Abstract
Here, we describe the isolation and characterization of the rhesus macaque homolog for human DC-SIGN, a dendritic cell-specific C-type lectin. mac-DC-SIGN is 92% identical to hu-DC-SIGN. mac-DC-SIGN preserves the virus transmission function of hu-DC-SIGN, capturing and efficiently transducing simian and human immunodeficiency virus to target CD4+ T cells. Surprisingly, however, mac-DC-SIGN plays no discernable role in the ability of rhesus macaque dendritic cells to capture and transmit primate lentiviruses. Expression and neutralization analyses suggest that this process is DC-SIGN independent in macaque, although the participation of other lectin molecules cannot be ruled out. The ability of primate lentiviruses to effectively use human and rhesus dendritic cells in virus transmission without the cells becoming directly infected suggests that these viruses have taken advantage of a conserved dendritic cell mechanism in which DC-SIGN family molecules are significant contributors but not the only participants.
Human and primate myeloid derived-dendritic cells (DC) are exceptional in their ability to transmit human and simian immunodeficiency viruses to target CD4+ T cell lymphocytes. The molecular basis for the potent virus transmission by this important subset of antigen-presenting cells has been the subject of intensive investigation since the initial observations by Steinman and colleagues in 1992 (1). Several models have been proposed to account for the mechanism of efficient HIV-1 and simian immunodeficiency virus (SIV) transmission from myeloid-derived DC. Because DC can be directly infected by HIV-1, it has been suggested that either these cells are very proficient in the production of HIV-1 or HIV-1 produced from DC have a higher specific infectivity (2–9). It has also been proposed that DC efficiently catalyze the infection of CD4+ T cells in a manner that is not dependent on HIV-1 production within the DC. Indeed, murine DC that are unable to support HIV-1 replication efficiently transmit the virus to human CD4+ T cells (1). Thus, presuming that HIV-1 may have taken advantage of a general property of DC, human DC analogously may present either cell membrane-captured or newly synthesized HIV-1 in a manner that promotes CD4+ T cell infection. Favored mechanisms for this model include (i) transmission via a virological synapse where the DC directs HIV-1 to the CD4/coreceptor complex present on T cells, (ii) ligand interactions between DC and T cells that create a microenvironment promoting virus transmission because of proximity of the respective cell membranes and a high local concentration of HIV-1, or (iii) an indirect mechanism in which DC-mediated stimulation of CD4+ T cells leading to cell activation renders them more susceptible to infection.
A search by Geijtenbeek et al. (10, 11) for novel intercellular adhesion molecule 3 (ICAM-3) ligands expressed on myeloid-derived DC unexpectedly revealed a molecular participant in DC-mediated virus transmission. A molecule identified by Curtis and colleagues (12) in a prospective screen for HIV-1 envelope (Env) adhesion receptors expressed in human placental tissue was rediscovered as a DC-expressed C-type lectin, which serves as an ICAM-3 ligand. This molecule was renamed dendritic cell-specific ICAM-3-grabbing nonintegrin (DC-SIGN). An initial supposition that DC-SIGN functions as a cis entry receptor on DC proved to be incorrect. However, based on the work of Steinman and others (1, 2, 6, 9, 13–16), it was then examined whether DC-SIGN could function as a trans adhesion receptor. Indeed, expression of DC-SIGN on monocytic cell lines conferred the cells with an ability to bind and efficiently transmit HIV-1 in a manner that was completely dependent on direct interactions between DC-SIGN and HIV-1 Env (11). Beyond a requirement for direct interaction between HIV-1 Env and the carbohydrate recognition domain of DC-SIGN for virus transmission function (17), little is currently known about the mechanism of transmission to target cells bearing HIV-1 receptors. Furthermore, it has not been determined whether DC-SIGN contributes to viral pathogenesis by (i) facilitating mucosal transmission of HIV/SIV through resident, patrolling immature DC or (ii) catalyzing sustained HIV/SIV replication in lymphoid tissue by mature DC.
SIV infection of macaques constitutes an important model for HIV-1 pathogenesis. Mucosal transmission of SIV by experimental infection of macaques can be monitored with regard to virus-associated target cell and tissue types within the mucosa and the subsequent dissemination of SIV (18–25). Macaque DC from blood, lymph nodes, mucosal tissue, and skin seem comparable to their human counterparts in biological function and ability to transmit virus (19, 26–28). Thus, the interaction of SIV with DC-SIGN-expressing cells observed by using this experimental system may be relevant in modeling HIV-1 transmission across the mucosa. Moreover, if mucosal transmission of SIV could be impaired by blocking DC-SIGN–SIV interactions, it would provide compelling evidence for a role of DC-SIGN in this process. We thus sought to isolate macaque DC-SIGN and examine its viability in SIV capture and transmission. mac-DC-SIGN cDNA was PCR amplified from macaque DC message. The predicted protein shares 92% identity with hu-DC-SIGN and is antigenically cross-reactive with antibodies that recognize hu-DC-SIGN. Not surprisingly, mac-DC-SIGN also binds ICAM-3. However, whereas mac-DC-SIGN functions to capture and transmit HIV-1 or SIV, it alone does not account for the virus transmission function of macaque DC. Despite being the homolog of hu-DC-SIGN, mac-DC-SIGN is poorly expressed in macaque DC as measured by protein and message analysis. Nonetheless, macaque DC potently transmit viruses to target CD4+ T cells, indicating that DC-SIGN-independent mechanisms for virus transmission can also be used by primate lentiviruses.
