Abstract
DNA microarrays were used to survey the adaptive genetic responses of Borrelia burgdorferi (Bb) B31, the Lyme disease spirochete, when grown under conditions analogous to those found in unfed ticks (UTs), fed ticks (FTs), or during mammalian host adaptation (Bb in dialysis membrane chambers implanted in rats). Microarrays contained 95.4% of the predicted B31 genes, 150 (8.6%) of which were differentially regulated (changes of ≥1.8-fold) among the three growth conditions. A substantial proportion (46%) of the differentially regulated genes encoded proteins with predicted export signals (29% from predicted lipoproteins), emphasizing the importance to Bb of modulating its extracellular proteome. For B31 cultivated at the more restrictive UT condition, microarray data provided evidence of a bacterial stringent response and factors that restrict cell division. A large proportion of genes were responsive to the FT growth condition, wherein increased temperature and reduced pH were prominent environmental parameters. A surprising theme, supported by cluster analysis, was that many of the gene expression changes induced during the FT growth condition were transient and largely tempered as B31 adapted to the mammalian host, suggesting that once Bb gains entry and adapts to mammalian tissues, fewer differentially regulated genes are exploited. It therefore would seem that although widely dissimilar, the UT and dialysis membrane chamber growth conditions promote more static patterns of gene expression in Bb. The microarray data thus provide a basis for formulating new testable hypotheses regarding the life cycle of Bb and attaining a more complete understanding of many aspects of Bb's complex parasitic strategies.
Borrelia burgdorferi (Bb), the etiological agent of Lyme disease (Lyme borreliosis), subsists within a complex enzootic life cycle that involves arthropod (Ixodes ticks) and mammalian (rodent) hosts (1). As Bb transitions between its two niches, dramatic physiological adaptations occur such as the reciprocal down-regulation of outer surface (lipo)protein (Osp)A and the up-regulation of OspC (2–6). However, the paucibacillary nature of Bb in both hosts has hampered a more thorough examination of the spirochete's differential gene expression profiles. To survey more broadly Bb's adaptive tendencies, various in vitro model systems have been used to mimic certain environmental cues. For example, elevated temperature, reduced pH, and an increase in Bb cell density, conditions that ostensibly mimic those during tick engorgement, have been shown to induce the reciprocal down-regulation of OspA, P22, Lp6.6 (group II proteins) and up-regulation of OspC, decorin-binding protein A (DbpA), OspF, Mlp-8, and the alternative σ factor RpoS (group I proteins; ref. 7). Recently, a novel regulatory pathway involving the control of RpoS by another alternative σ factor, RpoN, has been strongly implicated in the adaptive changes to these interdependent environmental factors (8). A model system also has been developed in which Bb is cultivated in dialysis membrane chambers (DMCs) implanted into rat peritoneal cavities to obtain spirochetes in a mammalian host-adapted state (9). However, studies on Bb differential expression under this or other growth conditions thus far have been practically restricted to examinations of only relatively few proteins at any given time.
A more complete analysis of the adaptive responses occurring during Bb's life cycle will be invaluable for understanding many aspects of the spirochete's transmission, virulence expression, and immune evasion and for the potential identification of new Lyme disease vaccine candidates. To this end, DNA microarrays (10) are being applied now to survey globally the adaptive gene responses of pathogenic bacteria (11–13). The application of microarrays for analysis of the Bb transcriptome is now feasible because of the availability of complete genomic sequence information (14, 15). In this study, we demonstrate the utility of applying DNA microarrays for the global analysis of gene expression patterns in Bb.
Materials and Methods
Bb and Growth Conditions.
The low-passage B31 strain of Bb (B31) used for genome sequencing (14, 15) was obtained (MedImmune, Gaithersburg, MD), and virulence was confirmed by intradermal injection of 1 × 103 borreliae in C3H/HeJ mice (16). Spirochetes from not more than three in vitro serial passages were cultivated at 37°C to the mid-logarithmic phase (≈5 × 106 B31 per ml) in complete Barbour–Stoenner–Kelly (BSK-H) medium (Sigma; ref. 17). For in vitro cultivation, the initial culture was diluted to 1 × 103 B31 per ml in either BSK-H medium held at 23°C and adjusted to pH 7.5 or in BSK-H medium prewarmed to 37°C and adjusted to pH 6.8 (7). Spirochetes from two independent cultivations at each growth condition were harvested at the late-logarithmic phase of growth (≈2 × 107 B31 per ml) for protein evaluation (Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org) and for total RNA extraction. To obtain Bb in a mammalian host-adapted state, spirochetes were diluted (as described above) in BSK-H medium and cultivated for 14 days in DMCs (9, 18); spirochetes from 20 DMCs (≈2 × 109) were harvested and pooled for protein evaluation and total RNA extraction. All animal experiments were approved by the University of Texas Southwestern Institutional Animal Care and Research Advisory Committee.
DNA and RNA Isolation.
