Abstract
Inactivation of the recently identified murMN operon in penicillin-resistant strains of Streptococcus pneumoniae was shown already to cause two major effects: elimination of branched-structured muropeptides from the cell wall and complete loss of penicillin resistance. We now show that cells with inactivated murMN also have a third phenotype: an increased susceptibility to lysis when exposed to low concentrations of fosfomycin, d-cycloserine, vancomycin, and nisin, indicating a wide-spectrum hypersensitivity to inhibitors of both early and late stages of cell wall biosynthesis. Mutants of murMN also lysed faster than the parental strain when treated with the detergent deoxycholate. Several different alleles of murM cloned in plasmid pLS578 and introduced into a murM deletion mutant of the penicillin-resistant strain Pen6 were able to reconstitute each one of the three mutant phenotypes: the highly branched cell wall structure, original high level of penicillin resistance, and normal sensitivity to lysis. In a penicillin-susceptible strain the same experiments caused increased concentration of cell wall branched peptides and suppression of sensitivity to antibiotic induced lysis. The observations suggest that the murMN operon plays a key role in the regulation of a stress-response pathway that can be triggered by perturbation of cell wall biosynthesis in S. pneumoniae.
Several penicillin-resistant clones of Streptococcus pneumoniae produce abnormal cell walls in which a high proportion of the stem peptide lysine residues carry short (seryl alanine or alanyl alanine) branches (1). The genetic determinants involved with the branching process, the murMN operon, were identified recently (2). Penicillin-resistant isolates with highly branched cell walls were shown to carry murM alleles that contained divergent sequences as compared with the murM genes of penicillin-susceptible pneumococci (3). The two proteins, MurM (407 amino acids) and MurN (411 amino acids), are involved with the addition of the first and second amino acid residues, respectively, to the cell wall precursor stem peptide at the lipid II stage of biosynthesis (4, 5). Inactivation of murMN by insertion-duplication mutagenesis did not interfere with growth of the bacteria but caused two phenotypes: elimination of branched peptides from the cell wall and complete loss of penicillin resistance (2). The purpose of the studies described in this communication was to follow up these earlier findings by the use of a deletion mutant of murM and complementation experiments. The surprising observation made was that inactivation of murM also caused a third phenotype: increased sensitivity of the bacteria to lysis by cell wall inhibitors.
Experimental Procedures
Strains and Growth Conditions.
All strains and plasmids used in this study are listed in Table 1. S. pneumoniae strains were grown in a casein-based semisynthetic medium (C + Y) at 37°C without aeration as described (11). S. pneumoniae strains containing pLS578 (10) or its derivatives were grown in the presence of 1 μg/ml tetracycline. Escherichia coli strains containing the pGEM-3Z plasmid or its derivatives were grown in LB medium in the presence of 100 μg/ml ampicillin (Sigma). S. pneumoniae and E. coli strains containing the pJDC9 plasmid (9) or the erm marker were grown in the presence of 1 μg/ml and 1 mg/ml erythromycin, respectively.
Table 1.
Relevant properties of the strains and plasmids used in this study
| Relevant characteristics | Reference | |
|---|---|---|
| S. pneumoniae strains | ||
| R36A | Penicillin-susceptible laboratory strain | (6) |
| Pen6 | Penicillin-resistant transformat of R6Hex with donor DNA from the penicillin-resistant strain 8249 | (7) |
| KY17 | Penicillin-resistant nontypable clinical isolate from a U.S. collection | (3, 8) |
| Pen6murMN | Pen6 mutant with murMN operon interrupted by insertion-duplication mutagenesis with pJDC9 | (2) |
| Pen6ΔmurM | Pen6 with murM gene deleted and replaced by erm gene from pJDC9 | (5) |
| Pen6murMR36A | Pen6 carrying murM allele of R36A in the chromosome | (4) |
| Pen6ΔmurMpM3N through pM18 | Transformants of Pen6ΔmurM carrying various constructs cloned into pLS578 | (5) |
| R36ApM1 through pM3N | Transformants of R36A carrying various constructs cloned into pLS578 | (5) |
| Escherichia coli strain | ||
| DH5α | RecA endA1 gyrA96 thi-1 hsdR17 supE44relA1 φ80 dlac ZΔ M15 | BRL |
| Plasmids | ||
| pJDC9 | E. coli plasmid EryR | (9) |
| pLS578 | S. pneumoniae plasmid TetR | (10) |
| pM1 through pM9N: | Plasmid pLS578, into which various murM alleles and constructs were cloned | (5) |
| pM1 | murM allele from strain R36A | (5) |
| pM2 | murM allele from strain DE1 | (5) |
| pM3 | murM allele from strain KY17 | (5) |
| pM3N | murM allele from strain KY17 plus the murN gene from strain R36A | (5) |
| pM9N | murM chimeric allele constructed from strains DE1 and R36A plus the murN gene from R36A | (5) |
| pM18 | murM mutant allele from strain KY17 with the residue at position 27 mutated from a glutamine to a glutamate, plus the murN gene from strain R36A | (5) |
In the experiments with antibiotic-induced or detergent-induced lysis, the strains were grown in C + Y until early exponential phase (OD590 of 0.1–0.2), at which time different concentrations of antibiotics were added, and the optical density of the cultures was followed.
