Abstract
Disease-related amyloid fibrils appear to share a common, but poorly understood, structure. We describe here the generation and preliminary characterization of two conformation-specific mAbs, WO1 and WO2, that bind to the amyloid fibril state of the Alzheimer's peptide Aβ(1–40) but not to its soluble, monomeric state. Surprisingly, these Abs also bind to other disease-related amyloid fibrils and amyloid-like aggregates derived from other proteins of unrelated sequence, such as transthyretin, islet amyloid polypeptide, β2-microglobulin, and polyglutamine. At the same time, WO1 and WO2 do not bind to the native protein precursors of these amyloids, nor do they bind to other kinds of protein aggregates. This new class of Abs associated with a fundamental amyloid-folding motif appear to recognize a common conformational epitope with little apparent dependence on amino acid side chain information. These Abs should contribute to the understanding of amyloid structure, assembly, and toxicity and also may benefit the development of diagnostic and therapeutic agents for amyloid diseases.
Amyloid fibrils are highly insoluble, ordered protein aggregates involved in a number of human diseases (1, 2), including Alzheimer's disease (3) and type II diabetes (4). Although the protein components of amyloid fibrils from various disease states differ considerably from each other in primary sequence, all amyloid fibrils share common features, including a high degree of β-sheet in a classical “cross-β” pattern, a fibrillar morphology in electron microscopy, and the ability to bind and alter the spectroscopic properties of heteroaromatic dyes Congo red and thioflavin T (ThT) (5, 6). Although these common properties suggest that amyloid fibrils must share deeper similarities at the molecular level, the extent of similarity between the polypeptide-folding patterns of different amyloids is unknown. Details of the nature of the amyloid fold remain obscure because of technical limitations to obtaining high resolution structural information on large, insoluble, heterodisperse aggregates.
Although mAbs have previously proved useful in the structural analysis of globular proteins, their use in the characterization of amyloid fibril structure has been limited. Most of the anti-fibril Abs generated in an immune response to fibrils tend to be directed at unstructured portions of the amyloidogenic peptide not involved in fibril structure. In a recent characterization of the Ab response in mice injected with amyloid β protein (Aβ) fibrils, it was found that the majority of the Abs are directed at the N-terminal 12 residues of the peptide and are capable of crossreacting strongly with the monomeric peptide (7). This agrees well with the results of limited proteolysis studies of Aβ fibrils indicating an exposed, unstructured N-terminal region in the aggregate (8). Thus, such Abs tell us about those parts of the amyloidogenic peptide that are not involved in fibril structure but little about the nature of fibril structure itself.
Identification of conformational epitopes in fibrils would therefore add an important new dimension to the structural information on fibrils available through Ab studies. Early experiments with rabbit polyclonal sera suggested that amyloid fibrils possess a nonnative structure and that Abs can be generated that are specific for the amyloidogenic conformation (9). More recently, conformational Abs have been reported that are specific for transthyretin (TTR) amyloid fibrils (10) or for the infectious form of mammalian prions (11).
In addition to the value of anti-fibril Abs as structural probes, the nature of the immune response to amyloid is of special interest because of recent reports of successful active and passive vaccine approaches to slowing and/or reversing amyloid plaque growth and/or its pathological consequences in mouse models of light chain amyloidosis (13, †), Alzheimer's disease (14, 15), and mammalian prion disease (16). Some Abs recognizing Aβ fibrils also appear capable of stimulating fibril disassembly (17) and/or preventing fibril assembly (18) in vitro.
We report here the results of hydridoma experiments specifically focused on generating conformation-specific Abs against Aβ fibrils (19). The results suggest the existence of a major conformational epitope present in many amyloid fibrils composed of diverse protein sequences.
Materials and Methods
Materials and General Methods.
Aβ(1–40) peptides, as well as the polyglutamine (polyGln) molecule NH2-KKQ42KK-COOH, were custom synthesized at the Keck Biotechnology Center, Yale University. Chemically synthesized, full-length human islet amyloid polypeptide (IAPP) was a gift from Per Westermark. Recombinant JTO5, an amyloidogenic Ig VL domain, was a gift of Jonathan Wall. Human β2-microglobulin (β2m), human TTR, and chicken lysozyme were purchased from Sigma, as were bovine collagen and acid-soluble calf collagen. The κ light chain IgM mAb was purchased from Calbiochem (catalog no. 401925), and an IgG mAb recognizing the 1–17 sequence of Aβ, MAB1560, was purchased from Chemicon. Gelatin was from Bio-Rad. Trifluoroacetic acid was from Pierce and 1,1,1,3,3,3-hexafluoro-2-isopropyl alcohol (HFIP) from Sigma.
Unless otherwise indicated, all quantitative experimental results shown are from measurements done in triplicate. Error bars in figures represent SDs.
Preparation of Solubilized Peptides and Amyloid Fibrils.
Each peptide and protein required customized protocols for fibril formation. With the exception of polyGln aggregates, all amyloid fibrils, as well as collagen and elastin, were sonicated on ice with a probe sonicator for five consecutive 30-sec pulses before immobilization onto plastic microtiter plates.
