Abstract
With the goal of identifying and characterizing traits of Enterococcus faecalis that play key roles in human disease, we identified an operon specifying synthesis of a capsular carbohydrate of the type most commonly expressed by clinical isolates. This surface-exposed carbohydrate consists of glycerol phosphate, glucose, and galactose residues, and its biosynthesis is encoded by a determinant that includes 11 ORFs. Insertional inactivation of genes in this pathway yielded mutants with enhanced susceptibility to phagocytic killing in vitro and compromised in the ability to persist in regional lymph nodes in vivo.
The commensal bacterium Enterococcus faecalis is found in the human gastrointestinal tract, but strains that are often resistant to multiple antibiotics are leading causes of hospital-acquired infection (1). As a result there is considerable interest in developing new strategies for treating enterococcal infections. Antibody to a capsular carbohydrate was recently shown to be essential for efficient, but serotype-specific, opsonization and killing of E. faecalis (2). Two studies examined the serologic diversity of E. faecalis strains (3, 4). The first study identified 11 types related to differences in cell surface carbohydrate (3). The second study proposed a new typing scheme capable of identifying 21 unique serotypes (4), with four serotypes accounting for 72% of all typeable isolates. However, the nature of the cell surface antigens forming the basis for this typing scheme was not examined.
With a view toward developing a scientific basis for deriving new antienterococcal therapies, the present study was designed to identify the pathway for serotype-specific carbohydrate biosynthesis, to determine its relationship to other invariant cell wall carbohydrates, and to determine the contribution of the serotype-specific carbohydrate to the biology of enterococcal infection.
Materials and Methods
Bacterial Strains and Culture.
Plasmid-free E. faecalis FA2–2 (5) was used for generating isogenic mutants by means of insertional inactivation, using the suicide vector p3ERM (6). E. faecalis was routinely cultured in brain heart infusion or Todd–Hewitt broth at 37°C without aeration. Escherichia coli strain XL1-Blue (Stratagene) was cultured aerobically in LB broth at 37°C. Antibiotics (Sigma) used for E. faecalis selection included erythromycin (50 μg/ml) and spectinomycin (500 μg/ml), and for E. coli, tetracycline (12.5 μg/ml), ampicillin (100 μg/ml), erythromycin (400 μg/ml), and spectinomycin (150 μg/ml). Isopropylthio-β-d-galactoside (IPTG) and 5-bromo-4-chloro-3-indoyl-β-galactoside (X-Gal) were used at 0.5 mM and 80 μg/ml, respectively, for induction and detection of β-galactosidase activity.
DNA Manipulation.
Recombinant DNA procedures were carried out as described (7). E. coli plasmid DNA was purified by using a Wizard Prep DNA purification kit (Promega). Restriction and modifying enzymes were purchased from New England Biolabs and Life Technologies, Gaithersburg, MD. Custom oligonucleotides listed in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org, were obtained from Integrated DNA Technologies, Coralville, IA. E. faecalis chromosomal DNA was prepared by using either a QIAMP Tissue Kit (Qiagen, Hilden, Germany) or the method of Pospiech and Neumann (8).
Preliminary Identification of the Serotype Carbohydrate Determinant.
Gene (cpsI), with sequence similarity to carbohydrate biosynthetic pathways from both Gram-negative and positive bacteria (Table 3, which is published as supporting information on the PNAS web site), was identified from among randomly cloned and sequenced fragments of the E. faecalis MMH594 (9) genome. cpsI sequences from M13mp18 (7) clone pCPS307 were subcloned as a 639-bp EcoRI/HindIII fragment into pBluescript II (SK), generating pCPSI. This 639-bp insert was radiolabeled by using the RadPrime Labeling Kit (Life Technologies) and used to probe a Southern blot of chromosomal DNA from E. faecalis strains MMH594, V583, and FA2–2. Once the presence of this gene on the chromosome of plasmid-free strain FA2–2 was established, FA2–2 was used in subsequent mutagenesis studies because it lacked the multiple antibiotic resistances that occur in more recently isolated clinical strains, including MMH594 (9) and V583 (10).
Genetic Identity of the Cell Surface Carbohydrate Determinants in Strains FA2–2 and V583.