Materials and Methods
Isolation of mac-DC-SIGN cDNA.
Peripheral blood mononuclear cells (PBMC) of rhesus macaque (Macaca mulatta) were Ficoll gradient-purified from peripheral blood and cultured in the presence of 500 units/ml IL-4 and 800 units/ml granulocyte–macrophage colony-stimulating factor (R & D Systems). Total RNA was isolated from the cultured cells with Trizol (Life Technologies, Grand Island, NY), primed with oligo(dT)12–18, and transcribed by using Superscript II reverse transcriptase (Life Technologies) according to the manufacturer's protocol. mac-DC-SIGN cDNA was amplified with the following primers: dc-sign1, 5′-AGT GGG GTG ACA TGA GTG AC-3′ and s-9m, 5′-GAA GTT CTG CTA CTC AGG AG-3′. Primers used to obtain cDNA were based on identity to macaque genomic sequence of DC-SIGN (genomic DNA from four individual macaques were sequenced before cDNA isolation). Amplification of the mac-DC-SIGN cDNA fragment was performed in two rounds of PCR: 38 cycles in the first round and 25 cycles for the second round at a 60°C annealing temperature. For the second round, a 1:50 dilution of the first-round amplificate was used as a template. The amplified DNA fragment was subcloned into the expression vector pcDNA3.1/V5-His/TOPO (Invitrogen).
Antibodies.
A panel of six monoclonal antibodies (mAbs) against hu-DC-SIGN was generated with R & D Systems; the mAbs were obtained by screening hybridoma supernatants of BALB/c mice immunized with NIH 3T3/hu-DC-SIGN cells for the ability to stain hu-DC-SIGN. All six mAbs react with human DC and are effective in neutralizing HIV-1 capture and transmission by these cells (L.W., T.D.M., R. Vazeux, D.U., and V.N.K., unpublished data).
Cell Culture.
Monocytic cell lines THP-1 and hu-DC-SIGN expressing THP-1/hu-DC-SIGN were provided by Doug Kwon and Dan Littman (New York University Medical Center). THP-1/mac-DC-SIGN cells were generated by electroporation of the THP-1 cell line with the pcDNA3.1-mac-DC-SIGN construct, followed by selection for resistance to G418, and then positively sorted with cross-reactive mAbs recognizing mac-DC-SIGN.
Immature human DC were derived from CD14+ bead-purified monocytes cultured in the presence of 500 units/ml IL-4 (R & D Systems) and 800 units/ml granulocyte–macrophage colony-stimulating factor (R & D Systems), refreshed every 2 days. At day 7, cells expressed high levels of HLA-DR, MHC class I, CD11b, CD11c, and ICAM-1, moderate levels of LFA-1 and CD86, and low levels of CD14. Human DC also expressed high levels of DC-SIGN.
For macaque DC, CD14+ cells were cultured in IL-4 and granulocyte–macrophage colony-stimulating factor as described (29). Cells were HLA-DR moderate/high, CD86 moderate, CD80 low, and CD25 and CD83 negative/low. Culture medium used for all macaque DC generation was supplemented with 1% human plasma.
Hut/CC chemokine receptor (CCR) 5 (Hut/CCR5) cells are the transformed human T cell line Hut78 stably transduced with CCR5. GHOST cells are HIV-indicator cells derived from human osteosarcoma cells (30).
All suspended and primary cells described above were maintained in RPMI medium 1640 (Life Technologies) supplemented with 10% FBS in addition to specific cytokines or antibiotic requirements as indicated. GHOST cells were grown in DMEM (Life Technologies) supplemented with 10% FBS and antibiotics.
Fluorescence-Activated Cell Sorter (FACS) Analyses.
To assess surface expression of mac-DC-SIGN with mAbs raised against hu-DC-SIGN, THP-1/mac-DC-SIGN cells were stained with a panel of six distinct DC-SIGN-reactive mAbs. For staining, 2 × 105 cells were incubated in ice-cold PBS containing 2% FBS (FACS buffer) and 2 μg/ml mAb in a total volume of 100 μl. After 30 min at 4°C, cells were washed with FACS buffer and recovered in 100 μl of FACS buffer containing 2 μg/ml phycoerythrin-conjugated goat anti-mouse IgG antibody (Caltag, South San Francisco, CA). Cells were incubated for 30 min at 4°C, washed with FACS buffer, and analyzed by using flow cytometry.
ICAM-3 Bead Adhesion Assay.
The fluorescent bead adhesion assay was performed as described by Geijtenbeek et al. (10). Adhesion of recombinant ICAM-3 (R & D Systems) to hu-DC-SIGN or mac-DC-SIGN was determined by measuring the percentage of THP-1/hu-DC-SIGN or THP-1/mac-DC-SIGN cells that bound ICAM-3-coated fluorescent beads by using flow cytometry.
Virus Stocks.
Single-round infectious pseudotyped HIV-1 stocks were generated by calcium phosphate cotransfections of HEK293T cells with the proviral vector plasmid NL-Luc-E−R− (HIV-luc) containing a luciferase reporter gene (31) and an expression plasmid for R5-tropic HIV-1JRFL, X4-tropic HIV-1HXB2, SIVMAC1A11, SIVMAC239, SIVMAC239MER, or SIVAGM Env. Viral stocks were evaluated by limiting dilution infection of GHOST cells.
HIV-1 Infection Assays.
