6 OD600 units and induced by supplementation with 1 mM l-arabinose for 4 h. After the cells were harvested, 6×His- or FLAG-tagged protein was purified by using Ni-NTA agarose (Qiagen, Chatsworth, CA) or anti-FLAG M2 affinity gel (Sigma-Aldrich) according to the manufacturer's instructions, concentrated by using a Microcon YM-10 centrifugal filter (Millipore) and dissolved in 50 mM Tris⋅HCl (pH 8), 100 mM NaCl, 10 mM EDTA, and 50% glycerol.
Methylation Assay.
Methylation assays were performed by measuring the transfer of radioactivity from methyl-3H-labeled SAM ([3H]SAM) into trichloroacetic acid (TCA)-insoluble material. Substrate, purified HemK (or other protein) and 1 μCi of [3H]SAM (15 Ci/mmol; TRK236, Amersham Pharmacia) were mixed in 10 μl of reaction buffer (20 mM Tris⋅acetate/10 mM magnesium acetate/50 mM potassium acetate/1 mM DTT, pH 7.9) and incubated for 1 h at 37°C. The reaction product was washed by two cycles of TCA precipitation and rinsed with acetone. Finally, the radioactivity of the TCA-insoluble fraction was determined by liquid scintillation counter.
Matrix-Assisted Laser Desorption Ionization–Time of Flight (MALDI-TOF) MS Analysis.
A sample containing 10–50 μg of one of the RFs was resolved by SDS/PAGE (8% acrylamide) and stained by Coomassie brilliant blue, and the band for RF was isolated. Each protein sample within the gel slice was digested by using endoproteinase Lys-C and trypsin, extracted from the gel, and subjected to MALDI-TOF MS analysis with a Voyager DE-PRO (Perkin–Elmer Applied Biosystems) essentially as described (14).
Results
Characterization of the hemK Knockout Strains.
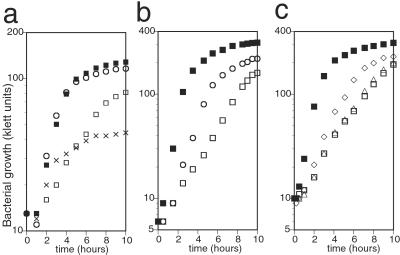
To elucidate the activity and cellular function of hemK, we constructed hemK knockout (ΔhemK) strains on several backgrounds. All of the strains showed a reduced growth rate, with a doubling time of 2–2.5 h at 37°C in LB broth, about four times longer than that of the respective parental strains. Because this deletion might directly affect the expression of upstream genes, hemA and prfA, in the same operon, or genes located downstream, b1213, ychA, and kdsA (see Fig. 1b), complementation by hemK was examined by introducing a plasmid, pAR-hemK, which express hemK under an inducible arabinose promoter. Growth of the transformant was restored thereby to the wild-type level, showing that the growth defect was caused by the loss of HemK (Fig. 2a). The level of HemK attained without inducer was sufficient for the complementation, and addition of excess arabinose (50 mg/liter) caused a negative effect on the growth rate. If hemK encodes an enzyme that acts in the porphyrin biosynthetic pathway, as proposed previously (1), supplying porphyrin in the medium should restore the growth defect. To test this, we introduced ΔhemK into a hemin-permeable background (15). Addition of hemin to the medium did not affect the growth of the resulting ΔhemK hemin-permeable strain (not shown), suggesting that the growth defect of ΔhemK is not primarily caused by a shortage of porphyrin. Next, we examined whether the DNA-(adenine-N6) MTase signature of HemK is related to the growth defect. Two proline codons of hemK, at codons 184 and 185 within the signature, were mutated to alanine codons in pAR-hemK, and the resulting plasmid, pAR-hemKm, was introduced into the ΔhemK strain. The mutated plasmid failed to complement the growth of the ΔhemK strain (data not shown), indicating a possible relationship between the growth defect in the hemK knockout cells and the putative MTase activity of HemK.
Figure 2.