The ability of lysozyme to trigger lysis prematurely in the stationary phase was tested by adding lysozyme at different concentrations to cultures of Pen6 and its murMN mutant at an OD590 of 0.2 and following the rate of decline of optical densities after the cultures entered the stationary phase.
DNA Techniques.
All routine DNA manipulations were performed by using standard methods (12, 13). Chromosomal DNA from S. pneumoniae was isolated as described (14). Plasmids were isolated by using the Wizard Plus Minipreps or Midipreps DNA purification system (Promega), and PCR products were purified by using the Wizard PCR Preps DNA purification system (Promega). Oligonucleotides were purchased from GIBCO/BRL Life Technologies. DNA sequencing was done at the Rockefeller University Protein/DNA Technology Center with the BigDye terminator cycle-sequencing method and either the 3700 DNA analyzer for capillary electrophoresis or the ABI Prism 377 DNA sequencers for slab-gel electrophoresis. Nucleotide and derived amino acid sequences were analyzed by using DNASTAR software.
Construction of S. pneumoniae Lacking murM.
To isolate a mutant of the penicillin-resistant strain Pen6 lacking the murM gene, we transformed Pen6 with a PCR fragment that included the 2-kb fragment from pJDC9, carrying the erm marker, flanked by the upstream and downstream regions of murM. The details of the preparation of this murM null mutant were described in a recent publication (5).
The transforming DNA used to prepare the murM null mutant of strain Pen6 was a PCR fragment of plasmid pZOO11, which contains the erm marker flanked by the upstream and downstream regions of murM, amplified with primers ZOO30 (5′-ATATTCTCTACGTTCAGAGG-3′) and ZOO45EC (5′-AGCGAATTCGGTTTTGACTACTACACGGC-3′) (5). The conditions used for the PCR reactions were: 94°C for 5 min; 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 2 min; and one final extension step of 72°C for 5 min. This PCR program was used for all amplifications except the extension time at 72°C, which was different depending on the size of the PCR fragment to be amplified.
The deletion of murM in the erythromycin-resistant Pen6ΔmurM mutant was confirmed by PCR amplification and subsequent sequencing.
Transformation of the murM Null Mutant (Pen6ΔmurM) and the Penicillin-Susceptible Strain R36A with Plasmid-Borne murM Alleles.
Several murM constructs were prepared (5), each in the S. pneumoniae plasmid pLS578, which is a plasmid with a strong promoter that can replicate in S. pneumoniae (10). Three S. pneumoniae strains were used as the source of murM alleles: the penicillin-susceptible laboratory strain R36A and two penicillin-resistant isolates, strain KY17 producing serine-rich branches in the cell wall and strain DE1 producing alanine-rich branches (5).
The murM alleles cloned in the pneumococcal plasmid pLS578 were introduced into the Pen6 murM null mutant (Pen6ΔmurM) or into R36A by genetic transformation by using previously described methods (15).
Cell wall preparation and analysis of the cell wall stem peptides were done by using a published method (16, 17) after mechanical breaking of the cells by glass beads (5). Cell wall material (2 mg) was suspended in 25 mM sodium phosphate buffer (pH 7.4) and treated with affinity-purified pneumococcal amidase (5–10 μg) at 37°C for 18–24 h with constant stirring. The products were dried, the precipitate was washed with acetone, and the peptides were extracted with acetonitrile/isopropanol/water (25:25:50) containing 0.1% trifluoroacetic acid (16, 17). After removal of the solvents by evaporation in a SpeedVac, the peptides were dissolved in 0.1% trifluoroacetic acid and separated on a Shimadzu LC-10AVP HPLC system (17).