Aβ peptides were solubilized and aggregated by a variation of the previously described protocol (8, 20). Amyloid fibrils were grown from Aβ(1–40) by incubating a 0.25 mg/ml disaggregated solution of the peptide in PBS containing 0.05% sodium azide (PBSA) at 37°C together with a seed consisting of 0.1% by weight of sonicated Aβ fibrils. The sample was incubated 5–7 days until fibril growth was judged complete by a ThT assay (21).
The polyGln peptide was dissolved and disaggregated as described previously (22) by using a 1:1 mixture of trifluoroacetic acid and 1,1,1,3,3,3-hexafluoro-2-isopropyl alcohol, and then aggregated by incubation of a 0.05 mg/ml solution in PBSA at 37°C for 2 wk until reaction was judged complete by the ThT assay (21). PolyGln aggregates prepared in this manner exhibit strong β-sheet spectra and a typical amyloid ThT response but exhibit ribbon morphology rather than a classical amyloid morphology; based on these and other criteria, we refer to the aggregates as “amyloid-like” (S. Chen, V. Berthelier, J. B. Hamilton, B.O.N., and R.W., unpublished data).
Human IAPP was solubilized and disaggregated by using 1:1 trifluoroacetic acid/1,1,1,3,3,3-hexafluoro-2-isopropyl alcohol (22). After removal of volatile solvents, the peptide was dissolved in 2 mM NaOH and centrifuged at 20,800 × g for 25 min. The supernatant was diluted 1:2 by using a 2 × PBS stock containing 0.1% sodium azide, pH 7.4, to a final concentration of ≈0.25 mg/ml. This solution was used immediately both to make amyloid fibrils and to fix monomers to microtiter plates. Fibrils were grown according to the protocol described above for Aβ.
Fibrils were grown from β2m and TTR in high salt and low pH as described (23, 24). Lysozyme fibrils were grown by a brief exposure to 65°C followed by incubation at 37°C in high salt at low pH (25).
All of the fibrils made as described above exhibited good amyloid fibril morphology by electron microscopy, with the exceptions that the IAPP aggregates appeared to be a mixture of classical amyloid fibrils and other organized structures (data not shown), and the polyGln aggregates appear to be protofilaments assembled into ribbons (26). All amyloid and amyloid-like aggregates exhibited typical ThT fluorescence (21, 27).
Preparation of Aggregated Carboxymethylated Proteins.
Reduction and alkylation of the disulfides of ovalbumin and human serum albumin (HSA) was accomplished by dissolving the native protein to a concentration of 0.5 mg/ml in an argon-purged buffer consisting of 0.1 M Tris⋅HCl, 6 M guanidine hydrochloride, 2 mM DTT, and 1 mM EDTA, pH 8.5. Reactions were incubated at 37°C for 1.5 h with occasional gentle mixing. Iodoacetic acid was then added to a final concentration of 15 mM, and the reaction was incubated in the dark at room temperature for 1 hr then dialyzed overnight at 4°C against PBSA. Complete cleavage of disulfide bonds was confirmed by mobility shifts in nonreducing SDS/PAGE (28). Complete modification of thiol groups was demonstrated by using 5,5′-dithiobis(2-nitrobenzoic acid) (29). The PBSA solution of reduced and alkylated proteins was subjected to several rounds of freezing and thawing, after which the amount of material collected in a pellet after centrifugation was 50% or more of the entire protein sample. These suspensions (without centrifugation) were used to immobilize modified proteins onto microtiter plates.
Hybridoma Isolation and mAb Purification.
Aβ(1–40) fibrils were sonicated as described previously (20). Five standard, female BALB/c mice were immunized with 50 μg/mouse/injection with sonicated fibrils. Two additional injections were given at 2-wk intervals. Bleeds were taken 1 wk after each injection and screened by using a modification of the microplate assay described below. After the third injection, two mice were killed and their spleens used to generate hybridoma cells. Initial screening of clones was performed by testing the ability of membranes containing uniformly deposited monomeric or fibrillar Aβ to bind Abs from an array of clonal supernatants; the bound Abs were detected with secondary Abs against murine Ig. The hybridoma experiments, including the membrane blot survey of initial hybridoma colonies, were conducted according to standard methods (30). Animal work, hybridoma creation, cloning, and preliminary screening were performed by the Hybridoma Development Facility at St. Louis University Health Center, St. Louis.
Clonal supernatants giving good binding to immobilized fibrils even in the presence of 80-fold weight excess monomeric Aβ were considered to be good candidates for conformation-specific Abs and were carried forward in the cloning process. mAbs were produced from stable hybridoma cell lines by growing the cells in high density culture by using CELLline incubator flasks (INTEGRA Biosciences). mAbs were purified from the accumulated Ab-containing supernatants by using a HiTrap IgM purification affinity column (Amersham Pharmacia) followed by a Sephacryl S-300 (Amersham Pharmacia) size exclusion chromatography column (PBSA, 4°C). Nonreducing SDS/PAGE analysis confirmed the IgM isotype of the Abs and showed that they were at least 90% pure.