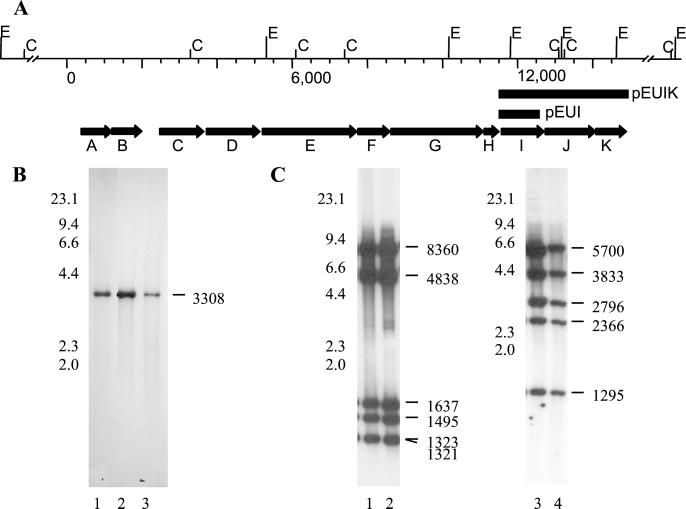
During the course of obtaining sequence information for the MMH594 cpsI gene and its flanking regions, the Institute for Genomic Research (TIGR) released preliminary sequence data for the genome of E. faecalis strain V583. The initially detected cpsI gene was mapped to a large contig of 33,741 bp (http://www.tigr.org). Restriction analysis, PCR amplification, and sequencing of each open reading within the cps determinant confirmed the identity throughout this region between strains V583 and FA2–2. Primers CpsL and CpsR, which are complementary to conserved flanking sequences, were used to amplify a 14,604-bp fragment (Fig. 1) from strain V583 (TaKaRa LA Taq kit, Panvera, Madison, WI). The 14,604-bp amplified fragment was used as a probe in a Southern blot with chromosomal DNA from V583 and FA2–2 (Fig. 1) for evidence of identity throughout this region of the chromosome. DNA sequence information was obtained by using standard dye-labeled chain termination reactions and was analyzed by using the Wisconsin Package version 9.0 (GCG). Gapped blastx (11) was used to search for homologous sequences in the National Center for Biotechnology Information database.
Figure 1.
(A) Genetic organization of the cps locus in E. faecalis strain V583. Restriction sites for EcoRI (E) and ClaI (C) are shown above the map. Solid bars mark regions used for complementing cpsI in strain HG101. The direction of transcription and relative size of each gene are designated by bold arrows. (B) Hybridization analysis of MMH594, V583, and FA2–2 total DNA, digested with HindIII and probed with radiolabeled cpsI. Lane 1, MMH594; lane 2, V583; lane 3, FA2–2. The relative position of λ HindIII size standard (kb) is shown to the left of lane 1, and the size (bp) of the HindIII fragment detected by the cpsI probe is shown to the right of lane 3. (C) Hybridization analysis of V583 and FA2–2 total DNA, digested with EcoRI (lanes 1 and 2) or ClaI (lanes 3 and 4), and probed with the radiolabeled cpsA-K fragment. Lanes 1 and 3, V583; lanes 2 and 4, FA2–2. The relative position of λ HindIII size standard (kb) is shown to the left of lanes 1 and 3 and the size (bp) of fragments hybridizing to the probe is shown to the right of lanes 2 and 4.
Insertional Inactivation.
To insertionally inactivate cpsI, the 639-bp EcoRI/HindIII fragment from pCPSI was subcloned into p3ERM to generate pLEH101. To insertionally inactivate a gene that appeared to constitute the 5′ end of a putative operon, cpsC, a 355-bp intragenic portion of the gene was amplified by using primers CpsC1 and CpsC2 and cloned into the SmaI site of p3ERM to generate pLEH303. To target the apparent 3′- most gene cpsK, a 594-bp intragenic PCR fragment from cpsK was generated by using primers CpsK1 and CpsK2 and cloned into the SmaI site of p3ERM to generate pLEH202. Plasmids pLEH101, pLEH202, and pLEH303 were introduced into E. faecalis strain FA2–2 by electroporation using a Bio-Rad Gene Pulser as described (12), and transformants were recovered on brain heart infusion agar plates containing erythromycin at 50 μg/ml after 48–72 h incubation at 37°C. Transformants were analyzed by Southern blot to confirm insertional inactivation of the intended targets on the chromosome of FA2–2 (not shown).