HIV-1 cell capture and transmission assays were performed as described (32). In brief, donor cells THP-1, THP-1/hu-DC-SIGN, THP-1/mac-DC-SIGN, or DC (2.5 × 105 cells) were incubated with HIV-1 (multiplicity of infection ≈0.1) in a total volume of 400 μl for 3 h to allow cellular adsorption of the virus. For the antibody-blocking assay, cells were preincubated with cross-reactive DC-SIGN mAb 507 or 526 (10 μg/ml) for 30 min at 37°C before virus addition. After 3 h, cells were washed with 1 ml of PBS and were cocultured with Hut/CCR5 targets (1 × 105 cells) in the presence of 10 μg/ml Polybrene in 1 ml of cell culture medium. Cell lysates were obtained after 2 days and analyzed for luciferase activity (Promega).
mac-DC-SIGN RNA Expression Analysis.
Total RNA from cultured human and macaque immature DC and PBMC was isolated with Trizol. Three micrograms of the isolated RNA was electrophoresed on a 1% agarose gel, transferred to Hybond-XL (Amersham Pharmacia) as described (33), and then incubated with a radioactively labeled 1.2-kb mac-DC-SIGN cDNA probe containing the presumptive full coding sequence. Probe hybridization was performed by using the ExpressHyb hybridization solution (CLONTECH).
Results
Isolation and Sequence Alignment of mac-DC-SIGN with hu-DC-SIGN.
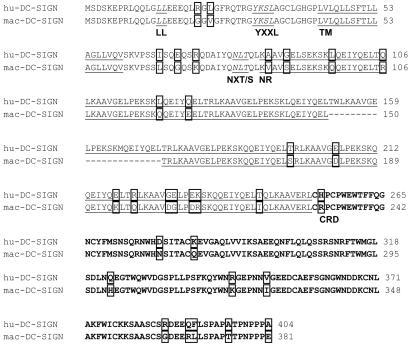
Alignment of the nucleotide and predicted amino acid sequence of hu-DC-SIGN and mac-DC-SIGN revealed 94% identity at the nucleotide level and 92% identity at the amino acid level. Alignment of the predicted protein sequences is shown in Fig. 1. The potential N-linked glycosylation sites (NXT/S) (34) and cytoplasmic functional domains (LL, dileucine motif; YXXL, internalization motif) (35–38) as well as the transmembrane domain of mac-DC-SIGN are identical to those of hu-DC-SIGN (see Fig. 1 legend). The neck region of mac-DC-SIGN contains six repeats of the 23-aa sequence KAAVGELXEKSKXQEIYQELTXL instead of the seven repeats found in hu-DC-SIGN. The numbers of different amino acids in the cytoplasmic tail, neck repeats, and carbohydrate recognition domains between hu-DC-SIGN and mac-DC-SIGN are 2, 15, and 11, respectively. The carbohydrate recognition domain region, which is required for virus and ICAM-3 binding, is 93% identical between hu-DC-SIGN and mac-DC-SIGN.
Figure 1.
Alignment of the presumptive amino acid sequence of hu-DC-SIGN and mac-DC-SIGN. Predicted N-linked glycosylation sites (NXT/S) (34) and cytoplasmic functional motifs (LL, dileucine motif; YXXL, potential internalization motif) (35–38) are annotated (italicized and underlined). The beginning of each of the following is marked: TM, transmembrane domain (underlined); NR, neck repeats (underlined); CRD, carbohydrate recognition domain (bold type). Nonconserved amino acids are boxed. Amino acids that precede the transmembrane domain represent cytoplasmic residues, and those that follow the transmembrane domain jut into the extracellular matrix.
Reactivity of mac-DC-SIGN with Anti-hu-DC-SIGN mAbs.
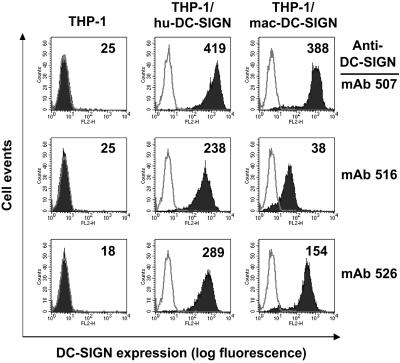
Given that mac-DC-SIGN is highly homologous to hu-DC-SIGN, we tested whether monoclonal antibodies raised against hu-DC-SIGN could recognize mac-DC-SIGN. To do this, we generated a stable THP cell line expressing cloned mac-DC-SIGN. THP-1/mac-DC-SIGN and THP-1/hu-DC-SIGN cells were stained separately with a panel of six mAbs recognizing hu-DC-SIGN. We found that three of the six mAbs (507, 516, and 526) are hu-DC-SIGN and mac-DC-SIGN cross-reactive (Fig. 2), and mAb 507 showed the highest affinity to both hu-DC-SIGN and mac-DC-SIGN. Surprisingly, despite the high conservation, the other three mAbs that recognize hu-DC-SIGN did not appreciably stain mac-DC-SIGN-expressing cells (data not shown).
Figure 2.
FACS analysis of reactivity of mac-DC-SIGN with anti-hu-DC-SIGN mAbs. Stable THP-1/mac-DC-SIGN and THP-1/hu-DC-SIGN cells were stained separately with six mAbs recognizing hu-DC-SIGN; three of the six mAbs (507, 516, and 526) are mac-DC-SIGN cross-reactive. On all histograms, the gray curve represents staining with an isotype control antibody, whereas the black curve represents DC-SIGN mAb staining. The mean fluorescence is shown in the top right corner of the histograms. One of three representative experiments is shown.