Results
Recovery of High Level Penicillin Resistance in the murM Null Mutant of Strain Pen6 by Complementation with Various murM Alleles Cloned in Plasmid pLS578.
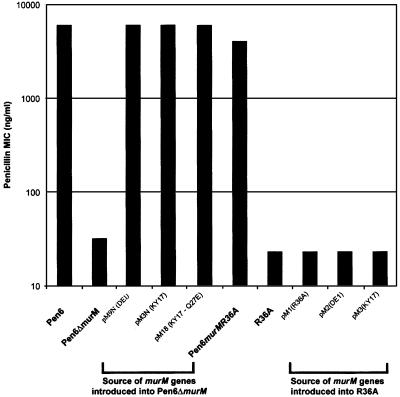
Loss of high level penicillin resistance in strain Pen6 upon inactivation of murMN by insertion-duplication mutagenesis has been described already (2). To rule out possible indirect and/or polar effects by the inserted plasmid (18, 19), a deletion mutant Pen6ΔmurM was constructed (5) to allow complementation. In Pen6ΔmurM, the murM gene was replaced by the erm marker from the plasmid pJDC9 (9) to prevent recombination with the plasmid-borne murM alleles introduced in the course of complementation experiments (18). Confirmation of the erythromycin-resistant Pen6 murM null mutant was obtained by sequencing the PCR fragment amplified with primers ZOO30 and ZOO45EC (which includes the murMN region). The penicillin minimum inhibitory concentration (MIC) of the parental strain Pen6 (6 μg/ml) was reduced to 0.032 μg/ml in both Pen6ΔmurM and the insertionally inactivated murMN mutant. Deletion of murM caused no affect on the growth rate or morphology of the cells. Complementation experiments were done to prove that the loss of penicillin resistance was caused by the inactivation of murM. The murMB5 allele from the penicillin-resistant strain KY17 (3, 4) was cloned together with the murN of R36A both under the control of the conserved promoter of plasmid pLS578, and the resulting construct pM3N was transformed into the Pen6ΔmurM mutant. After introduction of this plasmid into the deletion mutant the MIC increased to 6 μg/ml, indicating full recovery of parental level penicillin resistance (Fig. 1). Full recovery of penicillin resistance also was obtained in the chromosomally complemented Pen6murM36 and with pM9N in which the murM gene was a chimera: the first 250 amino acids were derived from the murM of the penicillin-susceptible strain R36A and the next 166 amino acids from the murM of the penicillin-resistant strain DE1 (5). The construct also carried murN from R36A. Complementation with pM18, a variant of pM3N in which the glutamine residue at position 27 in the murM gene was replaced by glutamic acid through site-directed mutagenesis, also resulted in full recovery of penicillin resistance. This substitution has no affect on the activity of murM (5). No increase in the penicillin MIC occurred when plasmid-borne murM alleles were introduced into the penicillin-susceptible strain R36A.
Figure 1.
Recovery of penicillin resistance in Pen6ΔmurM complemented with different murM alleles. The strain Pen6ΔmurM was transformed with plasmid pLS578 carrying different murM alleles and constructs. Several of the plasmid-borne murM alleles also were introduced into the penicillin-susceptible strain R36A. Penicillin MIC values were determined by the E test.