Complementation.
Expression vector pEU327 contains a xylose promoter that is constitutively expressed in E. faecalis (13). We used this vector to generate several chimeric plasmids for complementation of the insertionally inactivated cpsI gene in HG101. A 1,260-bp PCR product that contains the entire cpsI gene was generated by using primers CpsI3 and CpsI4 and cloned as a HindIII fragment into pEU327. The orientation of the insert with respect to the xylose promoter was established by restriction analysis using KpnI and SacI. The resulting construct, which placed cpsI under control of the xylose promoter, was termed pEUI. The cpsI-inactivated mutant, HG101, was transformed with pEUI and designated HG101 (pEUI). Because of the possibility of polar effects exerted by the insertion of a suicide vector into cpsI on downstream genes, an additional complementing plasmid harboring cpsI and the downstream genes cpsJ and cpsK was made by using primers CpsI5 and CpsK3. XhoI sites at both ends of the 3,466-bp fragment permitted cloning into the unique SalI site of pEU327. Orientation of the insert was confirmed by restriction with ApaI and SacI. One plasmid, possessing the insert oriented in the proper direction for transcription from the xylose promoter and designated pEUIK, was used to transform HG101, resulting in HG101(pEUIK).
Purification and Characterization of Cell Wall Polysaccharides.
E. faecalis strain FA2–2 was grown at 37°C without aeration to midlog in 10 liters of Todd–Hewitt broth supplemented with 1.0% glucose. For purification of cell wall polysaccharides, the method of Huebner et al. (2) was used with modification. After enzymatic digestion of the enterococcal cell wall, carbohydrates were precipitated by the addition of ethanol to a final concentration of 75%. The precipitable material was collected by centrifugation (22,000 × g, 20 min), washed with 75% ethanol, air-dried, and redissolved in 50 mM Tris⋅HCl (pH 8.0), 150 mM NaCl.
Crude carbohydrate was applied to a Sephacryl S-500 column (Amersham Pharmacia) and serotype carbohydrate-containing fractions were identified by immunodot blot, using serotype-specific antiserum (4) (kindly provided by S. Maekawa, Sapporo Medical College, Sapporo, Japan). To assess purity, chromatographic fractions were analyzed by electrophoresis through 10% polyacrylamide (33:1) in Tris-borate buffer (0.2 M Tris-base/0.2 M boric acid/20 mM EDTA, pH 8.3), with detection of polysaccharides using the cationic dye Stains-All (3,3′-dimethyl-9-methyl-4,5,4′5′-dibenzothiacarbocyanine) (14). Gels were stained for a minimum of 6 h in the dark and destained in distilled water before photography. Serologically reactive fractions were further purified by using DEAE Bio-Gel A (Bio-Rad) and eluted with a linear NaCl salt gradient (0–500 mM) in starting buffer of 10 mM NaOAc (pH 6.0).
For size determination, carbohydrates were labeled with N-methyl isatoic anhydride (15). The molecular size distribution of each purified carbohydrate fraction was determined by comparison to fluorescein-labeled dextran standards (Sigma). Dextran standards and labeled carbohydrate fractions were resolved on a Polysep 3000 column (7.8 × 300 mm; Phenomenex, Torrance, CA), eluted in 0.2 M sodium nitrate, and detected with UV absorbance at 350 nm.
Carbohydrate Compositional Analysis.
Preliminary data on carbohydrate composition was obtained at the Complex Carbohydrate Research Center at the University of Georgia, Athens and the Channing Laboratory at Harvard University, Boston. Further analysis of purified carbohydrate fractions was performed at the University of Oklahoma Center for Medical Glycobiology by using high-pH anion-exchange chromatography (MA-1; Dionex) with pulsed-amperometric detection (model PAD2; Dionex) (16). Samples were hydrolyzed with 2 M trifluoroacetic acid at 100°C for 5 h, before injection onto the column. Sugars were eluted with 120 mM sodium hydroxide at a flow rate of 0.4 ml/min. Phosphate determinations for each sample were performed as described (17).
