Abstract
Functional inactivation of the tumor suppressor protein p53 by accelerated ubiquitin/proteasome-dependent proteolysis is a common event in tumor progression. Proteasomal degradation is inhibited by the Gly-Ala repeat (GAr) of the Epstein–Barr virus nuclear antigen-1, which acts as a transferable element on a variety of proteasomal substrates. We demonstrate that p53 chimeras containing GAr domains of different lengths and positions within the protein are protected from proteolysis induced by the ubiquitin ligases murine double minute 2 and E6-associated protein but are still ubiquitinated and retain the capacity to interact with the S5a ubiquitin-binding subunit of the proteasome. The GAr chimeras transactivate p53 target genes, induce cell cycle arrest and apoptosis, and exhibit improved growth inhibitory activity in tumor cells with impaired endogenous p53 activity.
The p53 tumor suppressor protein is involved in a variety of cellular processes, including the regulation of the cell cycle and apoptosis (1, 2). Inactivation of p53, by mutations or through interaction with viral and cellular proteins, is one of the most frequent alterations observed in cancer (3), suggesting that restoration of wild-type p53 activity in tumor cells could be of immense therapeutic potential. Protein stability is a critical parameter of p53 function, which is regulated, in normal as well as malignant cells, by ubiquitin-dependent proteolysis. Targeting of p53 for ubiquitin-mediated proteolysis is accomplished by two ubiquitin ligases: the murine double minute 2 (Mdm2) oncoprotein (4, 5), and the E6-associated protein (E6-AP) (6). Mdm2 ubiquitinates p53 in the nucleus and facilitates nuclear export delivering polyubiquitinated p53 for destruction by cytoplasmic proteasomes (7). Recent evidence demonstrates that several proteins involved in oncogenesis, such as ARF (8), oncogenic ras (9), the retinoblastoma protein (10), and TSG101 (11), affect the stability of p53 by modulating Mdm2-mediated degradation. E6-AP acts as a p53 ubiquitin ligase only in concert with the E6 oncoproteins of the high-risk human papillomaviruses (HPV) types 16 and 18 (6, 12). Hence, formation of a trimolecular complex of E6, E6-AP, and p53 results in dramatic acceleration of p53 proteolysis.
Inactivation of p53 by high levels of Mdm2 or HPV E6 provides a clear growth advantage in vivo (13, 14), suggesting that a therapeutically active p53 protein should resist the activity of these ligases. In this context, it should be stressed that Mdm2 and E6-AP belong to distinct families of ubiquitin ligases (15). Mdm2 was recently shown to be a RING-finger ubiquitin ligase that binds to the transcriptional transactivation domain of p53 (16). In contrast, E6-AP interacts with the DNA-binding domain and is the classic example of HECT (homology to E6-AP C terminus) domain ubiquitin ligase (6). These explicit differences between the two ligases imply that simultaneous rescue of p53 from E6- and Mdm2-mediated degradation may be attained only by targeting common downstream events in the degradation pathway.
An interesting opportunity to achieve this goal may be offered by a recently discovered modulator of the ubiquitin-dependent proteolysis, the Gly-Ala repeat (GAr) of the Epstein–Barr virus (EBV) nuclear antigen-1 (EBNA1). This protein domain interferes with the proteasomal processing of EBNA1 (17), prolonging its half-life and abrogating the presentation of EBNA1 epitopes to major histocompatibility complex class I-restricted CD8+ T-lymphocytes (18, 19). The GAr acts as a cis-acting transferable element on different proteasome substrates (17, 20). The promiscuity of the effect implies that the GAr may be able to counteract the activity of a wide variety of ubiquitin ligases, thus providing an attractive tool for regulating the proteolysis of many substrates of potential interest for gene transfer therapy (21).
Here we report on the generation of a set p53-GAr chimeras that exhibit improved resistance to Mdm2- and E6-induced degradation in cotransfection assays and in human tumor cell lines with accelerated proteolysis of the endogenous p53. The p53-GAr chimeras are efficiently rescued from ubiquitin/proteasome-dependent degradation, resulting in increased steady-state levels of functionally active p53.
Materials and Methods
Construction of the p53-GAr Chimeras.