Bb genomic DNA was isolated from late-logarithmic phase B31 by using the Stratagene DNA extraction kit. Total RNA was isolated by using Trizol reagent (GIBCO) according to the manufacturer's protocol. To validate mRNA integrity, Northern blot analysis was carried out (19) by using PCR-generated probes for flaB, ospA, and ospC (18, 19); all RNA samples yielded single bands of predicted sizes (data not shown). The methods for Cy5 labeling of DNA and Cy3 labeling of cDNA (from mRNA) are provided in Supporting Materials and Methods.
Microarray Construction and Data Analysis.
A microarray consisting of 1,754 (95.4%) of the 1,838 predicted ORFs of B31 was constructed (Supporting Materials and Methods). Data acquisition and analysis was performed on a GenePix 4000B scanner using GENEPIX 3 software (Axon Instruments, Foster City, CA). The data were normalized (see Supporting Materials and Methods), and comparisons between conditions were performed. An error model with replicates was used to calculate fold change and uncertainty intervals for each gene. Hierarchical cluster analysis of log-normalized ratios was performed also (see Supporting Materials and Methods; ref. 20).
Real-Time Reverse Transcription (RT)-PCR.
Real-time quantitative RT-PCR was used to validate selected data from microarray experiments. Genome-directed primers (GDPs; Table 1, which is published as supporting information on the PNAS web site) were used to prime first-strand cDNA synthesis from DNA-free RNA samples; RT was performed as described (Supporting Materials and Methods) without the inclusion of Cy dye. The cDNA samples were diluted to 8.0, 0.8, and 0.08 ng/μl. Duplicate quantitative PCR assays were performed on 5 μl of each cDNA dilution using the SYBR Green Master mix with an ABI 5700 sequence detection system according to the manufacturer's protocol (Applied Biosystems); the gene-specific primers (Table 2, which is published as supporting information on the PNAS web site) were designed by using PRIMEREXPRESS software (Applied Biosystems). The relative quantitation method (ΔΔCT) was used to evaluate quantitative variation between the three Bb growth conditions relative to each gene examined. The amplicon of 16S rRNA was used as an internal control and normalizer for all data.
Results and Discussion
B31 Growth Conditions for Comparative Transcriptional Analyses.
Different B31 growth conditions were used to evaluate transcriptional differences representative of three major stages in the life cycle of Bb. Currently it is infeasible to obtain sufficient quantities of Bb from ticks for global gene expression studies. Thus, to imitate the unfed-tick (UT) condition, spirochetes were cultivated in BSK-H medium (pH 7.5) at 23°C (18, 21); under this condition, Bb replicates inefficiently, has altered (elongated) morphology, and achieves only modest cell densities (data not shown). To mimic the fed-tick (FT) condition, Bb was cultivated at 37°C to the late-logarithmic phase in BSK-H medium adjusted to pH 6.8 (18); at the end of cultivation, the pH of the medium was unchanged. Finally, mammalian infection by Bb represents a paucibacillary condition, under which only very limited numbers of spirochetes are in tissues. Thus, DMCs implanted into rat peritoneal cavities were used to obtain sufficient mRNA from Bb in a mammalian host-adapted state (9, 18).
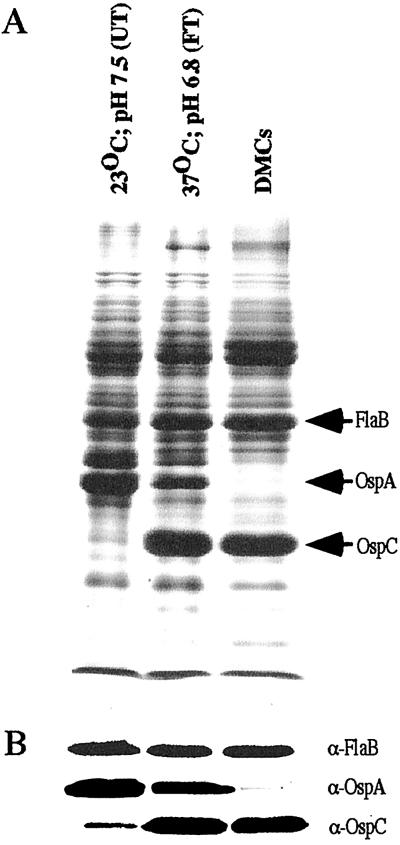
Knowledge of the OspA/OspC paradigm was exploited to validate that the RNA used in microarray analysis was derived from spirochetes suitably “adapted” to the various growth conditions. B31 cultured under UT conditions showed a high level of OspA expression and a very low expression of OspC as reported (ref. 7; Fig. 1 A and B). As expected, FT spirochetes showed decreased levels of OspA and markedly elevated levels of OspC (7). Finally, B31 cultivated in DMCs virtually lacked OspA but expressed high levels of OspC, which is consistent with previous studies (9). These protein profiles suggested that each of the Bb populations used as a source of mRNA for microarray analysis was adapted suitably.
Figure 1.
SDS/PAGE (Coomassie blue stain) (A) and immunoblot (B) of whole-cell lysates of B31 cultivated at the UT, FT, and DMC conditions. The arrows at the right denote the migration of FlaB (loading control), OspA, and OspC.