In Vivo Studies.
A s.c. infection model was used to assess the contribution of the serotype carbohydrate to the persistence of E. faecalis in vivo. Five- to 6-week-old female BALB/c mice (Harlan Sprague–Dawley) were injected s.c. with 200 μl of 5 × 107 colony-forming units (cfu) of FA2–2 or cpsI mutant HG101. On days 2 and 4 postinjection, draining abdominal lymph nodes were resected. Tissue samples were homogenized with a mini bead-beater (Bio-Spec Products, Bartlesville, OK) and serially diluted and plated onto brain heart infusion agar by track dilution (18). The mean numbers of recovered wild-type and mutant cfu were compared, and significance was determined by a Student's t test for two-group comparisons (19).
Opsonophagocytosis Assay.
An opsonophagocytosis assay was used to quantify the resistance to phagocytic killing of FA2–2, HG101, and HG103 in the absence of a specific antibody source. Neutrophils were isolated as described (20), using the LymphoPrep reagent (Life Technologies), and resuspended in 5 ml of Hanks' balanced salt solution (Life Technologies). Neutrophils were enumerated in a hemocytometer after trypan blue staining, and the final concentration was adjusted to 2 × 107 cells per ml. Human serum as a complement source was obtained from healthy volunteers and exhaustively adsorbed with E. faecalis strain FA2–2. Heat-treated normal rabbit serum was similarly adsorbed to remove antienterococcal antibodies. The phagocytosis assay was performed as described by Huebner et al. (2). The mean numbers of cfu surviving in the various samples were compared, and the overall significance of the differences was determined by a Student's t test.
Results
Preliminary Identification of a Carbohydrate Biosynthetic Pathway.
Sequence information from E. faecalis MMH594, a strain that caused multiple bacteremias and is associated with acutely terminal outcome (9), identified a number of genes potentially related to cell wall polysaccharide biosynthesis. One sequence in particular, i.e., that harbored by pCPS307 showed significant sequence similarity to the UDP-galactopyranose mutase in the type 33f capsule biosynthetic pathway of Streptococcus pneumoniae (21) and in the pathways for O-antigen biosynthesis in E. coli (22) and Klebsiella pneumoniae (23) (Table 3).
Southern blot confirmed the identity of the cloned fragment and flanking sequences on the chromosomes of MMH594, V583, and FA2–2 (Fig. 1). Using the released genome sequence information for strain V583 (www.tigr.org), we identified a large contig that included the putative UDP-galactopyranose mutase gene, designated cpsI. blastx analysis of sequence flanking cpsI revealed a block of genes possessing homologs related to polysaccharide biosynthesis (Fig. 1, Table 3). To confirm the sequence relationship between V583 and FA2–2, primers to each ORF in the cpsA-K region were designed, based on the V583 sequence, and used to amplify corresponding sequences from FA2–2. PCR products from strain FA2–2 of the predicted size were obtained in each case and sequenced in a single pass to confirm identity.
Generation and Analysis of the cpsI Mutant.
To examine the biological role for the cps determinant in E. faecalis strain FA2–2, and to determine the involvement of genes spanning the cpsC-K cluster in capsular polysaccharide biosynthesis, the suicide vector p3ERM was targeted to the cpsC, cpsI, and cpsK genes by inclusion of respective intragenic PCR fragments, and the fidelity of the resulting mutants was determined by Southern blot (data not shown).
In a preliminary report,§ we found the cpsI gene to be present in 60% of the clinical isolates examined. Based on the inferred function of the cpsI gene product, we hypothesized that a disruption in this gene would affect the production of a cell wall carbohydrate in a subset of E. faecalis isolates. To test this hypothesis, we performed slide agglutination with a previously characterized typing serum (4). Results from agglutination tests demonstrated specificity of the typing sera for the known serotypes and showed that strain FA2–2 was agglutinated by Maekawa type 2 antiserum. The cpsI mutant, HG101, failed to agglutinate with this antiserum (data not shown), confirming the prediction that cpsI relates to serotype antigen expression.
Purification of Enterococcal Carbohydrate Polymers.