Microtiterplate Assays of Fibril Binding.
Mouse sera and hybridoma supernatants were assayed for anti-fibril Abs as follows. First, 100 ng of sonicated Aβ(1–40) fibrils in 100 μl of PBSA was added to each well of a high-binding microtiter plate (Costar) and allowed to dry by incubating uncovered overnight in a 37°C oven. Plates were washed three times with PBSA containing 0.05% Tween 20 (the standard wash procedure for all subsequent steps of the protocol). Wells were blocked with 1% gelatin in PBSA at 37°C for 1 hr. Plates were then incubated with sera or hybridoma supernatants, with or without an 80-fold weight excess of monomeric Aβ(1–40) with respect to immobilized fibrils, for 1 hr at 37°C and then washed three times. The signal was developed by incubation with a biotinylated Ab, followed by treatment with a streptavidin conjugate.
For measuring the IgG response in mouse sera (Fig. 1A), the secondary Ab was a mixture of isotype-specific goat anti-mouse IgG Abs (Sigma ISO-2 kit) diluted into 1% gelatin, 0.05% Tween 20, and PBSA. The tertiary Ab was a biotinylated rabbit anti-goat Ig Ab (Vector Laboratories). After incubation with a streptavidin-horseradish peroxidase conjugate (Vector Laboratories), the signal was developed with 3,3′,5,5′-tetramethylbenzidine (TMB, Pierce no. 34021).
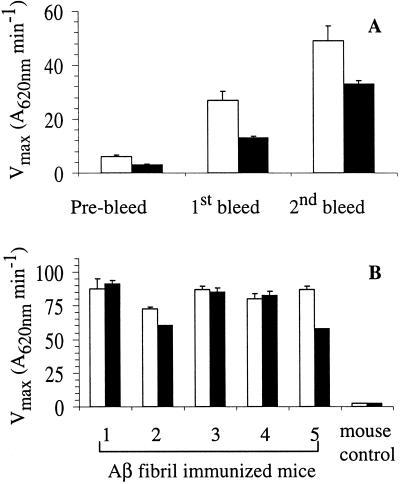
Figure 1.
Serum responses of mice immunized with Aβ(1–40) fibrils. Open bars show total Ig binding to 100 ng/well of amyloid fibrils immobilized onto microtiter plate wells. Closed bars show Ab binding to fibrils in the presence of a large excess of Aβ(1–40) monomer. (A) Increase in anti-amyloid IgG response over the course of the immunization treatment. (B) Total anti-amyloid IgG + IgM response of five immunized mice and a control at the completion of the immunization protocol.
For measuring the IgG + IgM response in mouse sera (Fig. 1B), the secondary Ab was a biotinylated goat anti-mouse Ig Ab (Sigma), and the signal was developed with streptavidin-horseradish peroxidase as described above. For measuring the IgG + IgM response of hybridoma culture supernatants (Fig. 4), the same secondary Ab was used, but the signal was developed by using a europium-streptavidin conjugate (EG & G Wallac) and counted by using time-resolved fluorescence (31) on a Wallac Victor (2) fluorescence microtiter plate reader.
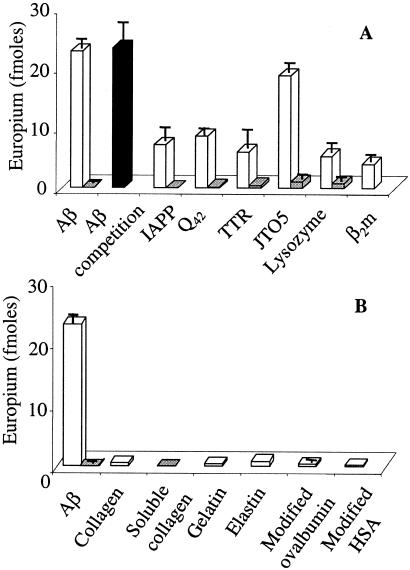
Figure 4.
Binding of WO1 to various aggregates. Open bars, binding to immobilized aggregate; gray bars, binding to immobilized monomeric form; and filled bar, binding to immobilized aggregate in the presence of an 80-fold weight excess monomeric F19P-Aβ(1–40). (A) Binding to various amyloids and their precursor proteins. (B) Binding to nonamyloid biological aggregates and nonnative globular protein aggregates. In these experiments, WO1 was used as a hybridoma supernatant diluted 1:2 in PBSA.
Characterization of Binding Properties of Purified mAbs.
For experiments featuring immobilized ligands, microtiter plate wells were coated either with amyloid fibrils or other aggregates (as described above), or with soluble precursor proteins. For the latter, proteins in 2 mM sodium phosphate buffer, pH 7.4, containing 0.05% sodium azide, were incubated uncovered overnight in a 37°C oven. The low salt minimizes aggregation during the coating process (data not shown). After washing, plates were blocked with 3% BSA in PBSA for 1 h at 37°C. For the assay, Ab solutions were incubated 2 h at 37°C in the wells with 3% BSA in PBSA containing 0.05% Tween 20. Binding was quantified by using a biotinylated secondary Ab as described above. In the case of Aβ fibrils, the ability of an 80-fold weight excess of monomeric peptide to inhibit Ab binding to fibrils was also assessed.