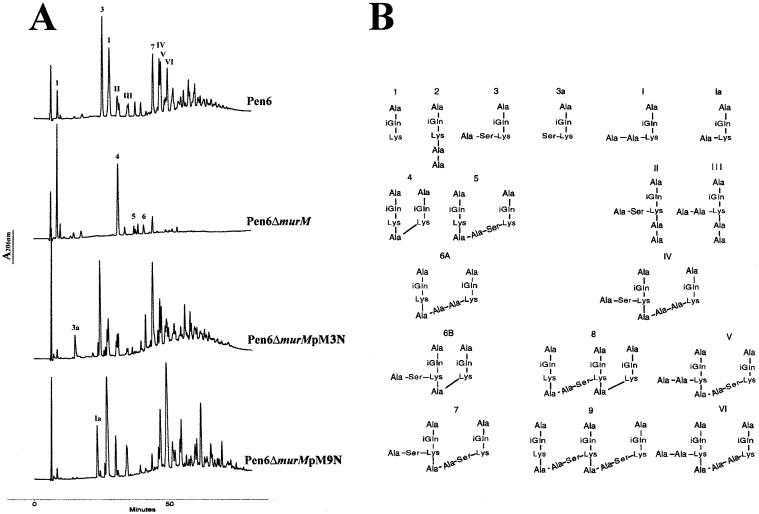
Analysis of the cell wall composition of Pen6ΔmurM showed a virtually complete disappearance of branched structured stem peptides from the cell wall peptidoglycan (Fig. 2). The composition of the peptidoglycan was indistinguishable from that of the already described Pen6murMN, which was obtained by insertion of a suicide plasmid in the murM gene (2). Cell wall analysis of the pM3N transformant showed a peptidoglycan rich in serine containing branched peptides, similar in composition to that of strain KY17 (5), the source of the murM allele in the plasmid (Fig. 2 and Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). The peptidoglycan of transformants carrying plasmid pM9N was rich in branched peptides in which alanine was the predominant first residue of the crossbridge, as expected from the specificity of the murM in strain DE1 (5).
Figure 2.
Cell wall composition of Pen6ΔmurM complemented with different murM alleles. (A) Cell walls from strain Pen6, its murM deletion mutant (Pen6ΔmurM), and two Pen6ΔmurM transformants, Pen6ΔmurMpM3N and Pen6ΔmurMpM9N complemented with murM alleles, were analyzed for peptidoglycan composition by HPLC as described in Experimental Procedures. (B) Structural assignments for the pneumococcal stem peptides. For determination of chemical structures see refs. 4 and 11.
Increased Sensitivity of murMN Mutants to Lysis by Inhibitors of Cell Wall Synthesis Other Than Penicillin.
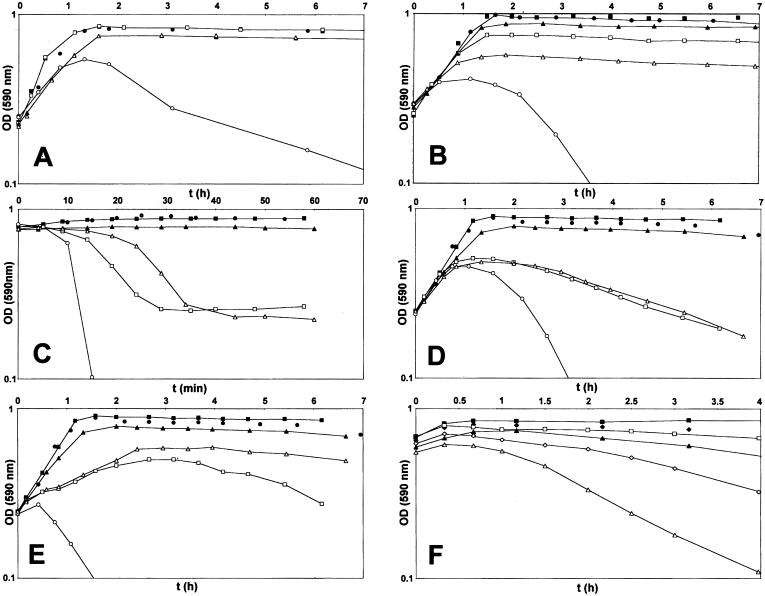
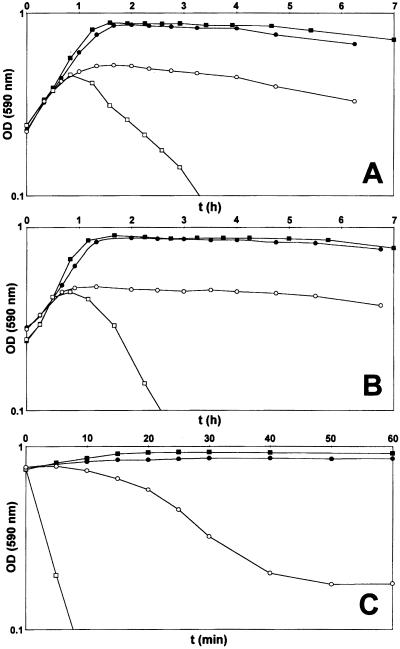
A careful scrutiny of the optical density profiles of cultures of Pen6 and its deletion mutant in the stationary phase of growth showed that spontaneous autolysis in the stationary phase, a characteristic property of S. pneumoniae, began earlier in the murM deletion mutant as compared with the parental strain Pen6. We followed up this observation by testing the lytic effect of a number of different cell wall inhibitors on cultures of strain Pen6 and its deletion mutant. Fig. 3 shows that each one of the five cell wall inhibitors tested and the detergent deoxycholate (DOC) caused unusually rapid lysis of cultures of the murM deletion mutant. In the case of cefotaxime, this lysis was related to the different MIC values of strain Pen6 (2.0 μg/ml) and its deletion mutant (0.03 μg/ml). However, the MIC values of Pen6 and Pen6ΔmurM for the rest of the drugs and DOC were either identical or differed only by a factor of 2. The abnormal lysis pattern was reversed in each case in complementation experiments in which the plasmid-borne murMN operon was introduced into the Pen6 deletion mutant (Fig. 3). Because in such plasmid transformants the majority of cell wall stem peptides had a branched structure (5), the results of the complementation experiments suggested a correlation between the proportion of branched peptides in the cell wall and the susceptibility of the bacteria to lysis