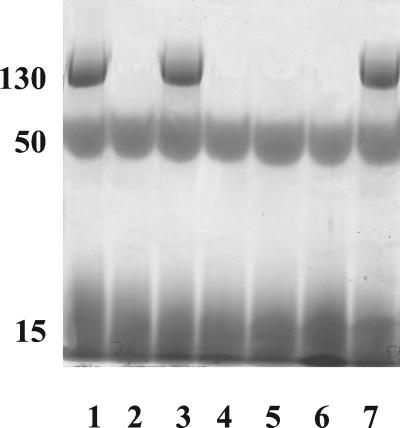
To determine that the Maekawa type 2 antiserum recognized a carbohydrate, we isolated total cell wall carbohydrates from FA2–2. Three distinct carbohydrate fractions were detected with the cationic dye, Stains-All (Fig. 2), and the size distribution of each fraction was determined by gel filtration (data not shown). Of the three carbohydrate fractions (130 kDa, 50 kDa, and 15 kDa), only the largest molecular mass polymer was detected by immunoblot with the type 2 antiserum (data not shown).
Figure 2.
Detection of variation in cell wall carbohydrate polymer species among cps mutants by electrophoresis through a 10% polyacrylamide gel with Stains-All detection. Lane 1, FA2–2; lane 2, HG101 (cpsI−); lane 3, HG103 (HG101 complemented with pEUIK); lane 4, HG102 (HG101 complemented with pEUI); lane 5, HG303 (cpsC−); lane 6, HG202 (cpsK−); lane 7, HG404 (insertional inactivation of an ORF 3′ to cpsK). The mean molecular size (kDa) of each carbohydrate, as assessed by gel filtration, is shown at the left.
Composition Analysis.
Compositional analysis of purified carbohydrate fractions was performed by high-pH anion-exchange chromatography–pulsed-amperometric detection (16), and the results, along with total phosphate determinations (17), are shown in Table 1.
Table 1.
Compositional analysis of E. faecalis FA2-2 cell wall polysaccharides
| Polysaccharide | Molar ratio
|
||||||
|---|---|---|---|---|---|---|---|
| Gro | Rha | GalN | GlcN | Gal | Glc | Pho | |
| 130 kDa | 1 | 1 | 4 | 2 | |||
| 50 kDa | 4 | 2 | 2 | 1 | 1 | 1 | |
| 15 kDa | 1 | 1 | 2 | ||||
Gro, glycerol; Rha, rhamnose; GalN, galactosamine; GlcN, glucosamine; Gal, galactose; Glc, glucose; Pho, phosphate.
Complementation of HG101 and Characterization of Additional cps Mutants.
As noted, an insertionally inactivated cpsI mutant did not react with the type 2 antiserum that recognized the parental strain. To control for polar effects that could confound interpretation of this result, we constructed several complementing plasmids by using the expression vector pEU327. Plasmids pEUI and pEUIK contain the cpsI gene and genes cpsI to cpsK, respectively. Each expression plasmid was introduced into the cpsI mutant, HG101, by electroporation, resulting in strains HG101(pEUI) and HG101(pEUIK). Carbohydrate polymers isolated from both strains were examined by PAGE, with Stains-All detection. Despite the presence of cpsI in trans, strain HG101 (pEUI) lacked the larger molecular weight serotype carbohydrate, which was, however, restored in strain HG101(pEUIK) (Fig. 2). Additionally, type 2 antiserum agglutinated HG101(pEUIK), but not HG101(pEUI) (data not shown). These results demonstrate that insertional inactivation of cpsI not only inactivated the intended target, but also affected expression of cpsJ and cpsK, providing genetic evidence that all are cotranscribed, and further that there are no cotranscribed genes 3′ to cpsK required for serotype carbohydrate expression.
To affix the 3′ end of the type 2-specific cell wall carbohydrate determinant, cpsK was directly insertionally inactivated, resulting in loss of the serotype-specific carbohydrate band (Fig. 2); disruption of the ORF immediately 3′ to cpsK had no effect. Similarly, cpsC was insertionally inactivated in strain HG303, which also lost the ability to synthesize this carbohydrate (Fig. 2), implicating the involvement of genes 5′ to cpsI up to cpsC in carbohydrate biosynthesis. Insertional inactivation of cpsA and cpsB, under conditions that yielded mutants in cpsC, cpsI, and cpsK, proved unsuccessful, indicating that cpsA and cpsB represent essential genes.