Binding of aggregates and soluble proteins to plastic by the above protocol was essentially quantitative. This was determined as follows. After overnight incubation as described, 50–150 μl of PBS was added to several wells and incubated 10 min at room temperature. Supernatants from several wells were pooled and assayed for the amount of recovered protein, either by a micro protein assay (Pierce MicroBCA) or by recovery of aggregates, solubilization, and quantitation by HPLC against a standard curve (8). Less than 5% of the applied protein was recovered by such analyses, consistent with greater than 95% fixation of aggregates to the plastic.
For experiments featuring immobilized Abs, 100 μl of 15 nM mAb solutions in PBSA were sealed and incubated for 1.5 h at 37°C. Plates were washed and blocked with a 3% BSA solution in PBSA by incubation at 37°C for 1 hr. Various concentrations of biotinyl-Aβ(1–40) were added to each well, and the plate was incubated for 4 h at 37°C and then quantified as before by using streptavidin-europium and time-resolved fluorescence. The N-terminally biotinylated Aβ molecule was prepared by alkylating a Cys−1 analog of Aβ(1–40) with PEO-iodoacetyl biotin (Pierce).
Results and Discussion
Conformation-specific Abs against the native states of many proteins have been described. Such Abs bind well to the native, folded state of the protein, and less well, or not at all, to the denatured protein or to isolated peptide fragments (32). To isolate Abs specific for conformational epitopes of the Aβ fibril, mice were injected with sonicated Aβ fibrils. These mice mounted a time-dependent serum response consisting of Abs capable of binding to the amyloid form of Aβ immobilized on microtiter plate wells (Fig. 1). Significantly, a major portion of the anti-fibril Ab population binds to Aβ fibrils despite the presence of a large excess of monomeric Aβ (Fig. 1). This suggests that a major portion of the Abs are directed against conformational epitopes that only exist in the fibril. All five mice injected with amyloid exhibit similar serum responses (Fig. 1B). Hybridoma fusions were generated from the spleens of responsive mice and resulting cells screened and cloned by using a variety of assays (Materials and Methods) to isolate stable cell lines producing Abs that bind to Aβ fibrils but not to Aβ monomers.
Based on results from screening hybridoma supernants, two stable cell lines, designated WO1 and WO2, were selected for further study. Reducing and nonreducing SDS/PAGE (not shown) of cell supernatants showed that both WO1 and WO2 have molecular weights in the 900-kDa range, consistent with the isotyping results on these supernatants showing that both Abs are of the IgM class. Light chains were isolated from the gels and subjected to amino acid sequence analysis. The light chain of WO1 exhibited the N-terminal sequence DIQMTQS, consistent with its being in the κ class of mouse light chains. WO2 appears to consist of a different light chain sequence because it exhibits a blocked N terminus, most consistent with a pyro-Glu residue derived from cyclization of a Gln side chain at position 1.
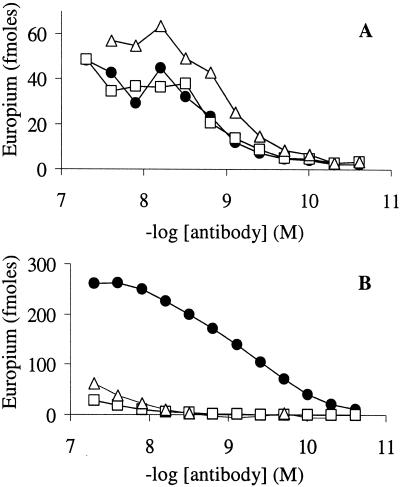
Fig. 2 compares the fibril-binding characteristics of WO1 to an anti-Aβ IgG mAb that recognizes a primary sequence epitope resident in the 1–17 sequence of Aβ. Fig. 2A shows that WO1 exhibits a saturable binding curve against immobilized Aβ fibrils with an EC50, or concentration of half-maximal binding, in the low nanomole range. Fig. 2A also shows that when a large weight excess of monomeric Aβ(1–40) is included in the incubation, strong binding to fibrils by WO1 is retained. (The apparent enhancement of binding of WO1 to amyloid fibrils in the presence of wild-type Aβ is probably due to higher fibril mass resulting from the extension of Aβ fibrils by the monomeric Aβ competitor during the incubation.) A soluble, proline-containing mutant incapable of making amyloid fibrils (33, 34), F19P-Aβ(1–40), also does not compete for WO1 binding to fibrils. In contrast, both mutant and wild-type monomeric Aβ molecules effectively compete for binding to fibrils by an Ab directed against a linear epitope of Aβ. Fig. 2B shows that an Ab directed at the linear 1–17 sequence of Aβ, MAB1560, binds well to immobilized Aβ fibrils. Fig. 2B shows that, in contrast to WO1 binding, MAB1560 binding to fibrils can be almost entirely eliminated when a large weight excess of monomeric, soluble Aβ is included in the binding incubation. In these experiments, WO1 and MAB1560 exhibit significantly different binding amplitudes to the same weight of Aβ fibrils; however, given the fact that different secondary Abs were used in these two panels, it is difficult to interpret this effect.