during treatment with cell wall inhibitors. To test this correlation further, the experiments were repeated with the penicillin-susceptible strain R36A, which is exquisitely sensitive to the lytic and bactericidal effect of cell wall inhibitors (20) and the cell wall of which is composed mainly of linear stem peptides (1, 17). Cultures of strain R36A and a transformant derivative carrying the plasmid pM18 were treated with fosfomycin, d-cycloserine, and nisin at concentrations close to their respective MIC values. In contrast to the rapid lysis of R36A, the plasmid transformants showed suppression or delay (nisin) of the lytic response to the antibiotics (Fig. 4). Analysis of the cell wall of the plasmid transformants showed that introduction of the plasmid-borne murMN gene caused major changes in the composition of the cell wall, which became highly enriched in stem peptides with a branched structure (Fig. 6 and Table 3, which are published as supporting information on the PNAS web site).
Figure 3.
(A–F) Hypersensitivity of the murM deletion mutant Pen6ΔmurM to the lytic effect of cell wall inhibitors. Inhibitors were added to cultures of strain Pen6, Pen6ΔmurM, and Pen6ΔmurM carrying plasmid pM18 in the early exponential phase (OD590 of 0.2), and their effects on growth and lysis were followed by monitoring the optical density. Of the antimicrobial agents tested, only cefotaxime had different MIC values for the three strains used; the MIC was 2 μg/ml for Pen6 and Pen6ΔmurMpM18 and 0.03 μg/ml for Pen6ΔmurM. For the rest of the agents the MIC values for the three strains were the same or differed by only a factor of 2. The concentration of various drugs used was as follows: A, cefotaxime (0.05 μg/ml); B, D-cycloserine (50 μg/ml, 1 × MIC); C, nisin (1 μg/ml, 2 × MIC); D, fosfomycin (50 μg/ml, 1 × MIC); E, vancomycin (0.4 μg/ml, 1 × MIC); F, DOC (50 μg/ml and 100 μg/ml, 1 and 2 × MIC). The symbols used for A–E are as follows. Cultures with antibiotic: □, Pen6; ○, Pen6ΔmurM, ▵, Pen6ΔmurMpM18. Controls without antibiotics: ■, Pen6; ●, Pen6ΔmurM; ▴, Pen6ΔmurMpM18. Symbols for F are as follows. ■ and □, Pen6 and Pen6murMN without antibiotics, respectively; ♦ and ⋄, Pen6 and Pen6murMN with 50 μg/ml DOC, respectively; ▴ and ▵, Pen6 and Pen6murMN with 100 μg/ml DOC, respectively.
Figure 4.
Suppression of the lytic response to various cell wall inhibitors in strain R36A overexpressing murMN. Antibiotics were added to early exponential phase cultures of strain R36A and its plasmid transformant R36ApM18, and the effects of antibiotics on growth and lysis were followed by optical density determinations. Antibiotics were used at concentrations close to their MIC values. A, penicillin (0.025 μg/ml, 1–2 × MIC); B, fosfomycin (50 μg/ml, 1 × MIC); C, nisin (2 μg/ml, 4 × MIC). The symbols used are as follows. Cultures with antibiotics: □, R36A; ○, R36ApM18. Control cultures without antibiotics: ■, R36A; ●, R36ApM18.