Clearance at a Site of Infection.
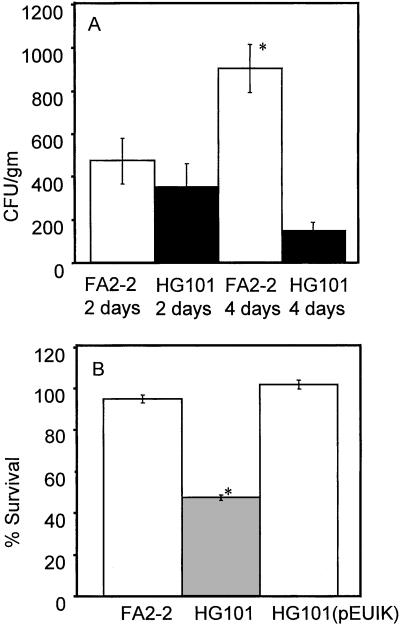
The cell surface localization of the FA2–2 carbohydrate antigen, as shown by the agglutination reaction, indicated that it was positioned at the interface between bacterium and host. The observation that this serotype was predominant among clinical isolates (24) suggested that it may play a role in infection. Therefore, the wild-type strain FA2–2 and its isogenic mutant HG101 were compared in a murine s.c. abscess model (25) (Fig. 3). HG101 was more readily cleared from the resulting abscess, as measured by a reduction in viable organisms recovered from the abdominal lymph nodes that drain this site.
Figure 3.
(A) Persistence in infection for FA2–2 and HG101. Persistence was monitored at 2 and 4 days. The mean from 13 animals is shown, with error bars representing SEM. * represent statistically significant differences (P < 0.01). (B) Opsonophagocytic killing of E. faecalis strains FA2–2, HG101, and HG103. Percent survival was calculated by dividing the cfu recovered after 90 min (t = 90) by the cfu at time 0 (t = 0), multiplied by 100. Experiments were performed in triplicate on two separate occasions, and the data from both experiments were pooled. Error bars for six independent measures show SEM. * represent statistically significant (P < 0.01) differences relative to cfu recovered from strains FA2–2 and HG103.
Opsonophagocytic Killing.
The observation that HG101 was more readily cleared from murine cutaneous infection sites indicated an increased susceptibility to host immune clearance. To test this hypothesis directly, we assessed susceptibility to human neutrophil-mediated killing in opsonophagocytosis assays for strains FA2–2, HG101, and the fully complemented strain, HG101 (pEUIK). The results showed that in the absence of specific antibody strains FA2–2 and HG101 (pEUIK) were not killed by neutrophils, whereas HG101 was reduced by 50% during the course of the experiment (Fig. 3).
Discussion
Enterococci are leading causes of hospital-acquired infection (26). The emergence of clinical isolates refractory to all antimicrobial regimens highlights the need for developing new therapeutic approaches. Immunotherapy and targeting the cell wall carbohydrate biosynthesis pathway represent viable alternatives. A recent report described the opsonic effect of antibodies directed to capsular antigens in a subset of E. faecalis and E. faecium isolates (2). The structure of the capsular antigen was shown to be a repeat structure of a kojibiose linked 1, 2 to glycerol phosphate. The determinant responsible for synthesis of this antigen has yet to be reported. Two reports by Xu and coworkers (27, 28) identified an E. faecalis genetic determinant from strain OG1RF capable of synthesizing a polysaccharide antigen in E. coli. However, the structure and composition of this antigen has not been determined, and this antigen was undetectable on the surface of the source strain (28). We therefore undertook the current study to reconcile these disparate observations and to establish a clear understanding of cell wall carbohydrate biosynthesis by E. faecalis.
Studies by Maekawa and coworkers (4, 24) showed that of typeable E. faecalis isolates (23% could not be typed), four serotypes (serotypes 1, 2, 4, and 7) accounted for 72%. Type 2, the most prominent, accounted for 30% of the typeable isolates. Using Maekawa's type 2 antiserum (4), we found that mutants in the polysaccharide biosynthetic pathway identified in the present study are no longer capable of synthesizing the agglutinating surface antigen recognized by type 2 serum. We also showed that in addition to the serotype polysaccharide antigen, E. faecalis strain FA2–2 produces additional polysaccharides of lower molecular weight.