Figure 2.
Binding of purified mAbs to Aβ fibrils in the presence and absence of excess monomeric Aβ. Aβ(1–40) fibrils were immobilized on microplate wells and the wells incubated with anti-Aβ Abs in the absence (●) or presence of monomeric Aβ wild-type (▵) or F19P mutant (□). (A) Binding of WO1. (B) Binding of the IgG mAb MAB1560 against a linear epitope (positions 1–17) of the Aβ(1–40) molecule.
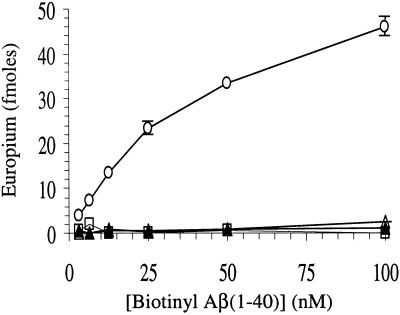
The experiments summarized in Fig. 2 provide indirect evidence that WO1 does not recognize monomeric Aβ. More direct evidence is the demonstration that neither WO1 (Fig. 4) or WO2 (not shown) bind appreciably to monomeric Aβ(1–40) immobilized onto microtiter plate wells. In addition, we find that monomeric Aβ(1–40) is not able to bind to immobilized WO1 and WO2. Thus, Fig. 3 shows that although immobilized MAB1560 effectively binds N-terminally biotinylated Aβ(1–40), immobilized WO1 and WO2, as well as a control IgM mAb, are completely ineffective at binding this peptide.
Figure 3.
Binding of monomeric, soluble biotinyl-Aβ to purified Abs immobilized on microplate wells. WO1 (▵); WO2 (▴); control IgM (□); and anti-Aβ(1–17) IgG MAB1560 (○).
Surprisingly, WO1 and WO2 are capable of binding not only to Aβ fibrils, but also to amyloid fibrils generated from other proteins. We generated amyloid fibrils from a number of amyloid precursor proteins (Materials and Methods). In each case the protein aggregates exhibit fibril or fibril-like structures in electron microscopy and a typical ThT fluorescence response (data not shown). Fig. 4A shows the binding of WO1 to equal weights of each of these amyloid fibrils immobilized onto microtiter plate wells. The figure shows that amyloid fibrils or amyloid-like aggregates composed of β2m, IAPP, TTR, polyGln (Q42), the Ig VL domain JTO5 (35), and lysozyme [in addition to Aβ(1–40)] are all capable of binding significant amounts of WO1. On a weight basis, WO1 binds different fibrils to different extents, exhibiting maximal binding to Aβ(1–40) fibrils. The binding differences observed may be due in part to differences in binding constants (see below) and/or to different numbers of epitopes per unit weight of these amyloid fibrils. WO2 exhibited similar pan-amyloid binding (not shown).
In contrast to its broad ability to bind to amyloid fibrils, WO1 exhibits little or no binding to the precursor proteins for each of these fibrils immobilized onto microtiter plate wells (Fig. 4A). This result suggests that the conformation recognized by WO1 is absent in the native precursor proteins. Fig. 4A also shows that WO1 binds to Aβ fibrils despite the presence of a high weight excess of soluble Aβ.
To probe the specificity of WO1 and WO2 for amyloid fibrils, we tested WO1 binding to different kinds of protein aggregates. Fig. 4B shows that WO1 does not bind appreciably to the fibrous proteins collagen and elastin, or to gelatin, the denatured form of collagen. Many globular proteins aggregate in response to disruption of their native states by chemical or physical stress, and these aggregates are often dominated by β-sheet structure (36). Fig. 4B shows, however, that WO1 does not bind to aggregates of ovalbumin or human serum albumin induced by denaturation via reduction and alkylation. Similar results were obtained for WO2 (not shown). These results show that the epitope(s) recognized by WO1 and WO2 is not a structural feature shared with amorphous protein aggregates, such as interchain β-sheet or diffuse patches of surface-exposed hydrophobicity. Thus, results to date suggest that the epitope recognized by these Abs is specific to the amyloid state of proteins.
To quantitatively assess the crossreactivity of WO1 and WO2 against other amyloid fibrils, we determined full binding curves for these Abs, as well as for a control κ light chain IgM, against amyloid fibrils composed of Aβ(1–40), JTO5, and IAPP. Fig. 5A shows that WO1 and WO2 exhibit saturable binding curves to immobilized Aβ(1–40) fibrils (the original immunogen) with EC50s of 2.8 nM and 1.3 nM, respectively. In contrast, the binding of a κ light chain control IgM to Aβ(1–40) fibrils exhibited a much weaker binding constant (121 nM) and a lower binding amplitude. The binding ability of this control IgM to Aβ fibrils is probably related to the general ability of globular proteins to bind nonspecifically to Aβ fibrils (B.O.N. and R.W., unpublished observations). Interestingly, a λ chain control IgM is much less effective at binding to Aβ fibrils compared to the control κ chain IgM (data not shown).