The human p53 ORF was PCR amplified from the pC53-SN plasmid [B. Vogelstein, Johns Hopkins University School of Medicine, Baltimore, MD (22)], by using the sense primer: 5′-GCGCTCGAGGCATGGAGGAGCCGCAGTCAG (XhoI underlined), and antisense primer: 5′-GCGCGCGGCCGCCTATGGTCGACCTGAGTCAGGCCCTTCTGTCTTG (NotI underlined, SalI double underlined, stop codon bold) that introduced flanking XhoI and NotI restriction sites, respectively, and then was cloned into pBK-CMV (Stratagene) or pCMS–enhanced green fluorescent protein (EGFP) (CLONTECH). An oligomer encoding the FLAG epitope was inserted into the 5′ NheI and HindIII sites resulting in pBK-CMV FLAGp53. Oligonucleotides encoding 25-aa-long GAr or glycine stretches flanked by 5′ SalI and 3′ XhoI restriction sites were generated as described previously (23). The repeats were inserted in the 5′ XhoI site or the 3′ SalI site of FLAGp53. The coding region of the 239-aa-long GAr of the prototype EBV EBNA1 was PCR-amplified and linked to the 3′ end of p53 by using the SalI and NotI sites. The pCOC-Mdm2X2 expression vector (24) and the HPV16-E6 expression vector pCB6+16E6 (25) were obtained from K. H. Vousden (National Cancer Institute, Frederick, MD). The Bp53-EGFP reporter plasmid contains the EGFP gene under the control of 13 copies of a p53-responsive element derived from a ribosomal gene cluster (V. J. Bykov, Karolinska Institute).
Tissue Culture, Transfection, and Colony Formation Assay.
The human osteosarcoma lines Saos-2 [p53 null (26),] and U2OS [p53 wild-type, Mdm2 overexpression (13)], and the human cervical carcinoma lines HeLa (p53 wild-type, HPV18 positive), CasKi and SiHa [p53 wild-type, HPV16 positive (14, 27)] were maintained in Iscove's modified Dulbecco's medium (Life Technologies, Grand Island, NY) supplemented with 10% FCS. Transient transfections of subconfluent monolayers were performed with Lipofectamine (Life Technologies). For colony formation assays, 1.9 105 transfected U2OS cells were split 16 h after transfection (8,000 cells per well) in medium containing 0.5 mg/ml of Geneticin (Sigma). The number of viable colonies was counted after 2 weeks selection.
Western Blot Analysis.
Total cell extracts were fractionated by SDS/PAGE, transferred to Protran BA85 nitrocellulose membranes (Schleicher & Schuell), and probed with primary anti-p53 mouse monoclonal antibody (D07, Dako) or anti-p21WAF/CIP1 mouse monoclonal antibody (Transduction Laboratories, Lexington, KY). After incubation with the appropriate peroxidase-conjugated secondary antibodies, complexes were visualized by enhanced chemiluminescence (Amersham Pharmacia–Pharmacia Biotech). For protein turnover determination, transiently transfected cells were incubated with cycloheximide (Sigma). Densitometry was performed by using a Personal Densitometer SI (Molecular Dynamics).
Flow Cytometry and Immunocytochemistry.
Flow cytometric analysis was performed by using a FACSort flow cytometer (Becton Dickinson) and cellquest software. For cell cycle analysis, cells were fixed and stained with the mouse anti-p53 antibody D07 and a FITC-labeled anti-mouse antibody (Dako) by using the Cytofix/Cytoperm kit (PharMingen) before incubation with propidium iodide (Sigma). For immunofluorescence staining, the cells were fixed in 4% paraformaldehyde and then incubated overnight with an anti-FLAG mouse monoclonal antibody (M5, Sigma).
Glutathione S-Transferase (GST) Pull-Down Assays.
The S5a and S8 ORFs were PCR-amplified from a human leukocyte cDNA library (CLONTECH) and cloned into pGEX-5X-1 (Amersham Pharmacia–Pharmacia Biotech). The GST-S5a and S8 fusion proteins were purified from Escherichia coli strain BL-21 (Amersham Pharmacia–Pharmacia Biotech). HeLa cells transiently transfected with FLAGp53 or FLAGp53-GA239/C were lysed at 4°C for 12 h in lysis buffer (1% digitonin/50 mM NaCl/50 mM Tris, pH 7.6) containing protease inhibitor mixture (Sigma) and 20 mM N-ethylmaleimide (Sigma). The lysates were cleared by centrifugation for 20 min at 4°C, 15,000 × g and diluted in binding buffer (50 mM Tris, pH 7.6/50 mM NaCL/0.01% Triton X-100/10% glycerol). Two micrograms of GST fusion protein was immobilized on 10 μl of glutathione Sepharose 4B gel (Amersham Pharmacia–Pharmacia Biotech) and incubated with HeLa cell lysates. After washing five times at 4°C in binding buffer, samples were resuspended in SDS sample buffer for separation by SDS/PAGE and immunoblotting with the anti-p53 antibody D07.
Results
Expression of p53-GAr Chimeras.