Adhesion of ICAM-3 to mac-DC-SIGN.
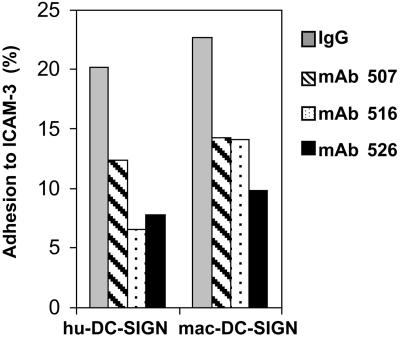
Cells expressing mac-DC-SIGN were assayed for their ability to bind human ICAM-3 affixed to fluorescent microbeads and used to stain cells by the method described by Geijtenbeek et al. (10). Adhesion of ICAM-3 to hu-DC-SIGN or to mac-DC-SIGN cells was similar, indicating that the amino acid differences between hu-DC-SIGN and mac-DC-SIGN are not critical for ICAM-3 adhesion. To test the specificity of interaction, bead binding was performed in the presence of hu-DC-SIGN-reactive mAbs. Cross-reactive DC-SIGN mAb 507, 516, and 526 (10 μg/ml) blocked adhesion of ICAM-3 to hu-DC-SIGN and mac-DC-SIGN (Fig. 3).
Figure 3.
Cross-reactive DC-SIGN mAbs 507, 516, and 526 (10 μg/ml) block adhesion of ICAM-3-coated fluorescent beads to THP-1/hu-DC-SIGN and THP-1/mac-DC-SIGN cells. ICAM-3 bead binding to THP-1 cells was uniformly less than 5% in repeat experiments. Mouse IgG was used as a background antibody control. One of four representative experiments is shown.
mac-DC-SIGN-Mediated HIV-1 Transmission.
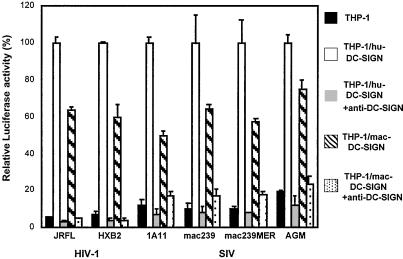
Previously developed DC-SIGN virus capture/transmission assays (11, 32) were used to evaluate whether these functions are preserved in mac-DC-SIGN. In agreement with the high conservation between the molecules, mac-DC-SIGN captured and transmitted HIV-1 or SIV to CD4+ T cells nearly as effectively as hu-DC-SIGN (Fig. 4). In this experiment, stable THP-1 cells expressing hu-DC-SIGN or mac-DC-SIGN were used as donor cells, and human T cells (Hut/CCR5) were used as targets. The relative expression levels of hu-DC-SIGN and mac-DC-SIGN seemed comparable by anti-DC-SIGN mAb 507 staining (Fig. 2) and ICAM-3 binding (Fig. 3). To test the ability of the cells to transmit virus, we compared the HIV-luc vector pseudotyped with two different HIV-1 envelopes (R5-tropic HIV-1JRFL and X4-tropic HIV-1HXB2) and four kinds of SIV envelopes (SIVMAC1A11, SIVMAC239, SIVMAC239MER, and SIVAGM). The luciferase transduction observed by using hu-DC-SIGN donor cells was normalized to 100% for the different virus stocks, and DC-SIGN-negative THP-1 cells were used as a background control in this assay. The results indicate that mac-DC-SIGN transmitted different enveloped primate lentiviruses 50–80% as efficiently as hu-DC-SIGN. Preincubation of cross-reactive DC-SIGN mAb 526 with donor cells significantly impaired virus transmission mediated by THP-1/hu-DC-SIGN or THP-1/mac-DC-SIGN cells, which indicates a direct participation by hu-DC-SIGN or mac-DC-SIGN in the process of virus capture and transmission. Mouse IgG was also used as background antibody controls in this assay, resulting in a similar infection level observed when antibodies were absent. In addition, evaluation of additional HIV-1 and SIV isolates as well as testing with the other two cross-reactive DC-SIGN mAbs (507 and 516) reveals similar effects on virus transmission (data not shown).
Figure 4.
Transmission of HIV-1 pseudotypes mediated by hu-DC-SIGN or mac-DC-SIGN. THP-1, THP-1/hu-DC-SIGN, or THP-1/mac-DC-SIGN donor cells were preincubated with DC-SIGN mAb 526 (10 μg/ml). Next, HIV-luc pseudotyped with Env from R5-tropic HIV-1JRFL, X4-tropic HIV-1HXB2, SIVMAC1A11, SIVMAC239, SIVMAC239MER, or SIVAGM was incubated with cells for 3 h at 37°C. Cells were washed and cocultured with Hut/CCR5 target cells. HIV-1 infection was determined after 2 days by measuring the transduction of luciferase activity. Each data set represents the mean of three separate wells of infected cells.
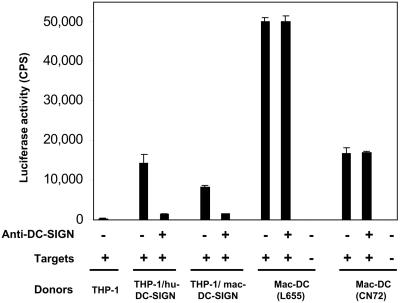
Transmission of SIV by Macaque DC.