Use of GDPs for cDNA Labeling.
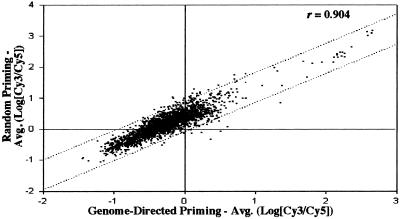
GDPs (22) were selected for the synthesis of Cy3-labeled cDNA, because they increase both the specificity and sensitivity of priming for a targeted mRNA population. To evaluate the use of GDPs for B31, a set of GDPs (Table 1) was constructed based on genome information (14, 15) and used to compare its cDNA labeling efficiency with that of the more conventional hexamer random-priming method. In an analysis of ≈3,400 features, cDNA labeling efficiencies between the two methods were substantially equivalent (Fig. 2). A majority of features (54.2%) hybridizing with GDP-derived cDNA displayed higher Cy3/Cy5 intensity ratios with an overall average log Cy3/Cy5 ratio being 2-fold higher than for random-primed cDNA. Based on these results, all subsequent experiments used GDPs.
Figure 2.
Correlation of Cy3/Cy5 fluorescence intensities of Cy3-labeled cDNA derived by either random priming or using B31-specific GDPs. The dotted lines represent data within 2 SDs of regression (95% confidence). r, correlation coefficient for the similarity between mean log ratios for each feature from the different labeling protocols.
Global Gene Expression Profiling.
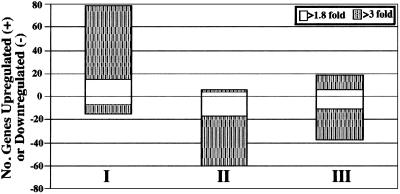
An evaluation of mRNA expression profiles of Bb cultivated under the UT, FT, and DMC conditions was conducted. Although mRNA changes as low as 1.5-fold may have biological relevance (23), the more stringent threshold of 1.8-fold used herein was derived statistically and thus believed to provide a conservative foundation for all interpretations. Using pairwise analysis among the three B31 growth conditions, globally, 8.6% (150) of Bb's genes were differentially expressed between the three conditions tested (Tables 3–5, which are published as supporting information on the PNAS web site). Of these 150 genes, the majority were differentially regulated by at least 3-fold (Fig. 3). When Bb was cultivated under FT conditions, 79 genes (4.5% of the genome) were up-regulated, whereas 15 genes (0.86% of the genome) were down-regulated (relative to UT spirochetes; Fig. 3 I). In contrast, B31 cultivated in DMCs up-regulated only 6 genes (vlsE, vlsE1, and four hypothetical genes) and down-regulated 60 genes (relative to the FT condition; Fig. 3 II), 30 of which had been up-regulated during the FT condition (Fig. 3 I). The sharp contrast between the number of genes up-regulated during the FT condition (Fig. 3 I) but down-regulated in DMC spirochetes (Fig. 3 II) would lend support to the idea that such differences are a reflection of Bb subsisting in the two very diverse niches. However, after further inspection of the gene expression profiles between B31 cultured under the UT and DMC conditions, DMC-cultured B31 showed an up-regulation of 19 genes and down-regulation of 38 genes (Fig. 3 III). From this comparison and an evaluation of hierarchical cluster analysis (Fig. 7, which is published as supporting information on the PNAS web site), it was surprising to find that despite dramatic changes in gene expression occurring between the UT/FT (Fig. 3 I) and FT/DMC (Fig. 3 II) conditions, growth in the mammalian environment (DMCs) ultimately returns Bb to a more homeostatic phase (i.e., more akin to the UT condition) (Fig. 3 III).
Figure 3.
Number of genes differentially expressed during various B31 culture conditions. The UT growth condition was used as a baseline reference. The bars reflect comparisons between the UT and FT (I), FT and DMC (II), and UT and DMC (III) growth conditions. The white areas denote gene expression changes of 1.8–3.0-fold, whereas shaded areas signify changes >3-fold. Individual gene listings are provided in Tables 3–5.
Concordance Between Microarray and Real-Time RT-PCR Data.
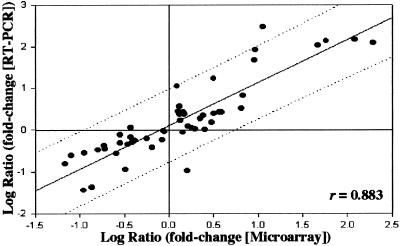
Real-time quantitative RT-PCR was used to corroborate selected values from microarrays. Among expression levels compared were those for ospA, ospC, dbpA, flaB, and 13 other randomly selected genes (Table 2) for which specific PCR primers could be designed. For all three Bb growth conditions (51 comparisons), there was a high degree of concordance (r = 0.883) between data from the two methodologies (Fig. 4).
Figure 4.
Correlation between microarray and real-time RT-PCR data. Log-transformed fold changes for 16 differentially expressed genes and one constitutively expressed gene (flaB; Table 2) were compared at each of the three (UT, FT, and DMC) growth conditions. The dotted lines represent data within 2 SDs of regression (95% confidence). r, correlation coefficient.