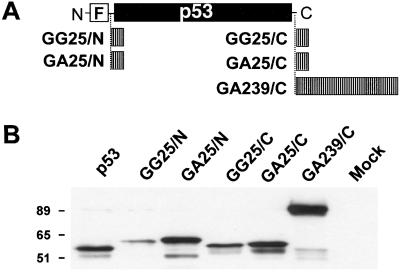
A set of GAr-containing chimeras was constructed to investigate the effect of the repeat on the turnover and function of the p53 tumor suppressor protein. A 239-aa-long GAr, derived from a natural EBV isolate (28), was inserted at the C terminus of p53 (Fig. 1A). In addition, a shorter 25-aa-long GAr or a control 25-aa-long glycine stretch was inserted at the N or C terminus of p53.
Figure 1.
Expression of p53-GAr chimeras in p53-negative Saos-2 cells. (A) Schematic representation of the FLAG-tagged (F) p53 chimeras containing N- or C-terminal GAr or GGr insertions. (B) Extracts of transiently transfected Saos-2 cells were examined by Western blotting with a monoclonal antibody specific for wild-type p53. Molecular weight markers are indicated.
Expression of the chimeras was investigated by transfection in the p53 negative osteosarcoma line Saos-2. Highest levels of expression were regularly detected in cells transfected with the p53-GA239/C chimeras followed by cells transfected with p53-GA25/N or p53-GA25/C, whereas the expression was consistently lower in cells transfected with wild-type p53 or the p53-GG25/N and p53-GG25/C chimeras (Fig. 1B).
The GAr Protects p53 from Mdm2- and HPV-E6-Induced Proteolysis.
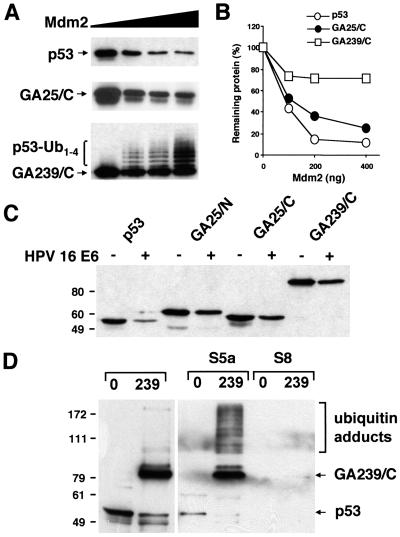
Expression of p53 was investigated by Western blot analysis of Saos-2 cells cotransfected with plasmids expressing the various p53 chimera and the ubiquitin ligase Mdm2. As expected, the steady-state levels of wild-type p53 were dramatically decreased in the presence of Mdm2 in a dose-dependent manner (Fig. 2 A and B). Insertion of a 25-residue GAr resulted in modest protection from Mdm2-induced degradation, whereas a more dramatic effect was achieved by insertion of a 239-residue-long repeat, in agreement with our earlier observation that the effect of the GAr is length-dependent (23). A distinct ladder of regularly spaced high-molecular weight species was detected in cells expressing the strongly stabilized p53-GA239/C, suggesting that the chimeras are still subject to Mdm2-induced ubiquitination. In accordance with the notion that ubiquitinated p53 is rapidly degraded by the proteasome, this ladder was not detected in cells expressing the wild-type molecule.
Figure 2.
The p53-GAr chimeras are resistant to Mdm2- and HPV-E6-induced degradation. (A) Saos-2 cells were transfected with 100 ng of the indicated p53-encoding plasmid together with 0, 100, 200, or 400 ng of an Mdm2-expression plasmid. Total cell extracts collected after 16 h were subjected to Western blot analysis with a monoclonal anti-p53 antibody. The ladder of high-molecular weight p53 species is indicated. (B) Densitometry of the Western blots shown in A. (C) Saos-2 cells were transfected with 100 ng of the indicated p53-encoding plasmid and 400 ng of control pcDNA3 (−) or HPV16-E6-expression plasmid (+). Experimental details as in A. The molecular weight markers are indicated. (D) The human 19S proteasome cap subunits S5a and S8 were expressed in bacteria as GST-fusion proteins, immobilized onto glutathione Sepharose, and analyzed in column-binding assays with lysates of HeLa cells transfected with wild-type FLAGp53 (0) or FLAGp53 containing a 239-aa GAr (239). Western blots were probed with a monoclonal anti-p53 antibody. A short exposure of 10% of the input is shown (Left). Species of p53 that interact with S5a and high-molecular weight p53 species are indicated.