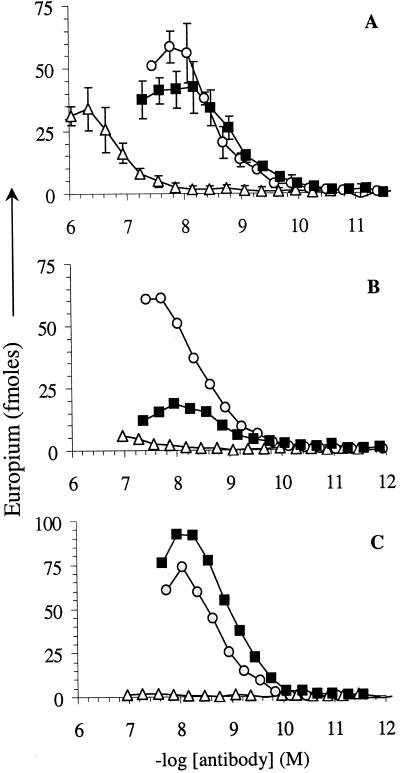
Figure 5.
Binding curves for purified WO1, WO2, and a control κ chain IgM against various amyloid fibrils immobilized onto microplate wells. WO1 (○); WO2 (■); and IgM control (▵). (A) Aβ(1–40) fibrils. (B) Ig light chain variable domain JTO5 fibrils. (C) IAPP fibrils. Data in B and C are from single replicate determinations.
Fig. 5B shows that WO1 and WO2 bind to JTO5 fibrils with EC50 values of 2.8 nM and 1.2 nM, respectively, values similar to those for binding to Aβ(1–40) fibrils. The amplitudes for WO1 and WO2 binding to JTO5 fibrils, although differing from each other by about a factor of three, are in the same range as the binding of these Abs to Aβ fibrils. The control κ light chain IgM exhibits essentially no binding to JTO5 fibrils. Fig. 5C shows that WO1 and WO2 also bind with similar amplitudes and binding constants (1.7 nM and 1.0 nM, respectively) to IAPP fibrils, whereas the control IgM binds negligibly. These data dramatically illustrate that WO1 and WO2 recognize what appears to be a universal amyloid epitope and that the ability to bind to amyloid fibrils is not shared by a control IgM. The Abs bind with similar strengths to amyloids other than the Aβ(1–40) amyloid immunogen. Although the IAPP peptide exhibits some amino acid sequence homology to Aβ(1–40) (P. Westermark, personal communication), JTO5 offers no significant homology. [This was confirmed by using the sim (37), lalign (38), and dotlet (39) sequence alignment programs (www.expasy.ch/tools) (data not shown)]. The nature of the common amyloid epitope, therefore, does not seem to depend on extensive amino acid side chain information. This point is made perhaps most dramatically by the fact that WO1 binds well to polyGln aggregates (Fig. 4A), which exhibit a number of amyloid-like features in their morphologies and biophysical properties (S. Chen, V. Berthelier, J. B. Hamilton, B.O.N., and R.W., unpublished data).
In their abilities to bind to multiple molecular species, WO1 and WO2 bear some resemblance to polyreactive Abs such as the anti-DNA Abs produced in certain autoimmune conditions. Although little is known about the basis for such polyreactive binding recognition, at least some anti-DNA Abs are thought to recognize repeat structures within DNA such as heteroaromatic bases or phosphate groups (40, 41). As highly ordered polymers, amyloid fibrils presumably also exhibit certain kinds of regular structural repeats that might serve as the basis for multidentate-binding recognition. The extent to which WO1 and WO2 binding to amyloid might depend on such multidentate binding is yet to be determined.
One possible contributor to a common amyloid structural epitope might be a unique array of H-bond donor and acceptor groups from the polypeptide backbone at the edge strands of the β-sheets on the ends of amyloid fibrils. If so, this configuration must be different from that of the edge strands of β-sheets in the native states of β-sandwich-based proteins such as the FV domain and TTR because these globular, native proteins do not bind WO1 and WO2 appreciably (Fig. 4). Another possibility is main chain elements involved in some unusual turn or chain reversal within the amyloid motif. Detailed structures of the epitopes of anti-protein Abs are normally characterized either by protein crystallography (42) or by mutational analysis of antigen fragments (43) or intact protein (44). Because amyloid has yet to be crystallized, and because WO1 and WO2 bind to many amyloids regardless of amino acid sequence, it is clear that the further structural analysis of the WO1/WO2 epitope(s) will be challenging.
Aβ(1–40) fibrils are not unique in their ability to stimulate production of generic anti-amyloid Abs in mice. For example, a mAb raised against an amyloidogenic Ig light chain fragment has been reported to recognize tissue amyloid deposits composed of the light chain variable domain, Aβ, and other amyloidogenic proteins (13). The reciprocity of these results with those described here further supports the existence of an epitope that is a universal signature of the amyloid fibril.