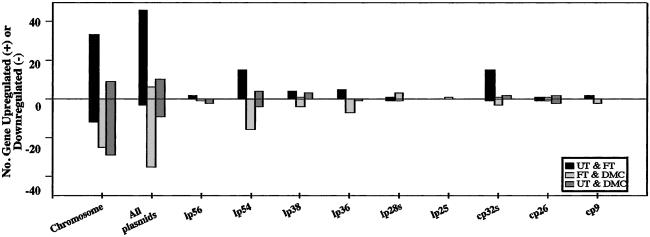
Distribution of Expression Patterns Among the B31 Replicons.
The complex genomic makeup of Bb includes a linear chromosome and numerous circular and linear plasmids (lps; refs. 14 and 15); the extent and complexity of the plasmids suggest that they are of enormous importance to the biology of Bb (15). There was nearly an equal distribution of the differentially expressed genes between the chromosome (n = 78) and the 21 plasmids (n = 72; Fig. 5). Seventeen plasmids demonstrated differential expression of their genes; unique genes were detected on lp56, lp54, lp38, lp36, and circular plasmid (cp)26, whereas only paralogous genes were detected on lp28-1, lp28-3, lp28-4, lp25, cp9, and all cp32 plasmids (Tables 3–5). Regulated genes were not detected on lp28-2, lp21, lp17, or lp5. The majority of the plasmid-encoded differentially expressed genes were on lp54, lp36, and the family of cp32 plasmids. A number of these genes were up-regulated under the FT condition (Fig. 5), once again suggesting that shifts in temperature, pH, and spirochete cell density likely are key sensory stimuli. Our data also imply that Bb genes on diverse replicons can be responsive as a group to a similar constellation of environmental signals. Although there are a number of possibilities to explain this collective responsiveness, regions of local DNA helicity among the diverse plasmids may be one plausible mechanism. For example, there is evidence that RpoS binding to DNA is affected adversely by negative supercoiling induced by GyrB (24). Topoisomerase IV (ParE) can relax DNA in a replication-independent manner to counteract the effects of GyrB and environmental changes (25), thereby facilitating the accessibility of RpoS to its docking sites. In our studies, parE was up-regulated under both the FT (3.3-fold) and DMC (2.4-fold) growth conditions (Tables 4 and 5). Thus, it is tempting to speculate that ParE activity may play an accessory role in the recently described Bb RpoN-RpoS pathway, wherein RpoN controls the expression of RpoS (8). In support of the notion that ParE serves an accessory role in the RpoN-RpoS pathway, cluster analysis revealed that dbpA, ospC, and nine other plasmid-encoded genes clustered with parE (Fig. 7).
Figure 5.
Number and distribution of differentially expressed genes among B31 genetic replicons. The UT growth condition was used as a baseline reference. The bars reflect comparisons between the UT and FT (black), FT and DMC (light gray), and UT and DMC (dark gray) growth conditions. Individual gene listings are provided in Tables 3–5.
lp25 has been closely associated with Bb infectivity (26–28). One lp25 gene, BBE16, a putative lipoprotein (15), was up-regulated 2.9-fold when B31 was cultivated in DMCs (compared with the FT condition; Table 4). Upstream of BBE16 and separated by only 16 bp is BBE17, which also was up-regulated 2.6-fold when B31 was cultivated in DMCs. Given the presence of a putative promoter upstream of BBE17 but none for BBE16 (data not shown), BBE17 and BBE16 may be operonic; their up-regulation during mammalian host adaptation warrants further investigation into their putative roles in Bb virulence expression.
lp36 also has been implicated in virulence because of its tendency to be present in infectious clones of Bb (27). This plasmid encodes a P35 homolog, BBK32 (15, 29). BBK32 was up-regulated 28-fold under the FT condition and down-regulated to near basal level in DMCs, consistent with previous observations that BBK32 is sharply up-regulated during the transmission of Bb into mammalian tissue but decreases to low levels by day 14 in mice (29). Another lp36 gene, BBK17, believed to encode adenine deaminase (AdeC), was up-regulated 16-fold in the FT condition (Table 3); this was the only nonchromosomal gene implicated in intermediary (purine) metabolism that was differentially regulated. Thus, lp36 and specifically BBK17 may be vital not only in intermediary metabolism but also to some poorly understood aspect(s) of Bb virulence.
lp54 encodes 76 genes including the ospAB and dbpBA operons. Consistent with the OspA/C expression paradigm, ospA was down-regulated ≈12-fold from the UT condition when B31 was cultivated in DMCs (Table 5; real-time RT-PCR showed a 4-fold decrease; Table 2). These levels of ospA down-regulation were less than anticipated given the degree of OspA reduction in B31 cultured within DMCs (Fig. 1). Although the explanation for the disparity between transcript and protein levels for OspA is unclear, one possibility is some form(s) of posttranscriptional regulation (30). dbpA was up-regulated dramatically (47-fold) when B31 was cultivated at the FT (37°C) condition (Table 3), which is consistent with the fact that dbpA is temperature-regulated (31, 32). In contrast, the expression of dbpA was 15-fold lower in the DMC versus the FT growth condition; this result is in accord with previous observations that dbpA likely is down-regulated within days postmammalian infection (33). In fact, the majority of plasmid genes up-regulated during the FT growth condition were subsequently down-regulated when spirochetes were cultivated in DMCs (Fig. 5). Thus it is possible that parameters of early Bb transmission likely trigger a number of plasmid-encoded gene responses that are only transiently necessary (FT). Hence, as noted earlier (Figs. 3 III and 7), both the tick (UT) and mammalian (DMCs) environments may largely represent more homeostatic phases in the life cycle of Bb.