To examine the role of mac-DC-SIGN in SIV transmission mediated by macaque DC, we used immature macaque DC as donors and cross-reactive mAbs to test the neutralization of SIV in a capture and transmission assay. Macaque DC derived from two donor animals (L655 and CN72) transmitted virus pseudotyped with SIVMAC1A11 Env at an equal or greater efficiency than THP-1/hu-DC-SIGN or THP-1/macDC-SIGN cells (Fig. 5). Unexpectedly, the efficient transmission of virus by macaque DC could not be blocked with the cross-reactive anti-DC-SIGN mAb 507. Results of another experiment using DC from two other monkey donors were consistent. In addition, a mixture of six hu-DC-SIGN mAbs, each of which can neutralize immature human DC transmission of virus (L.W., T.D.M., R. Vazeux, D.U., and V.N.K., unpublished data), was similarly ineffective on immature macaque DC pulsed with different pseudotypes (data not shown). Finally, use of soluble mannan, which impairs HIV-1 transmission from human DC, had almost no effect on virus transmission by macaque DC (10–20% inhibition). These data suggest that macaque DC transmit primate lentiviruses in a DC-SIGN-independent manner.
Figure 5.
Transmission of SIV to Hut/CCR5 cells by macaque DC is not blocked with anti-DC-SIGN. The virus capture and transmission assay was performed as described in Fig. 4. DC-SIGN mAb 507 (10 μg/ml) was preincubated with the DC before pulsing with HIV-luc pseudotyped with SIVMAC1A11 Env. DC isolated from two individual macaques (L655 and CN72) were use as donor cells, respectively. Each data set represents the mean of three separate wells of infected cells. CPS, counts per second.
Expression of mac-DC-SIGN in Primary Cells.
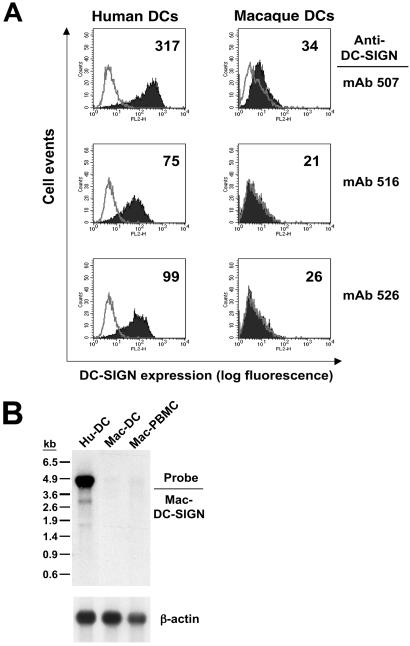
To better understand why primate lentivirus transmission by macaque DC was not impaired by pretreating with DC-SIGN-neutralizing mAbs, we quantitatively examined mac-DC-SIGN expression in primary cells. Immature macaque DC stained with the three cross-reactive DC-SIGN mAbs (507, 516, and 526) revealed that only one antibody (507) weakly stained these cells, indicating a low level of mac-DC-SIGN expression (Fig. 6A). This staining was performed with DC from the animal (L655) used in the previous virus capture/transmission experiment. Results with immature DC from 10 other monkey donors were similar. Parallel maturation of DC from several monkey donors did not reveal any increase in DC-SIGN expression.
Figure 6.
Expression of mac-DC-SIGN in vivo. (A) FACS analysis of mac-DC-SIGN expression on macaque DC stained with the cross-reactive DC-SIGN mAbs 507, 516, and 526. On all histograms, the gray curve represents staining with an isotype control antibody, whereas the black curve indicates DC-SIGN staining. The mean fluorescence is shown in the top right corner of the histograms. (B) Expression of mac-DC-SIGN mRNA. RNA isolated from hu-DC, mac-DC, or mac-PBMC was hybridized with a 1.2-kb mac-DC-SIGN cDNA probe. The human β-actin probe was used as a control for RNA loading.
To evaluate the expression of mac-DC-SIGN mRNA, total RNA isolated from immature human and macaque DC and macaque PBMC was hybridized with a 1.2-kb mac-DC-SIGN cDNA probe (Fig. 6B). Only RNA obtained from human DC strongly hybridized with the probe, revealing the 4.3-kb transcript for hu-DC-SIGN (32). A minimal RNA signal for mac-DC-SIGN was observed in macaque DC and PBMC. Indeed, mac-DC-SIGN was initially isolated by RT-PCR from macaque DC. Consistent with this result, reverse transcription of RNA from immature macaque DC followed by two rounds of PCR was necessary to obtain the 1.2-kb product of mac-DC-SIGN cDNA from a purified DC population (data not shown). These findings collectively indicate that macaque DC express very low levels of mac-DC-SIGN.
Discussion
We have described the cloning and characterization of mac-DC-SIGN, a molecule that shares many properties with its human counterpart, with the notable exception of its expression pattern. mac-DC-SIGN is very similar to hu-DC-SIGN, with the predicted protein displaying 92% identity to hu-DC-SIGN and 82% identity to the recently reported human L-SIGN/DC-SIGNR (liver/lymph node-specific ICAM-3-grabbing nonintegrin/DC-SIGN-related molecule; refs. 32, 38, 39). Given the strong conservation between macaque and human proteins, it is not surprising that mac-DC-SIGN can interact with human ICAM-3 and also capture and efficiently transmit primate lentiviruses. mac-DC-SIGN belies its namesake, however, as there is only minimal expression in myeloid-lineage DC. mac-DC-SIGN-reactive antibodies detected very little protein on the surface of macaque DC. This finding was supported by Northern blot analysis of DC-SIGN mRNA. Although DC-SIGN mRNA is expressed at high levels in human DC, very little mRNA is found in macaque DC.