Effect of hemK knockout and its suppressor mutation on bacterial growth. A fresh overnight culture of each strain was diluted to about 10 klett units (with filter no. 66) in 5 ml of medium and shaken at 37°C. The cell density was determined at intervals, and representative results of the experiments are shown. (a) The growth defect of the hemK knockout strain and complementation by hemK in an expression plasmid. CA293 (hemK+) and CK783 (ΔhemK) were transformed with pAR-hemK or its vector control, pAR3K. The growth of each transformant was examined in casamino acid broth. Essentially the same results were obtained in LB broth. Strains: ■, CA293 pAR3K; □, CK783 pAR3K; ○, CK783 pAR-hemK; x, CK783 pAR-hemK (for x, hemK was induced by adding 50 mg/liter arabinose to the medium at time 0). (b) The growth of the suppressor mutant was compared with that of the hemK+ and ΔhemK strains in LB broth. Strains: ■, LE392 (hemK+); □, LK783 (ΔhemK); ○, LLR201 (one of the suppressor mutants of ΔhemK). (c) The growth of ΔhemK strain transformed with wild-type or mutant prfB in a plasmid was compared with that of the hemK+ or ΔhemK strain in LB broth. Strains: ■, CA293 pMW218 (vector control); □, CK783 pMW218; ▵, CK783 pMW-prfB; ◊, CK783 pMW-prfB(A737G).
Isolation and Characterization of Suppressor Mutants.
Two suppressor mutant strains, LLR201 and HLR201, in which the growth defect was partially overcome, were obtained independently from the ΔhemK strain, LK783 (Fig. 2b). Plasmid libraries in pUC118 were constructed from EcoRI digests of the genomic DNA of each mutant and screened for clones that could rescue the growth deficiency. Three clones were isolated from each library and shown, by partial sequencing, to all contain an insert of about 3.2 kb from the same portion of the E. coli chromosome, at about 65.4 min. Subsequent subcloning revealed that a 1.6-kb PvuII–EcoRI fragment, including the 3′ half of recJ, the entire prfB, and the 5′ half of lysS, possessed the suppressor activity of each of these clones. The same 1.6-kb PvuII–EcoRI fragment was cloned from the parental strain (LK783), and the complete sequence of the fragment was compared with that of the mutant clones. A single mutation, adenine to guanine at position 737 of the prfB gene, resulting in substitution of threonine 246 of RF2 by alanine, was found in clones of both mutants and confirmed by direct nucleotide sequencing of PCR-amplified fragments from the chromosomal DNA. The sequence of the parental strain was no different from that of reference strain MG1655 in the database (16). Comparing the suppressor activity of the mutant gene on a low-copy-number plasmid with that of the wild-type gene showed that the T246A mutant exerted the suppressor effect, whereas the wild-type gene did not (Fig. 2c).
Effect of hemK Knockout on Translational Termination.
Because mutation T246A is known to enhance the efficiency of polypeptide chain termination by RF2 (17, 18), the phenotype of the suppressor mutant might reflect the defect of termination in ΔhemK cells. To test this, we measured the rate of read-through of nonsense codons at a given position in lacZ. A set of plasmids carrying lacZ with one of the three nonsense codons or an alanine codon (as a reference) at the seventh codon was introduced into hemK+ or ΔhemK cells, and the activity of β-galactosidase in each transformant was determined (Table 2). The rate of read-through, expressed as a percentage of the activity of the reference plasmid, at the UAG or UGA codon was enhanced in ΔhemK, showing that the function of RF1 or RF2, respectively, was affected. The rate of read-through at the UAA codon was not affected significantly, nor was the background rate (the rate of read-through of the Δ construct).
Table 2.
Rate of read-through at nonsense mutations in lacZ
| Strain/lacZ | Relative β-galactosidase
activity*
|
||||
|---|---|---|---|---|---|
| GGA | UAG | UAA | UGA | Δ | |
| hemK+ | 100 ± 3.7 | 0.35 ± 0.087 | 0.59 ± 0.10 | 0.65 ± 0.048 | 0.25 ± 0.091 |
| ΔhemK | 100 ± 1.5 | 1.5 ± 0.24 | 0.33 ± 0.0065 | 1.5 ± 0.10 | 0.12 ± 0.015 |
CA293 (hemK+) or CK783 (ΔhemK) was transformed with a set of pKK232-Z derivatives, in which GGA, UAG, UAA, or UGA was the seventh lacZ codon (shown in the lacZ row) or in which the segment from the 5′ nontranslated region to the seventh codon of lacZ was deleted (Δ, shown in the lacZ row), and grown on LB broth to 0.6–0.7 OD600. The β-galactosidase activity of the cells was assayed as described (13).