Next we tested whether the p53 chimeras were also resistant to proteolysis induced by the E6 protein of HPV16. Cotransfection of Saos-2 cells with wild-type p53 together with a 4-fold excess of HPV16-E6 resulted in a dramatic decrease of p53 expression, whereas p53-GAr chimeras containing 25- or 239-aa-long repeats were only modestly affected (Fig. 2C). The stabilization was not caused by a GAr-induced abrogation of the interaction between E6 and p53, because a GST-E6 fusion protein interacted equally well with wild-type p53- and GAr-containing chimeras in pull-down assays (data not shown). The stronger effect of the repeats in this experimental set-up is probably explained by the presence of physiological levels of the E6-AP ligase as compared with huge overexpression of Mdm2.
The accumulation of high-molecular weight p53-GAr chimeras prompted us to investigate whether these putative polyubiquitinated species can interact with the proteasome. Pull-down assays were performed by using as bait a GST-S5a fusion protein, because this is the only known polyubiquitin-binding subunit of the 19S cap (29). A GST-fusion construct of the S8 ATPase subunit of the 19S cap was included as control. Input extracts from transiently transfected HeLa cells contained mainly unmodified p53 or p53-GA239/C (Fig. 2D), but longer exposure typically revealed the presence of high-molecular weight species, especially in lysates of p53-GA239/C-expressing cells (not shown). The high-molecular weight p53-GA239/C species were strongly enriched in the GST-S5a pull-down, whereas there was no detectable interaction between S5a and wild-type p53 (Fig. 2D). Thus the polyubiquitinated p53-GA239/C can still interact with the S5a subunit. The small amount of unmodified wild-type p53 observed in the S5a pull-down was comparable to the signal obtained with several irrelevant proteins and is most likely because of nonspecific interaction. On the contrary, a significant amount of the unmodified p53-GA239/C polypeptide was retained in the GST-S5a pull-down. This interaction was abrogated by incubation with an excess of GAr peptides (not shown), which did not affect the binding of the high-molecular weight p53-GAr species, suggesting that the GAr may directly bind to the S5a subunit. As expected, the S8 subunit did not bind p53 or the p53 chimeras (Fig. 2D).
The p53-GAr Chimeras Retain the Functional Properties of Wild-Type p53.
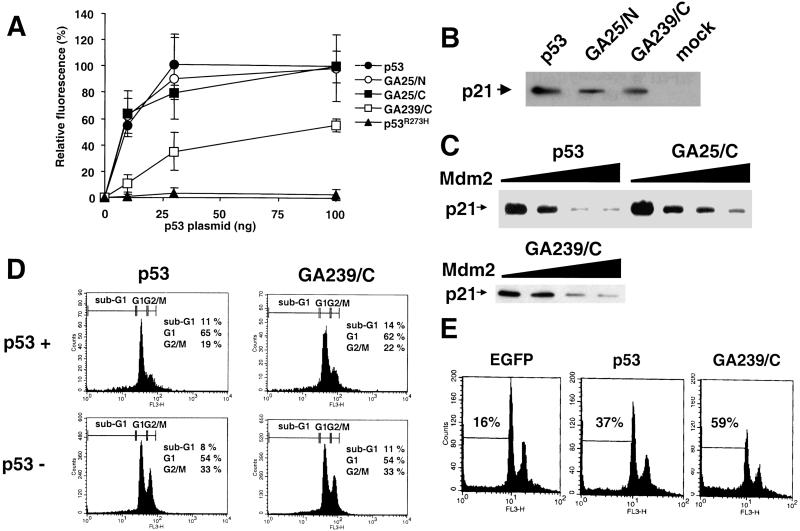
To investigate whether the GAr chimeras retain the transcriptional activity of wild-type p53, Saos-2 cells were cotransfected with p53-encoding plasmids and a reporter plasmid containing the EGFP gene under the control of a p53-responsive element. Expression of wild-type p53 resulted in a dose-dependent activation of transcription, whereas EGFP was not induced in cells expressing the DNA-binding mutant p53R273H (Fig. 3A) (30). Chimeras containing a 25-aa-long GAr were as active as wild-type p53, whereas the p53-GA239/C chimera showed an impaired transcriptional activity that reached ≈55% of the wild-type p53 effect at the highest plasmid concentration tested. The p53-target gene p21WAF/CIP1 was up-regulated in Saos-2 cells transfected with p53 or p53-GAr expressing plasmids, further confirming the transactivating competence of the chimeras (Fig. 3B).
Figure 3.