Infection analyses demonstrated that cloned mac-DC-SIGN captured and transmitted HIV-1 and SIV efficiently, and this effect could be neutralized by cross-reactive DC-SIGN mAbs. Consistent with our observation that macaque DC poorly expressed DC-SIGN, macaque DC were not impaired in virus capture or transmission by neutralizing antibody. It is unlikely that low levels of DC-SIGN expression in macaque DC contribute to their ability to transmit primate lentiviruses. Indeed, a recent study has indicated that high levels of DC-SIGN expression (60,000 molecules per cell) are required for efficient DC-SIGN virus transmission function (17). Moreover, cell lines overexpressing macaque DC-SIGN in our study were less potent in virus transmission than the macaque DC.
Use of an SIV Env pseudotyped HIV-1 vector was particularly beneficial in the macaque DC analysis, given that HIV-1 replication is blocked at an early stage in primary macaque cells (40, 41). Unlike analyses with human DC, single-round infection is detected only in the human T cell targets present in the coculture and not in the macaque DC because of the species-specific restriction. The lack of macaque DC infection is clearly shown by the direct infection challenge of the macaque DC with HIV-luc/1A11 that are not cocultured with Hut/CCR5 cells. In contrast, when Hut/CCR5 cells are placed in coculture, a potent transmission is observed. In this system, absolutely no neutralization of virus was observed with mAb 507 or even with a mixture of six distinct mAbs known to neutralize hu-DC-SIGN (data not shown), including the three cross-reactive DC-SIGN mAbs used in this study. These results are not fully inconsistent with DC-SIGN neutralization studies that we performed with human DC. Capture of HIV-1 by human DC and transmission to CD4+ T cells was never diminished completely by any of the six reactive DC-SIGN mAbs (60–80% inhibition) despite the robust neutralization observed by using DC-SIGN-engineered cell lines (L.W., T.D.M., R. Vazeux, D.U., and V.N.K., unpublished results). Collectively, our data strongly suggest that DC, particularly macaque DC, can transmit primate lentiviruses in a manner that is not strictly dependent on DC-SIGN. We know of no earlier formal demonstration that macaque DC can transmit virus without prior infection in a purely trans manner.
We have recently determined that human monocytes can be induced to express DC-SIGN by using cytokines released by CD4+ T helper type 2 (Th2) cells (L.W., T.D.M., R. Vazeux, D.U., and V.N.K., unpublished results). Rhesus monocytes and macrophages cultured under similar conditions may be useful in divining the expression of mac-DC-SIGN. FACS analysis of macaque PBMC indicated that a subset of non-DC reacted with the cross-reactive DC-SIGN mAbs; however, these cells have not been definitely identified. Similarly, immunohistochemical staining and/or in situ message analysis of macaque placental, liver, lymphoid, and mucosal tissues may provide greater sensitivity in identifying smaller candidate cell populations that express mac-DC-SIGN. Despite the minimal expression of mac-DC-SIGN in DC of the myeloid/monocyte lineage, it is possible that different DC populations express mac-DC-SIGN during their ontogeny. Thus, a role for mac-DC-SIGN in SIV pathogenesis in vivo cannot be ruled out.
However, the more significant point that these studies bring to light is that macaque DC in the absence of DC-SIGN expression are still completely competent in their ability to transmit virus to T cells. Precisely how these cells capture and transmit virus to target CD4+ T cells is currently not understood. Although we find that human DC transmission of HIV-1 can be inhibited by at least 60% by preincubation with soluble mannan, inconclusive mannan inhibition results were obtained with macaque DC. It would seem that conserved properties of DC in their interactions with CD4+ T cells have proven beneficial to primate lentiviruses in facilitating their dissemination to the T cell compartment. Prior studies have demonstrated that human or macaque DC transmission of HIV-1 or SIV to autologous CD4+ T cells is Env dependent (15, 26). Thus, examination of the normal biological roles of DC, such as antigen acquisition, antigen presentation, and T cell priming/stimulation, may provide insight into the processes that are also being usurped by the viral pathogen.
If a non-DC-SIGN family molecule(s) is involved in macaque DC transmission of primate lentiviruses, is there a human homolog(s) expressed on more than just myeloid-derived human DC, such as plasmacytoid DC (42), follicular DC, or CD1a+ Langerhans cells (10, 43), all of which are negative for DC-SIGN? Similarly, what is the preferred ICAM-3 ligand on myeloid-derived macaque DC (or do interactions between macaque T cells and DC not depend on ICAM-3)? Human monocyte-derived DC express at least three C-type lectin receptor molecules, including DC-SIGN, that are capable of binding HIV-1 Env (44). Whether these candidate molecules play a role in transmission similar to DC-SIGN has to be tested next. Clearly, the concept of a viral attachment receptor acting primarily in trans was previously unconsidered, and more likely, unrecognized in the rich experimental histories of prokaryotic and eukaryotic virus biology. Rather than being the exception to the rule, DC-SIGN may prove to be a useful paradigm as we begin to more completely understand cellular interactions that play a role in viral pathogenesis. Thus, despite the dispensability of DC-SIGN for efficient virus transmission from macaque DC, understanding how DC-SIGN confers this property on cells in which it is expressed may prove valuable in elaborating how cells, not just DC, promulgate HIV-1 infection of CD4+ T cells.