The average ± SD (of three experiments) β-galactosidase activity (Miller units) of each lacZ derivative is shown as the percentage of that of the reference gene (GGA) in the same strain.
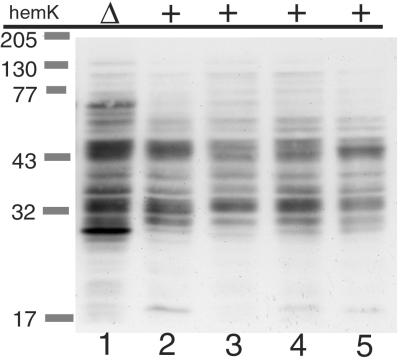
Next, we examined the induction of transfer-mRNA (tmRNA)-mediated tagging of proteins in the cell, which is triggered by the stalling or slowing down of the ribosome on mRNA (19–21). To detect endogeneous proteins tagged by tmRNA, a modified ssrA gene, which adds the FLAG peptide instead of the normal tmRNA tag peptide (unpublished data), was introduced to ΔssrA strains of various backgrounds, and the total cell extracts of the resulting strains were analyzed by immunoblotting (Fig. 3). Compared with the prototype strain, W3110 (Fig. 3, lane 2), prominent induction of several bands and a general increase in the level of tagged proteins were observed in the ΔhemK strain (Fig. 3, lane 1). No such marked alteration was observed in the other strains (Fig. 3, lanes 3–5).
Figure 3.
tmRNA-tagged proteins in ΔhemK and other cells were detected by immunoblot analysis. Chromosomal ssrA in each strain (lane 1, WK103; lane 2, W3110; lane 3, CA274; lane 4, LE392; lane 5, C600) was inactivated by transducing ΔssrA∷Km, and a plasmid encoding the FLAG-tagging ssrA variant was introduced. Then, cells grown in LB broth were harvested at OD600 of about 0.7, suspended in SDS sample buffer (200 μl/OD600-unit, and boiled for 5 min, and 10 μl of each cell extract was then analyzed by SDS/PAGE (12% acrylamide) and immunoblotting with anti-FLAG antibody.
Effect of hemK Knockout on the Expression and Modification of RF1.
One of the possible mechanisms by which this defect might affect translational termination in ΔhemK is an effect of HemK on the expression or activity of prfA/RF1, which is located next to hemK. We performed several experiments to test this hypothesis, but the results showed that the hemK knockout has little effect on the expression or cellular level of RF1, and the effect, if any, does not seem to account for the defect in ΔhemK (not shown).
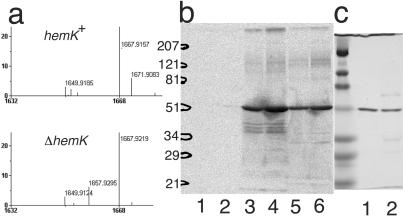
Recently, Dincbas-Renqvist et al. (18) reported the posttranslational modification (methylation) of RF2 at glutamine 252 within the highly conserved GGQ motif and suggested that the same modification occurs in RF1. To test this modification in the ΔhemK cells, FLAG-tagged RF1 was isolated from large-scale cultures transformed with a low-copy-number plasmid harboring the hemA-prfAflag operon (pMW-hApAFL, see Fig. 1) and analyzed by matrix-assisted laser desorption ionization MS after trypsin digestion. A fragment with m/z value 1657.9, which precisely matches the predicted mass of the 229–245 peptide (1657.8), was detected in the tryptic digest of RF1 from ΔhemK cells. Whereas the fragment was not detected in the sample from hemK+ cells, a new fragment with m/z value 1671.9 appeared (Fig. 4a), implying a single methylation of one of the residues (most probably glutamine 235 within the GGQ motif) in this fragment in the hemK+ cells.