The p53-GAr chimeras are transcriptionally active. (A) Saos-2 cells were cotransfected with 400 ng of a plasmid expressing EGFP under the regulation of a p53-responsive element and increasing amounts of the indicated p53-encoding plasmids. EGFP fluorescence was measured by flow cytometry 16 h after transfection. The mean fluorescence intensity of cells transfected with 100 ng of p53-encoding plasmid was standardized as 100%. Mean ± SD from three independent experiments. (B) Expression plasmids encoding wild-type FLAGp53 or the indicated chimeras were transfected into Saos-2 cells, and the induction of endogenous p21WAF/CIP1 was examined by Western blot analysis with a monoclonal anti-p21WAF/CIP1 antibody. (C) Saos-2 cells were cotransfected with 100 ng of FLAGp53-, FLAGp53-GA25/C-, FLAGp53-GA239/C-expressing plasmid, and 0, 100, 200, and 400 ng of Mdm-2-expressing plasmid. p21WAF/CIP1 expression was detected by Western blot analysis. (D) Cell cycle analysis of Saos-2 cells transfected with a FLAGp53- or FLAGp53-GA239/C-expressing plasmid. P53 expression and cell cycle distribution were detected after 72 h by staining with an anti-p53 antibody followed by propidium iodide staining. The cell cycle distribution of p53-expressing (p53+) and p53-negative (p53−) subpopulations is shown. The percentages of cells in the sub-G1, G1, and G2/M phases of the cell cycle are indicated. (E) Plasmids encoding EGFP alone or together with the indicated p53 variants were expressed in a Saos-2 clone yielding high transfection efficiency. The cells were stained with propidium iodide 20 h after transfection and analyzed by flow cytometry. The samples were gated for EGFP expression. The percentage of cells with sub-G1 DNA content is indicated. One representative experiment of three (C–E).
Mdm2 can inactivate p53 by binding to the transcriptional activation domain (31). We asked therefore whether the stable p53-GAr chimeras may remain transcriptionally active in the presence of Mdm2. Expression of Mdm2 reduced the induction of p21WAF1/CIP1 by p53, p53-GA25/C and p53-GA239/C (Fig. 3C). However, consistently higher levels of p21WAF1/CIP1 were detected in cells expressing the p53-GAr chimeras, which was more evident in the presence of 2- and 4-fold excess of the Mdm2 plasmid.
P53 controls cell proliferation and survival by inducing G1/G2 cell cycle arrest and apoptosis (1). We asked therefore whether the p53-GAr chimeras have retained these functional properties of p53. Expression of p53 or the chimeras in our commonly used Saos-2 cells resulted in a reduction in the percentage of cells in G2/M and increase of cells in G1 (Fig. 3D). The effect was specific, because cells transfected with the p53R273H mutant showed a cell cycle distribution identical to that of the p53-negative population (data not shown). However, apoptosis was induced only in a small proportion of the p53-expressing cells. It has been reported that, whereas low p53 expression induces cell cycle arrest, higher expression levels are required for induction of apoptosis (32). We turned therefore to a different Saos-2 clone that consistently yields higher transfection efficiency. In addition, wild-type p53 and the p53-GA239 chimera were subcloned in an EGFP-expressing plasmid to allow direct detection of the transfected cells. Expression of p53 induced a more then 2-fold increase in the percentage of apoptotic cells compared with the empty EGFP vector, and a further increase was observed in cells expressing the p53-GA239 chimera (Fig. 3E).
The p53-GAr Chimeras Are Stabilized in Cells with Accelerated p53 Turnover.
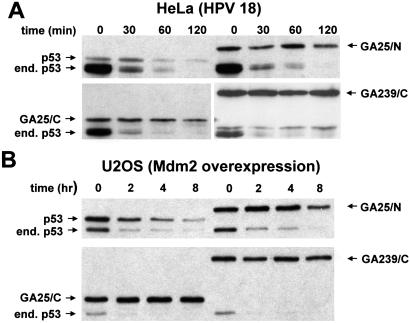
Accelerated p53 proteolysis was demonstrated in many tumor cell types that express wild-type p53. We asked therefore whether the p53-GAr chimeras are stabilized and functionally active also in tumor cells that carry oncogenic HPV strains or express levels of Mdm2 sufficient to inactivate the endogenous protein. Analysis of p53 turnover in the HPV18 positive cervical carcinoma line HeLa demonstrated that, whereas ectopic wild-type p53 had a half-life only slightly longer than the ≈20 min determined for the endogenous p53, chimeras containing 25- or 239-aa-long GAr domains had a significantly longer half-life (Fig. 4A). Similar results were obtained in two HPV16-positive cervix carcinoma cell lines, CasKi and SiHa (data not shown). The GAr chimeras exhibited a significantly prolonged half-life also in the U2OS osteosarcoma cell line (Fig. 4B) that expresses high levels of Mdm2 (13).