Abs such as WO1 and WO2 are important for a number of reasons. They may prove invaluable in improving our knowledge of the three-dimensional structures of amyloid fibrils. Such Abs will also allow us to monitor more closely the generation of the amyloid epitope during assembly in vitro and in vivo. Fibril-specific Abs may be useful as passive vaccines in anti-amyloid therapy (13, 15, 16, †). Furthermore, recognition that an amyloid fibril from one precursor protein can stimulate a pan-amyloid response in an animal suggests that amyloid fibrils composed of one constituent protein might serve as vaccines against other types of amyloid. Finally, Abs such as WO1 and WO2 may prove to be useful diagnostic reagents not only for Alzheimer's disease, but also for other protein aggregation diseases. The defining clinical test for the presence of amyloid in tissue continues to be birefringence after Congo red staining (12). However, as a technique that is inherently limited in resolution to the wavelength of visible light, Congo red birefringence is not capable of detecting deposits of small and/or disorganized fibrils if they do not exhibit macroscopic order. Abs such as WO1 and WO2 may thus prove to be more reliable probes for the presence of the amyloid-folding motif in tissue samples.
Acknowledgments
We gratefully acknowledge Charles Murphy for the amino acid sequence analysis of IgM light chains, Yesim Aydin Son for preliminary binding studies, and Dennis Wolfenbarger for help with preliminary immunization studies. We also thank Per Westermark for the gift of human IAPP, Jonathan Wall for the gift of soluble and fibrillar recombinant JTO5, Jonathan Wall and Alan Solomon for helpful discussions, and Fred Stevens for pointing out the limitations of Congo red birefringence.
Abbreviations
- PBSA
PBS containing 0.05% sodium azide
- polyGln
polyglutamine
- IAPP
islet amyloid polypeptide
- Aβ
amyloid β protein
- TTR
transthyretin
- β2m
β2-microglobulin
- ThT
thioflavin T
Footnotes
Hrncic, R., Wall, J., Schell, M., Macy, S. D., Wolfenbarger, D., Weiss, D. T. & Solomon, A. (1999) Blood 94, Suppl. 1, 310 (abstr. 4614).
References
- 1.Falk R H, Comenzo R L, Skinner M. N Engl J Med. 1997;337:898–909. doi: 10.1056/NEJM199709253371306. [DOI] [PubMed] [Google Scholar]
- 2.Martin J B. N Engl J Med. 1999;340:1970–1980. doi: 10.1056/NEJM199906243402507. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe D J. Science. 1997;275:630–631. doi: 10.1126/science.275.5300.630. [DOI] [PubMed] [Google Scholar]
- 4.Kahn S E, Andrikopoulos S, Verchere C B. Diabetes. 1999;48:241–253. doi: 10.2337/diabetes.48.2.241. [DOI] [PubMed] [Google Scholar]
- 5.Sipe J D. Annu Rev Biochem. 1992;61:947–975. doi: 10.1146/annurev.bi.61.070192.004503. [DOI] [PubMed] [Google Scholar]
- 6.Wetzel R. In: Methods in Enzymology. Abelson J N, Simon M I, editors. San Diego: Academic; 1999. p. 820. [Google Scholar]
- 7.Town T, Tan J, Sansone N, Obregon D, Klein T, Mullan M. Neurosci Lett. 2001;307:101–104. doi: 10.1016/s0304-3940(01)01951-6. [DOI] [PubMed] [Google Scholar]
- 8.Kheterpal I, Williams A, Murphy C, Bledsoe B, Wetzel R. Biochemistry. 2001;40:11757–11767. doi: 10.1021/bi010805z. [DOI] [PubMed] [Google Scholar]
- 9.Linke R P, Zucker-Franklin D, Franklin E C. J Immunol. 1973;111:10–23. [PubMed] [Google Scholar]
- 10.Goldsteins G, Persson H, Andersson K, Olofsson A, Dacklin I, Edvinsson A, Saraiva M J, Lundgren E. Proc Natl Acad Sci USA. 1999;96:3108–3113. doi: 10.1073/pnas.96.6.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korth C, Stierli B, Streit P, Moser M, Schaller O, Fischer R, Schulz-Schaeffer W, Kretzschmar H, Raeber A, Braun U, et al. Nature (London) 1997;390:74–77. doi: 10.1038/36337. [DOI] [PubMed] [Google Scholar]
- 12.Westermark G T, Johnson K H, Westermark P. Methods Enzymol. 1999;309:3–25. doi: 10.1016/s0076-6879(99)09003-5. [DOI] [PubMed] [Google Scholar]
- 13.Hrncic R, Wall J, Wolfenbarger D A, Murphy C L, Schell M, Weiss D T, Solomon A. Am J Pathol. 2000;157:1239–1246. doi: 10.1016/S0002-9440(10)64639-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schenk D, Barbour R, Dunn W, Gordon G, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, Khan K, et al. Nature (London) 1999;400:173–177. doi: 10.1038/22124. [DOI] [PubMed] [Google Scholar]