Figure 4.
Methylation of RFs by HemK. (a) Matrix-assisted laser desorption ionization–time of flight MS analysis of a tryptic digest of RF1 from CA293(hemK+) or CK783(ΔhemK). A section of the MS spectrum of the tryptic digest of RF1-FLAG is shown. The predicted m/z value for each fragment after correction for the isotopes is indicated. A peak matching the predicted mass of the 229–245 peptide (1657.8 m/z) was present in the sample from ΔhemK cells, but this peak was apparently shifted by 14 points (to 1671.9 m/z) in the sample from hemK+ cells. (b) SDS/PAGE analysis of in vitro-methylated RFs. The methylation reaction was performed on a 10-fold larger scale than described in Table 3 and the samples were subjected to SDS/PAGE (12% polyacrylamide). 3H-labeled protein was then visualized by autoradiography. In each reaction, 1 μg of 6×His-HemK was used as enzyme, and the substrate was: lane 1, 5 μg of BSA; lane 2, 10 μg of BSA; lane 3, 5 μg of RF1-FLAG; lane 4, 10 μg of RF1-FLAG; lane 5, 5 μg of 6×His-RF2; and lane 6, 10 μg of 6×His-RF2. The positions of the size markers are shown on the left. (c) One microgram of RF1-FLAG (lane 1) or 6×His-RF2 (lane 2) was analyzed by SDS/PAGE with the same size markers as in b and stained by using Coomassie brilliant blue.
HemK Methylates RF1 and RF2 in Vitro.
The above finding may imply that HemK protein, although possessing a DNA MTase signature, directly methylates RF1 (and RF2). To test this, 6×His-tagged HemK was purified from an overproducing strain, and its methyl-transferase activity to RF1 or RF2 was examined in vitro. When RF1-FLAG or 6×His-RF2 fusion proteins, produced in ΔhemK cells, were incubated with 6×His-HemK in the presence of [3H]SAM, radioactivity was incorporated into the TCA-insoluble fraction (Table 3). On the other hand, the incorporation was not detected when a mutant HemK in which the prolines in the DNA-(adenine-N6) MTase signature were altered to alanines (see above) was used instead of the wild-type HemK. To identify the molecule in which 3H was incorporated, the radiolabeled samples were separated by SDS/PAGE and subjected to autoradiography. The radioactive bands migrated at the same positions as the RF1-FLAG or 6×His-RF2 protein, respectively (Fig. 4 b and c). To confirm that the in vitro methylation also occurred in the GGQ motif, the methylation reaction was performed with unlabeled SAM and the samples were subjected to mass analysis. Whereas a fragment with m/z value 1658.0 or 1053.8, which corresponds with the unmethylated 229–245 fragment of RF1 or 246–256 fragment of RF2 (containing the GGQ motif) was detected in the samples not incubated with HemK, a fragment with about 14 points higher m/z value (1671.9 or 1067.7 m/z) was detected only in the sample that had been incubated with HemK (data not shown). These results clearly show that HemK directly methylates RF1 and RF2 proteins within the fragment containing the GGQ motif.
Table 3.
In vitro methylation of RF1 and RF2 by purified HemK or HemKP184A,P185A
| HemK (0.1 μg)/substrate (1 μg) |
3H incorporation (cpm)*
|
|
|---|---|---|
| RF1-FLAG | 6xHis-RF2 | |
| BSA† | 61 ± 20 | 45 ± 9 |
| HemK (wild type) | 1,701 ± 255 | 1,246 ± 89 |
| HemKP184A,P185A | 79 ± 22 | 47 ± 6 |
Incorporation of 3H into TCA-insoluble material was assayed as described in Materials and Methods. The average ± standard error from at least three experiments is shown.
BSA was used instead of HemK.
Global Effect of hemK Knockout on the Gene Expression.