Figure 4.
The p53-GAr chimeras are stable in tumor cells with accelerated p53 degradation. The HPV18-E6-positive cervix carcinoma cell line HeLa (A) or the Mdm2-overexpressing osteosarcoma cell line U2OS (B) was transiently transfected with FLAGp53 or the indicated FLAGp53-GAr chimeras. After 16 h, the cells were incubated with 60 μg/ml of cycloheximide, and cell extracts were harvested after the indicated incubation times. Expression of p53 was detected by Western blotting with a monoclonal anti-p53 antibody. The endogenous p53 (end.) and ectopic FLAGp53 products are indicated.
The growth inhibitory effect of p53 variants has been compared by monitoring the outgrowth under the selective condition of cells transfected with plasmids encoding p53 together with a resistance gene (33). We have used this strategy to ask whether the stable p53 chimeras could withstand functional inactivation under conditions of sustained p53 turnover as observed in malignant cells. In accordance with the known growth inhibitory activity of p53, transfection of the Mdm2 overexpressing U2OS cells with a standard amount of wild-type p53-expressing plasmid resulted in more then 80% decrease in the number of geneticin-resistant colonies compared with the empty vector control (Table 1). The growth inhibitory effect was further enhanced in cells transfected with the p53-GAr chimeras with a ranking order that appeared to correlate with their transcriptional activity (compare Table 1 and Fig. 3A). The increased growth inhibitory activity of the chimeras was further confirmed when the cells were transfected with a 50-fold lower amount of plasmids. Under these conditions, wild-type p53 induced only a modest decrease in the number of colonies compared with the empty vector. In contrast, the p53-GAr chimeras still induced ≈50% decrease in the number of colonies. Similar results were obtained when the colony formation assay was performed in HPV18-positive HeLa cells (not shown).
Table 1.
Colony formation assay
| 500 ng of plasmid
|
10 ng of plasmid
|
|||
|---|---|---|---|---|
| Number of colonies* | % inhibition | Number of colonies* | % inhibition | |
| Empty vector | 403 | 0 | 511 | 0 |
| FLAGp53 | 65 | 84 | 433 | 15 |
| FLAGp53-GA25/C | 9 | 98 | 281 | 45 |
| FLAGp53-GA239/C | 49 | 88 | 241 | 53 |
U20S cells were transfected with 500 or 10 ng of the indicated plasmids. After overnight recovery, the cells were split into duplicate wells of a 24-well plate (≈8,000 cells per well) and grown for 2 weeks in the presence of 0.5 mg/ml of Geneticin. The number of Geneticin-resistant colonies and the percentage of p53-induced inhibition compared to the empty vector control are indicated.
Mean of duplicates. One representative experiment of three performed by transfecting 500 ng of plasmids and one performed with 10 ng of plasmid are shown.
Discussion
The involvement of the cellular Mdm2 and the viral E6 in the accelerated turnover of p53 in many malignancies combined with our previous identification of a viral GAr that blocks proteasomal degradation in cis prompted us to investigate the possibility of generating stable p53 variants by introduction of this repeat. We anticipated that, if functional, such stabilized p53 molecules could be of potential interest for gene therapy applications, as they could have tumor suppressor activity in tumors that lack endogenous p53 as well as in a large number of tumors where p53 is functionally inactivated by accelerated degradation. Moreover, we expected that studies of the GAr in the context of p53 could yield new information on the mechanism of action of the viral inhibitor because of the well characterized degradation pathway of p53 and the simultaneous targeting by two unrelated ubiquitin ligases. We have indeed found that the p53-GAr chimeras are stable in the presence of the three identified oncogenes directly involved in ubiquitin/proteasome-dependent inactivation of p53: HPV16-E6, HPV18-E6, and Mdm2. Moreover, these p53 chimeras maintain the capacity to transcriptionally activate p53 target genes. Importantly, the chimeric p53 molecules are fully competent to induce cell cycle arrest and apoptosis and have improved growth inhibitory activity in tumor cell lines that express elevated levels of Mdm2 or harbor oncogenic HPV.