Acknowledgments
We thank Chad Borchert, Monica Tsang, and R & D Systems for help in developing monoclonal antibodies to DC-SIGN; Doug Kwon and Dan Littman for providing cells; and Genoveffa Franchini, Jeff Lifson, Jeff Rosio, and the Tulane Regional Primate Research Center for primary cells. We thank Jeff Lifson and members of the KewalRamani lab for scientific discussions. We thank Anne Arthur and Stephen Hughes for manuscript comments. Primary funding for this research was provided by the National Cancer Institute's intramural Center for Cancer Research, which supports the HIV Drug Resistance Program. This publication has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract NO1-CO-12400. Additional funding supporting this research was obtained from The Rockefeller Foundation and National Institutes of Health Grant AI40877.
Abbreviations
- DC
dendritic cell(s)
- ICAM-3
intercellular adhesion molecule 3
- hu-DC-SIGN
human DC-specific ICAM-3-grabbing nonintegrin
- mac-DC-SIGN
rhesus macaque DC-SIGN
- Env
envelope
- FACS
fluorescence-activated cell sorter
- PBMC
peripheral blood mononuclear cells
- SIV
simian immunodeficiency virus
- CCR
CC chemokine receptor
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY040319).
References
- 1.Cameron P U, Freudenthal P S, Barker J M, Gezelter S, Inaba K, Steinman R M. Science. 1992;257:383–387. doi: 10.1126/science.1352913. [DOI] [PubMed] [Google Scholar]
- 2.Cameron P U, Lowe M G, Crowe S M, O'Doherty U, Pope M, Gezelter S, Steinman R M. J Leukocyte Biol. 1994;56:257–265. doi: 10.1002/jlb.56.3.257. [DOI] [PubMed] [Google Scholar]
- 3.Cameron P, Pope M, Granelli-Piperno A, Steinman R M. J Leukocyte Biol. 1996;59:158–171. doi: 10.1002/jlb.59.2.158. [DOI] [PubMed] [Google Scholar]
- 4.Zaitseva M, Blauvelt A, Lee S, Lapham C K, Klaus-Kovtun V, Mostowski H, Manischewitz J, Golding H. Nat Med. 1997;3:1369–1375. doi: 10.1038/nm1297-1369. [DOI] [PubMed] [Google Scholar]
- 5.Blauvelt A, Asada H, Saville M W, Klaus-Kovtun V, Altman D J, Yarchoan R, Katz S I. J Clin Invest. 1997;100:2043–2053. doi: 10.1172/JCI119737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granelli-Piperno A, Delgado E, Finkel V, Paxton W, Steinman R M. J Virol. 1998;72:2733–2737. doi: 10.1128/jvi.72.4.2733-2737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubbert A, Combadiere C, Ostrowski M, Arthos J, Dybul M, Machado E, Cohn M A, Hoxie J A, Murphy P M, Fauci A S, Weissman D. J Immunol. 1998;160:3933–3941. [PubMed] [Google Scholar]
- 8.Patterson S, Robinson S P, English N R, Knight S C. Immunol Lett. 1999;66:111–116. doi: 10.1016/s0165-2478(98)00166-7. [DOI] [PubMed] [Google Scholar]
- 9.Granelli-Piperno A, Finkel V, Delgado E, Steinman R M. Curr Biol. 1999;9:21–29. doi: 10.1016/s0960-9822(99)80043-8. [DOI] [PubMed] [Google Scholar]
- 10.Geijtenbeek T B, Torensma R, van Vliet S J, van Duijnhoven G C, Adema G J, van Kooyk Y, Figdor C G. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 11.Geijtenbeek T B, Kwon D S, Torensma R, van Vliet S J, van Duijnhoven G C, Middel J, Cornelissen I L, Nottet H S, KewalRamani V N, Littman D R, et al. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 12.Curtis B M, Scharnowske S, Watson A J. Proc Natl Acad Sci USA. 1992;89:8356–8360. doi: 10.1073/pnas.89.17.8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankel S S, Wenig B M, Burke A P, Mannan P, Thompson L D, Abbondanzo S L, Nelson A M, Pope M, Steinman R M. Science. 1996;272:115–117. doi: 10.1126/science.272.5258.115. [DOI] [PubMed] [Google Scholar]
- 14.Granelli-Piperno A, Moser B, Pope M, Chen D, Wei Y, Isdell F, O'Doherty U, Paxton W, Koup R, Mojsov S, et al. J Exp Med. 1996;184:2433–2438. doi: 10.1084/jem.184.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankel S S, Steinman R M, Michael N L, Kim S R, Bhardwaj N, Pope M, Louder M K, Ehrenberg P K, Parren P W, Burton D R, et al. J Virol. 1998;72:9788–9794. doi: 10.1128/jvi.72.12.9788-9794.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinman R M, Inaba K, Turley S, Pierre P, Mellman I. Hum Immunol. 1999;60:562–567. doi: 10.1016/s0198-8859(99)00030-0. [DOI] [PubMed] [Google Scholar]
- 17.Pöhlmann S, Baribaud F, Lee B, Leslie G J, Sanchez M D, Hiebenthal-Millow K, Munch J, Kirchhoff F, Doms R W. J Virol. 2001;75:4664–4672. doi: 10.1128/JVI.75.10.4664-4672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J, Pope M, Brown C, O'Doherty U, Miller C J. Lab Invest. 1998;78:435–451. [PubMed] [Google Scholar]