Finally, we performed DNA chip experiments (see additional Methods, which are published as supporting information on the PNAS web site, www.pnas.org), expecting to detect alterations of gene expression caused by the loss of methylation of the RFs and yet-unidentified HemK target(s). Total RNA was isolated from exponentially growing cultures of two ΔhemK strains from different backgrounds (CK783 or WK103), and the amount of RNA for every gene was compared with that from the respective parental strains. The data can be seen in Table 4, which is published as supporting information on the PNAS web site) In fact, the expression of numerous genes was affected: of 3,275 genes for which reliable data were obtained, 116 genes were repressed by less than a half and 144 genes were induced more than 2-fold in both ΔhemK strains. As expected, genes related to translation, such as ribosomal proteins, were generally repressed, but this may have been caused by the growth rate control (22). Regarding other genes, the expression of genes for energy metabolism was particularly interesting. In the hemK knockout cells, genes for aerobic respiration, such as those for cytochrome bo3 oxidase (cyoA-E), and NADH and succinate dehydrogenases (nuoA-N and sdhA-D) were consistently repressed (by about 1/3 on average) whereas genes for anaerobic respiration, such as nitrate reductases and DMSO reductase (narK-I, narU-V, and dmsA-C) were induced (about 4-, 2-, and 2-fold, respectively) as if the cells had been grown in an anaerobic environment. Because other slow-growing strains did not show a similar pattern of gene expression (see Discussion), this shift to anaerobiosis seems to be a unique feature of ΔhemK cells.
Discussion
Our in vitro and in vivo findings revealed that HemK methylates both of the class 1 release factors (RF1 and RF2) within the tryptic fragment that contains the GGQ motif. Because the glutamine residue (Q252 in the GGQ motif) of RF2 is methylated at the N5 position (18), HemK should be responsible for this modification. The GGQ motif is not only conserved among all class 1 RFs found in eubacteria, eukaryotes, and archaea (23), but is also known to be important for the hydrolysis of peptidyl-tRNA at the termination of translation (24, 25), and in vitro studies have shown that loss of the methylation of Q252 decreases the activity of RF2 of E. coli (18). Accordingly, global deficiency of translation termination is expected in hemK knockout cells, in which neither RF1 nor RF2 is methylated. Several lines of evidence support this prediction. First, the growth of ΔhemK cells is impaired, at least partially, by the defect in translation termination. This conclusion is based on the fact that the T246A mutation in RF2 partially rescues the growth defect of ΔhemK strains. Because this mutation is known to increase the activity of RF2 (17, 26) and because the effect of the T246A mutation is independent of that of Q252 methylation (18), the suppression by the T246A mutation is considered to be a result of partial restoration of RF2 activity that have been low because of the lack of HemK. This defect in cell growth was observed under all of the conditions and backgrounds examined, and thus is more serious than that caused by the lack of RF3, which also causes a universal translational defect, but causes a growth defect only under certain circumstances (27). Second, translational read-through at the UAG and UGA codons, which are recognized by RF1 and RF2, respectively, is increased in the absence of HemK. This induction is likely to be caused by prolonged pausing of ribosomes at these stop codons and indicates that the activity of both RF1 and RF2 is affected by the absence of the methylation. We do not yet know why the rate of read-through at UAA was not altered significantly. Because the methylation of RF2 affects the activity of RF2 at UAA stop codons in vitro (18), this result may reflect the UAA-specific redundancy of RFs (both RF1 and RF2 recognize UAA) in vivo. Alternatively, it may not be general to all UAA codons, but may rather be specific to the context, because variability in the rate of read-through has been seen previously for different stop codons (28). Third, tmRNA-mediated tagging of cellular proteins is substantially induced. In particular, the amount of several tagged protein species, which were also tagged in the hemK+ strains, is markedly increased. Because some of the major endogenous targets of tmRNA were tagged at their natural stop codons (21), this result may indicate that tagging at these sites is enhanced by a termination deficiency.