Paralogous Families.
Gene redundancy (numerous paralogous families) is a salient yet incompletely understood feature of Bb genetic organization (14, 15). The paralogous families potentially complicate the interpretation of microarray data in at least two ways. First, whereas the primer pairs from Sigma-Genosys were intended to amplify specifically each gene of B31, some uncertainty regarding their actual specificities for the paralogous ORFs remained. To examine this uncertainty to some extent, sequencing of amplicons from 21 paralogous ORFs was performed. In every case, the primer pairs specifically amplified their respective genes (data not shown), suggesting that target specificity was engendered largely by the set of B31 primer pairs.
There was the added possibility of cross-hybridization among related gene family members, thereby masking true differential expression patterns for individual genes. Cross-hybridization could have been a limitation in our analyses, given that except in one case (family 161; Table 3), multiple genes of any given paralogous family were regulated similarly at a particular growth condition. However, of the 49 paralogous families in which a subset of member genes showed regulation, 30 families were represented by only a single gene (i.e., 19 families had more than one gene represented). Furthermore, among the 14 genes of the P35 family (family 54), although five members were differentially regulated similarly as a group, their ranges varied greatly [3–75-fold up-regulation during the FT condition (relative to the UT condition); 6–31-fold down-regulation in DMCs (relative to the FT condition) (Tables 3–5)]. Taken together, these data suggest that the arrays, at least to some degree, were capable of distinguishing expression level differences among members of the same paralogous family. Nonetheless, further experiments are warranted for a more precise analysis of differential regulation among paralogs.
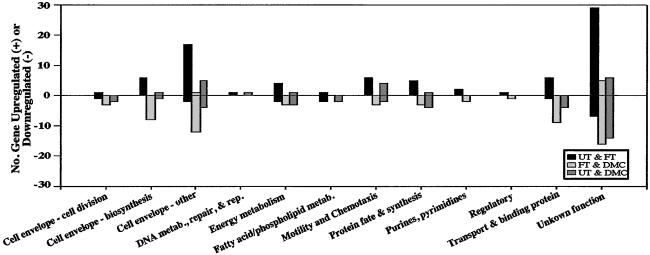
Functional Classification of Differentially Expressed Genes.
Differential responses of genes within putative functional classes were assessed according to B31 genome annotation (14, 15). Given the small B31 genome size (≈1.5 megabases) and the putative absence of many components of key metabolic pathways (14), it perhaps is not inordinate that genes with essential functions, in general, tended not to show large adaptive changes (Tables 3–5). A logical extension of this observation is that Bb optimally utilizes its limited housekeeping genes even during more static phases of growth. Cluster analysis supported this view in that the majority of the essential genes grouped within the two largest clusters represented by either constitutively high or low levels of gene expression across all conditions (data not shown).
Cultivation of Bb under the FT condition (37°C), where robust spirochete replication occurs, generally led to increased expression of genes associated with the cell envelope, protein synthesis, motility/chemotaxis, transport/binding proteins, and energy metabolism (Fig. 6); these included genes involved in the glycolytic and phosphoenolpyruvate/carbohydrate phosphotransferase system pathways as well as others involved in central intermediary metabolism. Additionally, under both FT and DMC conditions, there were increases in the expression of the V-type ATPase operon (genes BB0090–6; ref. 14) likely responsible for proton efflux (proton motive force) across the cell membrane; such efforts to maintain membrane potential during the more robust periods (FT condition) of cell division are further suggestive that the UT condition represents a more static phase analogous to the restricted growth of Bb in UTs (21).
Figure 6.
Number of differentially expressed genes grouped by putative protein functions. The UT growth condition was used as a baseline reference. The bars reflect comparisons between the UT and FT (black), FT and DMC (light gray), and UT and DMC (dark gray) growth conditions. Individual gene listings are provided in Tables 3–5.