Discussion
In a previous communication we described loss of antibiotic resistance in penicillin-resistant isolates of S. pneumoniae in which the murMN operon was inactivated by insertion-duplication mutagenesis (2, 4). The observations described in this communication confirm this finding; the penicillin MIC of the murMN deletion mutant Pen6ΔmurM was reduced from 6.0 μg/ml to 0.03 μg/ml, i.e., to the MIC range for penicillin-susceptible pneumococci. Normal levels of resistance were recovered in transformants of Pen6ΔmurM that received functional murM alleles cloned into a plasmid that can replicate in pneumococci. These experiments together provide unequivocal evidence for the critical importance of the murMN operon for penicillin resistance.
In the complementation experiments with the particular constructs used, full recovery of penicillin resistance was accompanied also by an increase in the proportion of branched peptides in the cell wall of transformants. On the other hand, the chemical composition of the branched peptides in various transformants differed depending on the source of the particular murM allele (3, 5). Serine-rich branched peptides were produced in transformants that received murM from strain KY17, whereas transformants with the murM strain from DE1 produced alanine-rich branches. It therefore seems that recovery of optimal penicillin resistance, although requiring functional murM (and murN) genes, does not depend on the reintroduction of the same murM allele that was resident originally in the resistant strain Pen6. The situation may be different in the case of strains with a very high level of resistance. In recent communications, Smith and Klugman (21, 22) confirmed and extended the role of murM in conferring penicillin resistance in a strain of S. pneumoniae that had an extremely high MIC value of 12 μg/ml. They found that transformation of penicillin resistance above a certain high MIC value was not possible by using only the cloned resistant penicillin-binding protein genes as donor DNA but also needed a portion of the murM gene from the resistant bacteria.
Our results also make it clear that the murMN operon, although necessary, is not sufficient to cause resistance to penicillin, because strain R36A transformed with the murM alleles from resistant strains remained fully susceptible to penicillin despite the fact that the cell walls of the transformants became greatly enriched for branched muropeptides (Fig. 1).
An unexpected property of murMN mutants described in this communication is the hypersensitivity of the bacterium to a variety of cell wall inhibitors. The most striking feature of this new phenotype was its lack of specificity; inhibitors of both early stages in cell wall synthesis (fosfomycin and d-cycloserine) as well as inhibitors of the late stages (vancomycin and nisin) induced a rapid loss of optical density in cultures of Pen6ΔmurM at antibiotic concentrations close to the MIC values. The agents also included DOC, a detergent that can induce autolysis of pneumococci by an unknown mechanism.
Rapid and extensive loss of viability followed by lysis is the most frequent response of S. pneumoniae and other bacteria to treatment with β-lactam antibiotics and other wall inhibitors. In the case of β-lactams the lethal pathway is initiated by the acylation of penicillin-binding proteins and terminates in the autolytic suicide of the cells. Earlier work from our laboratory has shown clearly that the mechanism of the killing and lytic effects of penicillin involved bacterial factors beyond the primary targets of penicillin (i.e., the penicillin-binding proteins; ref. 23). One of these factors is LytA (24), a pneumococcal murein hydrolase primarily responsible for autolytic phenomena in this bacteria. A second factor(s) that seems to control sensitivity of pneumococci to the cidal effect of penicillin has also been described (20). Unique bacterial mutants, so-called “penicillin-tolerant mutants” in which the lytic action alone or together with the bactericidal action of penicillin were suppressed, have been isolated in the laboratory from pneumococci (25) and also from E. coli (26) and Staphylococcus aureus (27).
Pen6 and Pen6ΔmurM were grown in the presence of sub MIC concentrations of penicillin. Little if any difference was observed in the susceptibility of cell walls isolated from these bacteria to hydrolysis by LytA in vitro (data not shown). Furthermore, inactivation of murMN also caused rapid loss of viability of the penicillin-treated bacteria under conditions when the activity of LytA was inhibited (20, 28).