- 19.Stahl-Hennig C, Steinman R M, Tenner-Racz K, Pope M, Stolte N, Matz-Rensing K, Grobschupff G, Raschdorff B, Hunsmann G, Racz P. Science. 1999;285:1261–1265. doi: 10.1126/science.285.5431.1261. [DOI] [PubMed] [Google Scholar]
- 20.Hu J, Miller C J, O'Doherty U, Marx P A, Pope M. AIDS Res Hum Retroviruses. 1999;15:1305–1314. doi: 10.1089/088922299310205. [DOI] [PubMed] [Google Scholar]
- 21.Margolis L, Glushakova S, Chougnet C, Shearer G, Markham P, Robert-Guroff M, Benveniste R, Miller C J, Cranage M, Hirsch V, Franchini G. Virology. 2000;275:391–397. doi: 10.1006/viro.2000.0528. [DOI] [PubMed] [Google Scholar]
- 22.Hu J, Gardner M B, Miller C J. J Virol. 2000;74:6087–6095. doi: 10.1128/jvi.74.13.6087-6095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Rompay K K, Miller M D, Marthas M L, Margot N A, Dailey P J, Canfield D R, Tarara R P, Cherrington J M, Aguirre N L, Bischofberger N, Pedersen N C. J Virol. 2000;74:1767–1774. doi: 10.1128/jvi.74.4.1767-1774.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crotty S, Miller C J, Lohman B L, Neagu M R, Compton L, Lu D, Lu F X, Fritts L, Lifson J D, Andino R. J Virol. 2001;75:7435–7452. doi: 10.1128/JVI.75.16.7435-7452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenier J L, Miller C J, Lu D, Dailey P J, Lu F X, Kunstman K J, Wolinsky S M, Marthas M L. J Virol. 2001;75:3753–3765. doi: 10.1128/JVI.75.8.3753-3765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ignatius R, Isdell F, O'Doherty U, Pope M. J Med Primatol. 1998;27:121–128. doi: 10.1111/j.1600-0684.1998.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 27.Ignatius R, Wei Y, Beaulieu S, Gettie A, Steinman R M, Pope M, Mojsov S. AIDS Res Hum Retroviruses. 2000;16:1055–1059. doi: 10.1089/08892220050075318. [DOI] [PubMed] [Google Scholar]
- 28.Pope M, Elmore D, Ho D, Marx P. AIDS Res Hum Retroviruses. 1997;13:819–827. doi: 10.1089/aid.1997.13.819. [DOI] [PubMed] [Google Scholar]
- 29.O'Doherty U, Ignatius R, Bhardwaj N, Pope M. J Immunol Methods. 1997;207:185–194. doi: 10.1016/s0022-1759(97)00119-1. [DOI] [PubMed] [Google Scholar]
- 30.Cecilia D, KewalRamani V N, O'Leary J, Volsky B, Nyambi P, Burda S, Xu S, Littman D R, Zolla-Pazner S. J Virol. 1998;72:6988–6996. doi: 10.1128/jvi.72.9.6988-6996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Connor R I, Chen B K, Choe S, Landau N R. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 32.Bashirova A A, Geijtenbeek T B, van Duijnhoven G C, van Vliet S J, Eilering J B, Martin M P, Wu L, Martin T D, Viebig N, Knolle P A, et al. J Exp Med. 2001;193:671–678. doi: 10.1084/jem.193.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chomczynski P. Anal Biochem. 1992;201:134–139. doi: 10.1016/0003-2697(92)90185-a. [DOI] [PubMed] [Google Scholar]
- 34.Lanctot P M, Leclerc P C, Escher E, Leduc R, Guillemette G. Biochemistry. 1999;38:8621–8627. doi: 10.1021/bi9830516. [DOI] [PubMed] [Google Scholar]
- 35.Trowbridge I S, Collawn J F, Hopkins C R. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- 36.Francis M J, Jones E E, Levy E R, Martin R L, Ponnambalam S, Monaco A P. J Cell Sci. 1999;112:1721–1732. doi: 10.1242/jcs.112.11.1721. [DOI] [PubMed] [Google Scholar]
- 37.Bates E E, Fournier N, Garcia E, Valladeau J, Durand I, Pin J J, Zurawski S M, Patel S, Abrams J S, Lebecque S, et al. J Immunol. 1999;163:1973–1983. [PubMed] [Google Scholar]
- 38.Soilleux E J, Barten R, Trowsdale J. J Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- 39.Pöhlmann S, Soilleux E J, Baribaud F, Leslie G J, Morris L S, Trowsdale J, Lee B, Coleman N, Doms R W. Proc Natl Acad Sci USA. 2001;98:2670–2675. doi: 10.1073/pnas.051631398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shibata R, Sakai H, Kawamura M, Tokunaga K, Adachi A. J Gen Virol. 1995;76:2723–2730. doi: 10.1099/0022-1317-76-11-2723. [DOI] [PubMed] [Google Scholar]
- 41.Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patterson S, Rae A, Hockey N, Gilmour J, Gotch F. J Virol. 2001;75:6710–6713. doi: 10.1128/JVI.75.14.6710-6713.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soilleux E J, Coleman N. Blood. 2001;98:1987–1988. doi: 10.1182/blood.v98.6.1987. [DOI] [PubMed] [Google Scholar]
- 44.Turville S G, Arthos J, Mac Donald K, Lynch G, Naif H, Clark G, Hart D, Cunningham A L. Blood. 2001;98:2482–2488. doi: 10.1182/blood.v98.8.2482. [DOI] [PubMed] [Google Scholar]