- 15.Bard F, Cannon C, Barbour R, Burke R L, Games D, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, et al. Nat Med. 2000;6:916–919. doi: 10.1038/78682. [DOI] [PubMed] [Google Scholar]
- 16.Peretz D, Williamson R A, Kaneko K, Vergara J, Leclerc E, Schmitt-Ulms G, Mehlhorn I R, Legname G, Wormald M R, Rudd P M, et al. Nature (London) 2001;412:739–743. doi: 10.1038/35089090. [DOI] [PubMed] [Google Scholar]
- 17.Solomon B, Koppel R, Frankel D, Hanan-Aharon E. Proc Natl Acad Sci USA. 1997;94:4109–4112. doi: 10.1073/pnas.94.8.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solomon B, Koppel R, Hanan E, Katzav T. Proc Natl Acad Sci USA. 1996;93:452–455. doi: 10.1073/pnas.93.1.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Nuallain B, Wetzel R. Protein Sci. 2001;10:87–88. [Google Scholar]
- 20.Kheterpal I, Zhou S, Cook K D, Wetzel R. Proc Natl Acad Sci USA. 2000;97:13597–13601. doi: 10.1073/pnas.250288897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeVine H. Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 22.Chen S, Wetzel R. Protein Sci. 2001;10:887–891. doi: 10.1110/ps.42301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colon W, Kelly J W. Biochem. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 24.McParland V J, Kad N M, Kalverda A P, Brown A, Kirwin-Jones P, Hunter M G, Sunde M, Radford S E. Biochemistry. 2000;39:8735–8746. doi: 10.1021/bi000276j. [DOI] [PubMed] [Google Scholar]
- 25.Krebs M R, Wilkins D K, Chung E W, Pitkeathly M C, Chamberlain A K, Zurdo J, Robinson C V, Dobson C M. J Mol Biol. 2000;300:541–549. doi: 10.1006/jmbi.2000.3862. [DOI] [PubMed] [Google Scholar]
- 26.Chen S, Berthelier V, Yang W, Wetzel R. J Mol Biol. 2001;311:173–182. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- 27.Naiki H, Higuchi K, Hosokawa M, Takeda T. Anal Biochem. 1989;177:244–249. doi: 10.1016/0003-2697(89)90046-8. [DOI] [PubMed] [Google Scholar]
- 28.Plunkett G, Ryan C A. J Biol Chem. 1980;255:2752–2755. [PubMed] [Google Scholar]
- 29.Zahler W L, Cleland W W. J Biol Chem. 1968;243:716–719. [PubMed] [Google Scholar]
- 30.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 31.Diamandis E P. Clin Biochem. 1988;21:139–150. doi: 10.1016/0009-9120(88)90001-x. [DOI] [PubMed] [Google Scholar]
- 32.Laver W G, Air G M, Webster R G, Smith-Gill S J. Cell. 1990;61:553–556. doi: 10.1016/0092-8674(90)90464-p. [DOI] [PubMed] [Google Scholar]
- 33.Wood S J, Wetzel R, Martin J D, Hurle M R. Biochemistry. 1995;34:724–730. doi: 10.1021/bi00003a003. [DOI] [PubMed] [Google Scholar]
- 34.Teplow D B, Lomakin A, Benedek G B, Kirschner D A, Walsh D M. In: Alzheimer's Disease: Biology, Diagnosis and Therapeutics. Iqbal K, Winblad B, Nishimura T, Takeda M, Wisniewski H M, editors. New York: Wiley; 1997. pp. 313–321. [Google Scholar]
- 35.Wall J, Schell M, Murphy C, Hrncic R, Stevens F J, Solomon A. Biochemistry. 1999;38:14101–14108. doi: 10.1021/bi991131j. [DOI] [PubMed] [Google Scholar]
- 36.Fink A L. Folding Des. 1998;3:R9–R23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 37.Huang X, Miller W. Adv Appl Math. 1991;12:337–357. [Google Scholar]
- 38.Duret L, Gasteiger E, Perriere G. Comput Appl Biosci. 1996;12:507–510. doi: 10.1093/bioinformatics/12.6.507. [DOI] [PubMed] [Google Scholar]
- 39.Junier T, Pagni M. Bioinformatics. 2000;16:178–179. doi: 10.1093/bioinformatics/16.2.178. [DOI] [PubMed] [Google Scholar]
- 40.Fermand J P, Danon F, Brouet J C. Clin Exp Immunol. 1985;59:467–474. [PMC free article] [PubMed] [Google Scholar]
- 41.Eilat D, Anderson W F. Mol Immunol. 1994;31:1377–1390. doi: 10.1016/0161-5890(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 42.Padlan E A, Silverton E W, Sheriff S, Cohen G H, Smith-Gill S J, Davies D R. Proc Natl Acad Sci USA. 1989;86:5938–5942. doi: 10.1073/pnas.86.15.5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Getzoff E D, Geysen H M, Rodda S J, Alexander H, Tainer J A, Lerner R A. Science. 1987;235:1191–1196. doi: 10.1126/science.3823879. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Lipschultz C A, Mohan S, Smith-Gill S J. Biochemistry. 2001;40:2011–2022. doi: 10.1021/bi0014148. [DOI] [PubMed] [Google Scholar]