Overall, B31 genes associated with cell division tended to be down-regulated in the FT and DMC growth conditions (Fig. 6), an observation that seemingly was counterintuitive. However, this categorization may be misleading given that certain gene products serve to inhibit cell division. For example, during the UT growth condition, B31 replicated more slowly and displayed a morphology 2–3 times longer than when cultivated at the FT or DMC condition (data not shown). Of potential relevance to this phenotype was the increased expression of the minD ortholog ylxH-1 (BB0269), which was up-regulated 2.6–3.3-fold in the UT condition (relative to the FT and DMC growth conditions, respectively). In other bacteria, MinD is a cell-division inhibitor that, when overexpressed, leads to cell elongation (34). Although the regulation of other cell-division genes may contribute to the elongated phenotype of B31 grown at room temperature, it is possible that YlxH-1 (MinD) restricts Bb replication until growth conditions become more favorable. Additionally, these more favorable conditions may induce cell-division enhancers that act either as proteases for or suppressors of cell-division inhibitors such as the HslVU dimer (35) or ChpAI (36), respectively. The B31 homologs of hslV (BB0296) and chpAI (BBA07) showed increased expression levels during the FT and DMC conditions (Tables 4 and 5), implying that inhibitory mechanisms for stasis in Bb (UT condition) are extant to impede cell division until appropriate growth stimuli (e.g., elevated temperature and/or reduced pH) are encountered.
A number of genes have been associated with the maintenance of protein stability during periods of bacterial stasis or oxidative stress; among these genes is trxA, which encodes thioredoxin A (37). When B31 was cultivated at the suboptimal UT condition, trxA (BB0061) increased 2- and 3-fold relative to the FT and DMC growth conditions, respectively. In other bacteria, trxA is regulated by (p)ppGpp levels that in turn are governed via the activities of at least two enzymes, RelA and SpoT (37, 38). RelA has not been noted yet in B31, but SpoT, which can have overlapping function with RelA (38), has been ascribed (BB0198; ref. 14). At the UT growth condition, spoT expression was up-regulated 1.8- and 2.0-fold relative to the FT and DMC growth conditions, respectively. These data combined with the previously noted genes implicated in inhibition and derepression of cell division (ylxH-1, hslVU, and chpAI) suggest that the steady-state existence of Bb in UTs (21) may be likened to the bacterial stringent response induced by either limited nutrients or other stress conditions (38). During a stringent response in Bb (i.e., in UTs), it is conceivable that there is (i) a conservation of energy that assists in cell viability and (ii) a state of cellular readiness that minimizes lags in responsiveness to new sensory information (signifying growth and transmission). Although this notion remains hypothetical with present knowledge, it is supported further by the observed up-regulation of Bb genes involved in energy metabolism, protein synthesis, and cell-envelope biogenesis during the FT growth condition (Fig. 6).
With regard to the cell envelope serving as the principal interface with the host, 17 genes encoding envelope-associated proteins were up-regulated in B31 cultivated at the FT condition (cell envelope-other; Fig. 6). Among these were genes encoding the lipoproteins DbpA, DbpB, OspC, five members of the Erp family, and three members of the Mlp family. Except for OspC, these genes subsequently tended to be down-regulated when B31 was cultivated in DMCs, which is consistent with the notion that a more tempered gene response occurs as Bb adapts to the mammalian environment. Genes associated with cell-envelope biogenesis and transport/binding tended to be down-regulated by B31 cultivated in DMCs (compared with the FT condition), again suggesting a somewhat more controlled growth state perhaps representative of the paucibacillary nature of mammalian infection.
Genes associated with motility/chemotaxis generally were up-regulated during the FT growth condition (Fig. 6). More specifically, for both chemotaxis (che) operons of B31, 3 of 5 genes (flaA, cheA-2, cheX; ref. 39) and 2 genes (cheW-2, cheA-1) of a second incompletely characterized operon (14, 40) were up-regulated in both the FT and DMC growth conditions. Accessory genes encoding methyl-accepting chemotaxis proteins (mcp4 and mcp5) also were up-regulated under the FT condition. Thus it was not surprising that the majority of these motility/chemotaxis genes clustered with genes associated with mammalian host-transmission events (e.g., ospC and dbpA; Fig. 7). The combined data are consistent with the idea that motility and chemotaxis may foster Bb dissemination processes in both feeding ticks (midgut to salivary glands) and mammals (spread from dermal tissues) but are less important before transmission events (i.e., within UTs).
Evidence of adaptive changes in B31 regulatory genes was sparse in microarrays (Fig. 6). This in part may be a reflection that only a few homologs of bacterial regulatory proteins have been noted in B31 (14). For example, there is no known heat-shock σ factor in B31 (14) even though a heat-shock response in Bb has been reported (41). In this study, the heat-shock genes groES, dnaK-2, and hslV were up-regulated during the FT growth condition, suggesting the presence of the relevant regulator(s). However, groES, dnaK-2, and hslV clustered into three different groups (Fig. 7), suggesting that they may be regulated by different networks. Another possibility for the apparent lack of regulatory gene involvement is that Bb may be able to exploit only relatively small changes in the expression of its regulatory genes to affect downstream targets. A third possibility may be analogous to the situation for rpoN, which is constitutively expressed and posttranscriptionally regulated (42). Consistent with the notion of posttranscriptional regulation, the levels of rpoN mRNA did not vary in response to the diverse growth conditions. Additionally, in Bb, RpoN seems to regulate via a novel pathway the expression of rpoS (8). However, we did not observe significant changes in rpoS mRNA levels under any growth condition, a finding that was surprising given that RpoS levels rise in response to elevated temperature and reduced pH (the FT condition; ref. 7). The simplest explanation for this result is posttranscriptional regulation over RpoS, as has been noted in other bacteria (43).