The function of HemK identified here raises the question of how it leads to the suppression of the light sensitivity of a protoporphyrin IX-accumulating mutant (hemH) (1). The result of our microarray analysis might give a hint regarding the answer to this question. In ΔhemK, many genes related to aerobic respiration were repressed, whereas those for anaerobic respiration were simultaneously induced, indicating the dependence of these cells on anaerobic respiration while growing under aerobic conditions. Although we do not know whether the shift to anaerobic respiration was a result of the translational defect or the demethylation of another unidentified HemK substrate(s), it was not caused solely by the slow growth rate, because a shift to anaerobic respiration is not a common feature of slow-growing mutants (T.O., unpublished results). For example, both aerobic and anaerobic respiration are repressed in hupA− hupB− cells, which lack histone-like protein HU and have a doubling time of about 3 h. In that case, the cells may keep using oxygen as a electron acceptor without inducing an alternative pathway, unlike in the case of ΔhemK. Because the production of an intolerable amount of active oxygen in the cell is the cause of the light sensitivity of the hemH mutant, and because growth under conditions of anaerobic respiration rescues the light sensitivity (29), a shift to anaerobic respiration by the hemK mutation may lead to a similar phenotype.
SAM-dependent MTases are a large family of proteins, which accept a wide variety of substrates, such as nucleic acids, proteins, and many small molecules, and transfer the methyl group from SAM to nitrogen, oxygen, or carbon atoms. Although analyses of crystal structures have revealed a common folding pattern of MTases that accept different types of substrates (30, 31), the conservation of the primary sequences among them is usually low. Therefore, our finding that HemK, which shows high similarity to the γ subclass of DNA-(adenine-N6) MTases throughout the primary sequence of the catalytic domain (6), exhibits protein MTase activity was surprising. The group of DNA-(adenine-N6) MTase is a fairly large family and includes a few DNA-(cytosine-N4) and RNA-(guanine-N2) MTases (32, 33), but no member of the family has been reported to methylate proteins. Considering the structural characteristics of the class 1 RFs, i.e., molecular mimicry of tRNA (34), the substrate recognition and catalytic mechanism of the HemK are very interesting from a molecular evolutionary standpoint.
Because the class 1 RFs are essential and their structure is highly conserved in all eubacteria and genome-containing organelles (23, 35), it is likely that conserved HemK homologs play the same role as in E. coli. Consistent with this idea, at least one HemK homolog is encoded in all of the eubacterial and eukaryotic genomes that have been completely sequenced. By examining the substrates and structures of HemK homologs and other closely related proteins, such as E. coli YfcB (GenPept accession no. AAC75390) or Mesorhizobium loti mlr6740 product (NP_107190), we will be able to obtain more knowledge about the evolution, catalytic mechanism, and cellular function of the newly identified protein MTase.
Supplementary Material
Acknowledgments
We thank Dr. Shigeru Taketani (Kyoto Institute of Technology) for his valuable discussions. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan and Japan Science and Technology Corporation.
Abbreviations
- MTase
methyl transferase
- RF
release factor
- SAM
S-adenosyl-l-methionine
- tmRNA
transfer-mRNA
- TCA
trichloroacetic acid
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 1104.
References
- 1.Nakayashiki T, Nishimura K, Inokuchi H. Gene. 1995;153:67–70. doi: 10.1016/0378-1119(94)00805-3. [DOI] [PubMed] [Google Scholar]
- 2.Elliott T. J Bacteriol. 1989;171:3948–3960. doi: 10.1128/jb.171.7.3948-3960.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, et al. Nature (London) 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 4.Presecan E, Moszer I, Boursier L, Cruz Ramos H C, de la Fuente V, Hullo M F, Lelong C, Schleich S, Sekowska A, Song B H, et al. Microbiology. 1997;143:3313–3328. doi: 10.1099/00221287-143-10-3313. [DOI] [PubMed] [Google Scholar]