Our finding that p53-GAr chimeras have improved growth inhibitory activity in cells that overexpress Mdm2 has interesting implications in view of the complex mechanism of action of this ligase. Although ubiquitination and proteolysis are believed to play a pivotal role in the inactivation of p53 (34), other inhibitory functions of Mdm2 have been described (35). Mdm2 was shown to promote the nuclear export of ubiquitinated p53 (7), which would be sufficient to prevent the activation of p53 target genes. Interestingly, we observed exclusive nuclear localization for each of the chimeras and could not demonstrate cytosolic relocalization even in the presence of Mdm2 concentrations sufficient to induce a dramatic accumulation of polyubiquitinated adducts (data not shown). It is thus possible that the GAr acts on events, between ubiquitination and proteasomal degradation, that are required for both nuclear export and proteolysis. Another plausible explanation is that the p53-GAr chimeras, once transported to the cytosol, are rapidly shuttled back to the nucleus after deubiquitination. Besides targeting p53 for proteolysis and nuclear export, Mdm2 inhibits p53 also by steric blockage of the transcriptional activation domain of p53 (31). Importantly, in a colony formation assay with U2OS cells, which display elevated levels of Mdm2, the p53-GAr chimeras had improved growth inhibitory potential compared with wild-type p53, arguing that proteasome-independent inhibition of p53 by Mdm2 is of minor importance under physiologic conditions (34).
The finding that the GAr could protect p53 from degradation induced by two ubiquitin ligases that recognize independent targeting signals suggests that the inhibitory domain may interfere with a common event downstream of ubiquitination. This was earlier inferred from the observation that EBNA4-GAr chimeras are efficiently ubiquitinated in a cell free system (17) and by the accumulation of ubiquitinated IκBα-GAr chimeras in cell extracts treated with inhibitors of deubiquitination enzymes (20). It is noteworthy that, even though the ubiquitination of these substrates might have been unaffected, only unconjugated chimeras were regularly detected in cell lysates, which led us to speculate that the GAr chimeras may be caught in a perpetual cycle of ubiquitination and deubiquitination that could preclude interaction with the proteasome. By coexpressing the p53-GAr chimeras with high amounts of Mdm2, we observed accumulation of polyubiquitinated GAr containing proteins in cells. This accumulation supports the notion that the inhibitory domain acts on a postubiquitination event.
We have previously shown that the stabilizing effect of the GAr is inversely proportional to the strength of the degradation signal (23). By monitoring the degradation of p53 chimeras containing GAr domains of different lengths in cells that express increasing amounts of Mdm2, we have now provided direct evidence that the rate of ubiquitination is a critical parameter in determining the length of the repeat required for the inhibitory effect. We have earlier speculated that long repeats may strengthen the interaction of ubiquitinated GAr-containing substrates with a putative partner, in a way analogous to the requirement of multiple ubiquitin recognition signals to trigger progressive degradation (23). An interesting possibility is that such interaction may alter the binding of the polyubiquitinated substrate to the proteasome, which prompted us to investigate whether polyubiquitinated p53 could bind to S5a, the only identified ubiquitin-binding subunit of the regulatory particle (29). By using a coimmunoprecipitation strategy, we were earlier unable to demonstrate interaction of polyubiquitinated IκBα-GAr chimeras with the proteasome, whereas a weak interaction was demonstrated for wild-type IκBα under the same conditions (20). It is noteworthy that, to promote the chain of events that lead to proteolysis, all interactions of the proteasome with ubiquitinated substrates must be relatively weak and restricted in time. We have therefore reexamined this question by using a more sensitive GST pull-down strategy (36) and succeeded in demonstrating interaction of polyubiquitinated p53-GA239/C with the S5a subunit. These findings are consistent with the possibility that, although not precluding the binding of ubiquitinated substrates to the proteasome, the GAr may affect the outcome of this interaction, leading to the rapid release of functionally unharmed proteins. This model is particularly attractive in the light of recent findings suggesting that a specific binding site for misfolded proteins in the 19S cap may trigger refolding rather then degradation of the substrate (37, 38).
In conclusion, we have now provided compelling evidence that the EBV GAr can act as a selective inhibitor of ubiquitin-dependent proteolysis that counteracts a broad range of targeting signals and ubiquitin ligases. The demonstration that p53-GAr chimeras remain functionally competent suggests that insertion of the viral repeat could provide a convenient strategy for stabilization of a wide variety of proteins that are of potential interest for gene replacement therapies.
Acknowledgments
We gratefully acknowledge the excellent technical support of Marianne Jellne, Anthony Chen, and Niloofar Rasti. We thank Karen H. Vousden, Bert Vogelstein, and Vladimir J. Bykov for the kind gifts of reagents. This investigation was supported by grants awarded by the Swedish Cancer Society, the Swedish Foundation for Strategy Research, the Swedish Research Council, the Petrus och Augusta Hedlunds Stiftelse, and the Karolinska Institute. S.H. and N.P.D. were supported by fellowships awarded by the European Commission Training and Mobility Program (ERBFMRXCT960026). A.L. is a fellow of a Joint MSc/Ph.D. Program between the Medical Academy of Latvia (AML) and the Karolinska Institute (KAMP).