Genes Encoding Putative Signal Sequences.
Globally, 46% of the differentially regulated genes of B31 were predicted to encode either signal peptidase I (n = 25) or signal peptidase II (n = 44) leader sequences. This proportion of putative signal peptide-encoding, differentially regulated genes supports the contention that differential expression of the extracellular proteome is of paramount importance to Bb's adaptive response(s). Additionally, that 29% of the changing expression patterns represented putative lipoprotein genes likely underscores the dynamic interplay between lipoprotein expression and transitions in the life cycle of Bb.
Summary and Future Implications.
The goal of this study was to examine globally the transcription profile of Bb and use the information as a basis for establishing new hypotheses regarding the molecular pathogenesis of Lyme disease. By using B31 growth conditions believed to simulate three key phases in the life cycle of Bb, several valuable insights were garnered. First, a substantial percentage of the differentially regulated genes theoretically encoded export signals, and thus they likely contribute to the extracellular proteome's involvement in host-parasite interactions during strategic Bb life cycle transitions. Second, although there has been general acceptance that Bb in UTs represents a form of spirochetal quiescence (21), microarray data provided herein now implicate the bacterial stringent response (38) and factors that restrain cell division (34–36) during this Bb life cycle phase. Third, a large proportion of genes was responsive to the FT growth environment, wherein increased temperature and reduced pH were prominent environmental conditions analogous to those encountered in feeding ticks (7). Fourth, many of the profound changes in gene expression induced during the FT growth condition were largely ameliorated as B31 adapted to the mammalian host, suggesting that once Bb gains entry and adapts to mammalian tissues, fewer differentially regulated genes are exploited for this phase of survival. Cluster analysis also supported that despite the disparity between the tick and mammalian hosts, Bb subjected to conditions analogous to those of UTs and mammalian tissues seemed to induce gene expression profiles more consistent with homeostatic mechanisms. Finally, the microarray data can be used as a guide to select genes encoding putative membrane proteins and lipoproteins that are differentially regulated during varying growth conditions; these will provide a framework for new avenues of investigation into the membrane biology of Bb. Taken together, the microarray data hold promise for formulating new testable hypotheses regarding the life cycle of Bb and attaining a more complete understanding of many aspects of Bb's complex parasitic strategies.
Supplementary Material
Acknowledgments
We thank investigators in the Microarray Core Facility at University of Texas Southwestern for advice and technical assistance, Xiaofeng Yang, Kayla Hagman, and Jeanne Sheffield for help with implanting DMCs, Molly Isbell for aiding in statistical analysis, and Anette Hübner, Vanessa Sperandio, Eric Hansen, and Leon Eidels for critical review of the manuscript. This work was supported by Public Health Service Grant AI-45538 and by a grant (Advanced Technology Program) from the Texas Higher Education Coordinating Board. A.M.T. was funded by grants from the National Institutes of Health and Defense Advanced Research Planning Agency to Stephen Albert Johnston.
Abbreviations
- Bb
Borrelia burgdorferi
- Osp
outer surface (lipo)protein
- Dbp
decorin-binding protein
- DMC
dialysis membrane chamber
- B31
Bb strain B31
- RT
reverse transcription
- GDP
genome-directed primer
- UT
unfed tick
- FT
fed tick
References
- 1.Steere A C. Hosp Pract. 1993;28:37–44. doi: 10.1080/21548331.1993.11442776. [DOI] [PubMed] [Google Scholar]
- 2.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Silva A M, Telford S R, Brunet L R, Barthold S W, Fikrig E. J Exp Med. 1996;183:271–275. doi: 10.1084/jem.183.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilmore R D, Piesman J. Infect Immun. 2000;68:411–414. doi: 10.1128/iai.68.1.411-414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwan T G, Piesman J. J Clin Microbiol. 2000;38:382–388. doi: 10.1128/jcm.38.1.382-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohnishi J, Piesman J, de Silva A M. Proc Natl Acad Sci USA. 2001;98:670–675. doi: 10.1073/pnas.98.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Goldberg M S, Popova T G, Schoeler G B, Wikel S K, Hagman K E, Norgard M V. Mol Microbiol. 2000;37:1470–1479. doi: 10.1046/j.1365-2958.2000.02104.x. [DOI] [PubMed] [Google Scholar]