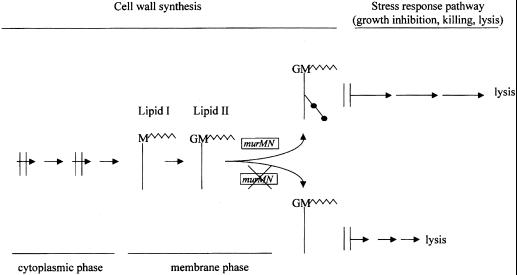
The profound influence of cell wall branching on lytic phenomena described in this communication suggests that the murMN operon occupies a critical position in the stress-response pathway triggered by perturbation of cell wall synthesis in S. pneumoniae (23). As a model we propose that the relevant activity of murMN modulating the lytic phenotype is the production of Lipid II carrying the dipeptide substituents on the ɛ amino group of the lysine residues in the cell wall precursor stem peptide (Fig. 5). The presence of the extra dipeptide residues in lipid II should increase the hydrophilic nature of the molecule, and the accumulation of these heptapeptides in the plasma membrane may suppress the assembly of the hypothetical holin-like proteins that were postulated to initiate the irreversible effects of cell wall inhibitors in pneumococci (20) and other bacteria (29–31).
Figure 5.
Model for the postulated role of the murMN operon in the regulation of bacterial response to perturbation of cell wall biosynthesis. The model assumes that the critical regulatory component that influences the lytic/cidal response of pneumococci to inhibitors of cell wall biosynthesis is the structure of lipid II. In bacteria with functional murMN, lipid II is composed of bactoprenol-linked disaccharide heptapeptides that cause a delay in the hypothetical membrane injury caused by interruption of cell wall biosynthesis (see double vertical lines in model). In bacteria with inhibited murMN the structure of lipid II is a bactoprenol-linked linear disaccharide pentapeptide that is unable to delay the membrane injury that initiates the bacterial stress response leading to the suicidal activation of factors including LytA.
In a previous communication it was proposed that the mechanism of irreversible effects (killing and lysis) by cell wall inhibitors involved some injury to the plasma membrane, the integrity of which is essential for the survival of the bacterium. The same injury then would allow the release of autolytic enzymes to the cell wall and initiate lysis of the bacteria. This model is formally analogous to models proposed for the mechanism of lysis by bacteriophages in which the release of phage progeny is preceded by membrane injury through the insertion of a holin protein into the bacterial membrane (32, 33). Our model also is analogous to the proposed mode of action of nisin at the level of lipid II (34).
It is interesting that the activity of the same murMN operon seems to be involved not only in penicillin resistance but in antibiotic tolerance as well. It is conceivable that the step that defines the MIC of a cell wall active agent may not only depend on the affinity of the drug targets for the drug but also on the status of a response pathway that translates the inhibition of target molecules to its cell biological consequences, i.e., the inhibition of the target cell. Lowering the sensitivity threshold of response to the perturbation of wall synthesis may become the rate-limiting step in drug susceptibility. The evidence about the involvement of murMN with both the penicillin MIC and sensitivity to lysis by a variety of wall inhibitors suggests that murMN may be a key component of such a sensory circuit.
Supplementary Material
Acknowledgments
We are grateful to Dr. Sandford Lacks for plasmid pLS578. Partial support for these studies was provided by a grant from the National Institutes of Health RO1 AI37275 and by the Irene Diamond Foundation. S.F. was supported by Grant BD/9071/96 from PRAXIS XXI from Fundação para a Ciência e Tecnologia.