Abbreviations
- EBNA1
EBV nuclear antigen 1
- EBV
Epstein–Barr virus
- E6-AP
E6-associated protein
- GAr
glycine-alanine repeat
- GST
glutathione S-transferase
- HPV
human papilloma virus
- Mdm2
murine double minute 2
- EGFP
enhanced green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Vousden K H. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Lane D, Levine A J. Nature (London) 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 4.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 5.Honda R, Tanaka H, Yasuda H. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 6.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 7.Boyd S D, Tsai K Y, Jacks T. Nat Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Xiong Y, Yarbrough W G. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 9.Ries S, Biederer C, Woods D, Shifman O, Shirasawa S, Sasazuki T, McMahon M, Oren M, McCormick F. Cell. 2000;103:321–330. doi: 10.1016/s0092-8674(00)00123-9. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh J K, Chan F S, O'Connor D J, Mittnacht S, Zhong S, Lu X. Mol Cell. 1999;3:181–193. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Liao J, Ruland J, Mak T W, Cohen S N. Proc Natl Acad Sci USA. 2001;98:1619–1624. doi: 10.1073/pnas.98.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 13.Florenes V A, Maelandsmo G M, Forus A, Andreassen A, Myklebost O, Fodstad O. J Natl Cancer Inst. 1994;86:1297–1302. doi: 10.1093/jnci/86.17.1297. [DOI] [PubMed] [Google Scholar]
- 14.Scheffner M, Munger K, Byrne J C, Howley P M. Proc Natl Acad Sci USA. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 16.Fang S, Jensen J P, Ludwig R L, Vousden K H, Weissman A M. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 17.Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci M G. Proc Natl Acad Sci USA. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blake N, Lee S, Redchenko I, Thomas W, Steven N, Leese A, Steigerwald-Mullen P, Kurilla M G, Frappier L, Rickinson A. Immunity. 1997;7:791–802. doi: 10.1016/s1074-7613(00)80397-0. [DOI] [PubMed] [Google Scholar]
- 19.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen P M, Klein G, Kurilla M G, Masucci M G. Nature (London) 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 20.Sharipo A, Imreh M, Leonchiks A, Imreh S, Masucci M G. Nat Med. 1998;4:939–944. doi: 10.1038/nm0898-939. [DOI] [PubMed] [Google Scholar]
- 21.Powis S H. Nat Med. 1998;4:887–888. doi: 10.1038/nm0898-887. [DOI] [PubMed] [Google Scholar]
- 22.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 23.Dantuma N P, Heessen S, Lindsten K, Jellne M, Masucci M G. Proc Natl Acad Sci USA. 2000;97:8381–8385. doi: 10.1073/pnas.140217397. . (First Published July 11, 2000; 10.1073/pnas.140217397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barak Y, Gottlieb E, Juven-Gershon T, Oren M. Genes Dev. 1994;8:1739–1749. doi: 10.1101/gad.8.15.1739. [DOI] [PubMed] [Google Scholar]
- 25.Crook T, Ludwig R L, Marston N J, Willkomm D, Vousden K H. Virology. 1996;217:285–292. doi: 10.1006/viro.1996.0115. [DOI] [PubMed] [Google Scholar]
- 26.Chandar N, Billig B, McMaster J, Novak J. Br J Cancer. 1992;65:208–214. doi: 10.1038/bjc.1992.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meissner J D. J Gen Virol. 1999;80:1725–1733. doi: 10.1099/0022-1317-80-7-1725. [DOI] [PubMed] [Google Scholar]
- 28.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, et al. Nature (London) 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 29.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 30.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 31.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, Ko L J, Jayaraman L, Prives C. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 33.Lin J, Jin X, Page C, Sondak V K, Jiang G, Reynolds R K. Cancer Res. 2000;60:5895–5901. [PubMed] [Google Scholar]
- 34.Yap D B, Hsieh J K, Lu X. J Biol Chem. 2000;275:37296–37302. doi: 10.1074/jbc.M004359200. [DOI] [PubMed] [Google Scholar]
- 35.Freedman D A, Wu L, Levine A J. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Y, Varshavsky A. Proc Natl Acad Sci USA. 2000;97:2497–2502. doi: 10.1073/pnas.060025497. . (First Published February 25, 2000; 10.1073/pnas.060025497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strickland E, Hakala K, Thomas P J, DeMartino G N. J Biol Chem. 2000;275:5565–5572. doi: 10.1074/jbc.275.8.5565. [DOI] [PubMed] [Google Scholar]
- 38.Braun B C, Glickman M, Kraft R, Dahlmann B, Kloetzel P M, Finley D, Schmidt M. Nat Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]