- 8.Hubner A, Yang X, Nolen D M, Popova T G, Cabello F C, Norgard M V. Proc Natl Acad Sci USA. 2001;98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akins D R, Bourell K W, Caimano M J, Norgard M V, Radolf J D. J Clin Invest. 1998;101:2240–2250. doi: 10.1172/JCI2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 11.Wilson M, DeRisi J, Kristensen H-H, Imboden P, Rane S, Brown P O, Schoolnik G K. Proc Natl Acad Sci USA. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sperandio V, Torres A G, Giron J A, Kaper J B. J Bacteriol. 2001;183:5187–5197. doi: 10.1128/JB.183.17.5187-5197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smoot L M, Smoot J C, Graham M R, Somerville G A, Sturdevant D E, Lux Migliaccio C A, Sylva G L, Musser J M. Proc Natl Acad Sci USA. 2001;98:10416–10421. doi: 10.1073/pnas.191267598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, et al. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 15.Casjens S, Palmer N, van Vugt R, Huang W M, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson R J, et al. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 16.Barthold S W, Beck D S, Hansen G M, Terwilliger A A, Moody K D. J Infect Dis. 1990;162:133–138. doi: 10.1093/infdis/162.1.133. [DOI] [PubMed] [Google Scholar]
- 17.Pollack R J, Telford S R, Spielman A. J Clin Microbiol. 1993;31:1251–1255. doi: 10.1128/jcm.31.5.1251-1255.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X, Popova T G, Hagman K E, Wikel S K, Schoeler G B, Caimano M J, Radolf J D, Norgard M V. Infect Immun. 1999;67:6008–6018. doi: 10.1128/iai.67.11.6008-6018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porcella S F, Popova T G, Akins D R, Li M, Radolf J D, Norgard M V. J Bacteriol. 1996;178:3293–3307. doi: 10.1128/jb.178.11.3293-3307.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Silva A M, Fikrig E. Am J Trop Med Hyg. 1995;53:397–404. doi: 10.4269/ajtmh.1995.53.397. [DOI] [PubMed] [Google Scholar]
- 22.Talaat A M, Hunter P, Johnston S A. Nat Biotech. 2000;18:679–682. doi: 10.1038/76543. [DOI] [PubMed] [Google Scholar]
- 23.Hughes T R, Marton M J, Jones A R, Roberts C J, Stoughton R, Armour C D, Bennett H A, Coffey E, Dai H, He Y D, et al. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 24.Kusano S, Ding Q, Fujita N, Ishihama A. J Biol Chem. 1996;271:1998–2004. doi: 10.1074/jbc.271.4.1998. [DOI] [PubMed] [Google Scholar]
- 25.Diebler R W, Rahmati S, Zechiedrich E L. Genes Dev. 2001;15:748–761. doi: 10.1101/gad.872301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Kodner C, Coleman L, Johnson R C. Infect Immun. 1996;64:3870–3876. doi: 10.1128/iai.64.9.3870-3876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purser J E, Norris S J. Proc Natl Acad Sci USA. 2000;97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labandeira-Rey M, Skare J T. Infect Immun. 2001;69:446–455. doi: 10.1128/IAI.69.1.446-455.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fikrig E, Feng W, Barthold S W, Telford S R, Flavell R A. J Immunol. 2000;164:5344–5351. doi: 10.4049/jimmunol.164.10.5344. [DOI] [PubMed] [Google Scholar]
- 30.Jonsson M, Bergstrom S. Microbiology. 1995;141:1321–1329. doi: 10.1099/13500872-141-6-1321. [DOI] [PubMed] [Google Scholar]
- 31.Cassatt D R, Patel N K, Ulbrandt N D, Hanson M S. Infect Immun. 1998;66:5379–5387. doi: 10.1128/iai.66.11.5379-5387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagman K E, Yang X, Wikel S K, Schoeler G B, Caimano M J, Radolf J D, Norgard M V. Infect Immun. 2000;68:4759–4764. doi: 10.1128/iai.68.8.4759-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanson M S, Cassatt D R, Guo B P, Patel N K, McCarthy M P, Dorward D W, Hook M. Infect Immun. 1998;66:2143–2153. doi: 10.1128/iai.66.5.2143-2153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szeto J, Ramirez-Arcos S, Raymond C, Hicks L D, Kay C M, Dillon J-A R. J Bacteriol. 2001;183:6253–6264. doi: 10.1128/JB.183.21.6253-6264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang M S, Lim B K, Seong I S, Seol J H, Tanahashi N, Tanaka K, Chung C H. EMBO J. 2001;20:734–742. doi: 10.1093/emboj/20.4.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim C-J, Daws T, Gerami-Nejad M, Fuchs J A. Biochim Biophys Acta. 2000;1491:1–6. doi: 10.1016/s0167-4781(00)00026-9. [DOI] [PubMed] [Google Scholar]
- 38.Cashel M, Gentry D R, Hernandez V J, Vinella D. In: Escherichia coli and Salmonella. Niedhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1458–1496. [Google Scholar]
- 39.Ge Y, Charon N W. FEMS Microbiol Lett. 1997;153:425–431. doi: 10.1111/j.1574-6968.1997.tb12606.x. [DOI] [PubMed] [Google Scholar]
- 40.Trueba G A, Old I G, Saint Girons I, Johnson R C. Res Microbiol. 1997;148:191–200. doi: 10.1016/S0923-2508(97)85239-4. [DOI] [PubMed] [Google Scholar]
- 41.Cluss R G, Boothby J T. Infect Immun. 1990;58:1038–1042. doi: 10.1128/iai.58.4.1038-1042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buck M, Gallegos M-T, Studholme D J, Guo Y, Gralla J D. J Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hengge-Aronis R. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.