Abbreviations
- MIC
minimum inhibitory concentration
- DOC
deoxycholate
References
- 1.Garcia-Bustos J, Tomasz A. Proc Natl Acad Sci USA. 1990;87:5414–5419. doi: 10.1073/pnas.87.14.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filipe S R, Tomasz A. Proc Natl Acad Sci USA. 2000;97:4891–4896. doi: 10.1073/pnas.080067697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filipe S R, Severina E, Tomasz A. J Bacteriol. 2000;182:6798–6805. doi: 10.1128/jb.182.23.6798-6805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filipe S R, Pinho M G, Tomasz A. J Biol Chem. 2000;275:27768–27774. doi: 10.1074/jbc.M004675200. [DOI] [PubMed] [Google Scholar]
- 5.Filipe S R, Severina E, Tomasz A. J Biol Chem. 2001;276:39618–39628. doi: 10.1074/jbc.M106425200. [DOI] [PubMed] [Google Scholar]
- 6.Avery OT, Macleod C M, McCarty M. J Exp Med. 1944;79:137–157. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zighelboim S, Tomasz A. Antimicrob Agents Chemother. 1980;17:434–442. doi: 10.1128/aac.17.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corso A, Severina E P, Petruk V F, Mauriz Y R, Tomasz A. Microb Drug Resist. 1998;4:325–337. doi: 10.1089/mdr.1998.4.325. [DOI] [PubMed] [Google Scholar]
- 9.Chen J D, Morrison D A. Gene. 1988;64:155–164. doi: 10.1016/0378-1119(88)90489-1. [DOI] [PubMed] [Google Scholar]
- 10.Lacks S A, Ayalew S, de la Campa A G, Greenberg B. Mol Microbiol. 2000;35:1089–1098. doi: 10.1046/j.1365-2958.2000.01777.x. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Bustos J F, Chait B T, Tomasz A. J Bacteriol. 1988;170:2143–2147. doi: 10.1128/jb.170.5.2143-2147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1996. [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Marmur J. J Mol Biol. 1961;3:208–218. doi: 10.1016/s0022-2836(61)80023-5. [DOI] [PubMed] [Google Scholar]
- 15.Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein L S, Piccoli L, Simon D, Morrison D A. J Bacteriol. 1996;178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Bustos J F, Chait B T, Tomasz A. J Biol Chem. 1987;262:15400–15405. [PubMed] [Google Scholar]
- 17.Severin A, Tomasz A. J Bacteriol. 1996;178:168–174. doi: 10.1128/jb.178.1.168-174.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez P, Espinosa M, Stassi D L, Lacks S A. J Bacteriol. 1982;150:692–701. doi: 10.1128/jb.150.2.692-701.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiraby J G, Fox M S. Proc Natl Acad Sci USA. 1973;70:3541–3545. doi: 10.1073/pnas.70.12.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreillon P, Markiewicz Z, Nachman S, Tomasz A. Antimicrob Agents Chemother. 1990;34:33–39. doi: 10.1128/aac.34.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith A M, Klugman K P. Microbial Drug Resist. 2000;6:105–110. doi: 10.1089/107662900419401. [DOI] [PubMed] [Google Scholar]
- 22.Smith A M, Klugman K P. Antimicrob Agents Chemother. 2001;45:2393–2396. doi: 10.1128/AAC.45.8.2393-2396.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomasz A. Annu Rev Microbiol. 1979;33:113–137. doi: 10.1146/annurev.mi.33.100179.000553. [DOI] [PubMed] [Google Scholar]
- 24.Ronda C, Garcia J L, Garcia E, Sanchez-Puelles J M, Lopez R. Eur J Biochem. 1987;164:621–624. doi: 10.1111/j.1432-1033.1987.tb11172.x. [DOI] [PubMed] [Google Scholar]
- 25.Tomasz A, Albino A, Zanati E. Nature (London) 1970;227:138–140. doi: 10.1038/227138a0. [DOI] [PubMed] [Google Scholar]
- 26.Kitano K, Tomasz A. J Bacteriol. 1979;140:955–963. doi: 10.1128/jb.140.3.955-963.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomasz A, de Vegvar M L. Eur J Clin Microbiol. 1986;5:710–713. doi: 10.1007/BF02013310. [DOI] [PubMed] [Google Scholar]
- 28.Filipe, S. R., Severina, E. & Tomasz, A. (2001) Microb. Drug Resist., in press. [DOI] [PubMed]
- 29.Groicher K H, Firek B A, Fujimoto D F, Bayles K W. J Bacteriol. 2000;182:1794–1801. doi: 10.1128/jb.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunskill E W, Bayles K W. J Bacteriol. 1996;178:5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayles K W. Trends Microbiol. 2000;8:274–278. doi: 10.1016/s0966-842x(00)01762-5. [DOI] [PubMed] [Google Scholar]
- 32.Garrett R, Fusselman R, Hise J, Chiou L, Smith-Grillo D, Schultz J, Young R. Mol Gen Genet. 1981;182:326–331. doi: 10.1007/BF00269678. [DOI] [PubMed] [Google Scholar]
- 33.Heinrich B, Lubitz W, Plapp R. Mol Gen Genet. 1982;185:493–497. doi: 10.1007/BF00334146. [DOI] [PubMed] [Google Scholar]
- 34.Wiedemann I, Breukink E, van Kraaij C, Kuipers O, Bierbaum G, de Kruijff B, Sahl H. J Biol Chem. 2000;276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.