- 5.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 6.Bujnicki J M, Radlinska M. IUBMB Life. 1999;48:247–279. doi: 10.1080/713803519. [DOI] [PubMed] [Google Scholar]
- 7.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 8.Altman S, Brenner S, Smith J D. J Mol Biol. 1971;56:195–197. doi: 10.1016/0022-2836(71)90094-5. [DOI] [PubMed] [Google Scholar]
- 9.Bachmann B J. In: Derivations and Genotypes of Some Mutant Derivatives of Escherichia coli K-12. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2460–2488. [Google Scholar]
- 10.Nishihara K, Kanemori M, Kitagawa M, Yanagi H, Yura T. Appl Environ Microbiol. 1998;64:1694–1699. doi: 10.1128/aem.64.5.1694-1699.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakahigashi K, Yanagi H, Yura T. J Bacteriol. 2001;183:5302–5310. doi: 10.1128/JB.183.18.5302-5310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inokuchi H, Ozeki H. Virology. 1970;41:701–710. doi: 10.1016/0042-6822(70)90434-4. [DOI] [PubMed] [Google Scholar]
- 13.Miller J H. A Short Course in Bacterial Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 14.Hellman U, Wernstedt C, Gonez J, Heldin C H. Anal Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- 15.Nakayashiki T, Inokuchi H. Mol Gen Genet. 1997;255:376–381. doi: 10.1007/s004380050509. [DOI] [PubMed] [Google Scholar]
- 16.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 17.Uno M, Ito K, Nakamura Y. Biochimie. 1996;78:935–943. doi: 10.1016/s0300-9084(97)86715-6. [DOI] [PubMed] [Google Scholar]
- 18.Dincbas-Renqvist V, Engstrom A, Mora L, Heurgue-Hamard V, Buckingham R, Ehrenberg M. EMBO J. 2000;19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keiler K C, Waller P R, Sauer R T. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 20.Roche E D, Sauer R T. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roche E D, Sauer R T. J Biol Chem. 2001;276:28509–28515. doi: 10.1074/jbc.M103864200. [DOI] [PubMed] [Google Scholar]
- 22.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frolova L Y, Tsivkovskii R Y, Sivolobova G F, Oparina N Y, Serpinsky O I, Blinov V M, Tatkov S I, Kisselev L L. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Guen L, Santos R, Camadro J M. FEMS Microbiol Lett. 1999;173:175–182. doi: 10.1111/j.1574-6968.1999.tb13499.x. [DOI] [PubMed] [Google Scholar]
- 25.Song H, Mugnier P, Das A K, Webb H M, Evans D R, Tuite M F, Hemmings B A, Barford D. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 26.Wilson D N, Guevremont D, Tate W P. RNA. 2000;6:1704–1713. doi: 10.1017/s135583820000131x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikuni O, Ito K, Moffat J, Matsumura K, McCaughan K, Nobukuni T, Tate W, Nakamura Y. Proc Natl Acad Sci USA. 1994;91:5798–5802. doi: 10.1073/pnas.91.13.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grentzmann G, Brechemier-Baey D, Heurgue-Hamard V, Buckingham R H. J Biol Chem. 1995;270:10595–10600. doi: 10.1074/jbc.270.18.10595. [DOI] [PubMed] [Google Scholar]
- 29.Nakahigashi K, Nishimura K, Miyamoto K, Inokuchi H. Proc Natl Acad Sci USA. 1991;88:10520–10524. doi: 10.1073/pnas.88.23.10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schluckebier G, O'Gara M, Saenger W, Cheng X. J Mol Biol. 1995;247:16–20. doi: 10.1006/jmbi.1994.0117. [DOI] [PubMed] [Google Scholar]
- 31.Fauman E B, Blumenthal R M, Cheng X. In: Structure and Evolution of AdoMet-Dependent Methyltransferases. Cheng X, Blumenthal R M, editors. Singapore: World Scientific; 1999. pp. 1–38. [Google Scholar]
- 32.Bujnicki J M, Radlinska M. Nucleic Acids Res. 1999;27:4501–4509. doi: 10.1093/nar/27.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bujnicki J M. FASEB J. 2000;14:2365–2368. doi: 10.1096/fj.00-0076com. [DOI] [PubMed] [Google Scholar]
- 34.Kjeldgaad, M. & Nyborg, J. (2002) Cell, in press.
- 35.Ito K, Ebihara K, Uno M, Nakamura Y. Proc Natl Acad Sci USA. 1996;93:5443–5448. doi: 10.1073/pnas.93.11.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strohmaier H, Remler P, Renner W, Hogenauer G. J Bacteriol. 1995;177:4488–4500. doi: 10.1128/jb.177.15.4488-4500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.