Compositional analysis of cell-wall polysaccharides demonstrated that the Maekawa type 2 antiserum-agglutinated carbohydrate contained glycerol, phosphate, glucose, and galactose. The presence of glycerol phosphate in this carbohydrate was expected from the inferred function of the cpsC gene product, which in primary sequence is highly related to glycerophosphotransferases (Table 3). The overall composition of the polymer shows some relation to the carbohydrate purified by Huebner et al. (2), which contains glucose, glycerol, and phosphate in a 2:1:2 ratio. The composition of the serotype carbohydrate expressed by strain FA2–2 characterized in the present study consists of glucose, galactose, glycerol, and phosphate in a 4:1:1:2 ratio. Importantly, type 2 antiserum failed to agglutinate strain 12030 used in the previous study (2).
The carbohydrate polymer of ≈50 kDa identified in the present study contains rhamnose, glucosamine, galactosamine, glucose, galactose, and phosphate in a 4:2:2:1:1:1 ratio. The presence of rhamnose in this polysaccharide is predicted by the pathway described previously by us and others (27, 29). Examination of the sequence relatedness of this pathway in strains OG1RF and V583 revealed complete sequence conservation with the exception of the presence of an insertion sequence (IS) element in strain V583 (28). However, analysis of the V583 sequence (www.tigr.org) shows the insertion to occur in the junction between orfde10 and orfde11. As orfde11 and orfde12 are predicted to be on a separate transcript (28), the insertion of an IS element upstream of orfde11 is unlikely to abrogate the functional expression of this determinant in V583. We previously observed the conservation of the rmlA gene in all E. faecalis isolates tested, whereas the cpsI gene was found to be present in 60% (12/20). These data support the model that the pathway described by Xu et al. (27) represents an invariant component of the cell wall of all E. faecalis strains.
The composition of the ≈15-kDa polysaccharide consists of glucose, glycerol, and phosphate in 1:1:2 ratio. Based on its size and chemical composition, this polymer likely represents an integral cell wall teichoic acid. As no genetic pathway has been elucidated for its synthesis, the extent to which it is essential in cell wall structure remains an open question.
Comparison of an isogenic mutant in the serotype 2 determinant with the parental strain in a murine cutaneous infection model demonstrated that the mutant was more readily cleared from the infection site. Furthermore, this mutant was reduced by roughly 50% over controls in an in vitro phagocytic killing assay using human neutrophils. These data demonstrate an important contribution of the serotype polysaccharide in the biology of this organism. The reduction in viable numbers for the mutant in both in vivo and in vitro phagocytic killing experiments, as well as the surface accessibility of this carbohydrate as evident in agglutination reactions, demonstrates that this polysaccharide is accessible on the surface and functions as a capsule.
Based on (i) the ubiquity of the intermediate molecular weight polysaccharide pathway as shown by PCR analysis and sequence identity between V583 and OG1RF (28); (ii) the inability of specific antibodies to detect this polysaccharide on the surface of E. faecalis OG1RF (27); (iii) the demonstration of capsular material containing a repeat structure of glycerolphosphate linked 2, 1 to a kojibiose residue (2); and (iv) the failure of antiserum directed to this capsular material to kill strain FA2–2 in opsonophagocytosis assays (2), along with the failure of the strain used in that study to react with type 2 antiserum in the current study, the following model is proposed for the organization of carbohydrates in the cell walls of E. faecalis strains (Fig. 4).
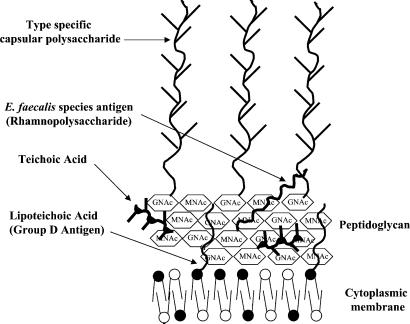
Figure 4.
Schematic representation of a model for the organization of cell wall polymers in the cell wall of E. faecalis.
The 50-kDa rhamnopolysaccharide appears to be an invariant component of the E. faecalis cell wall that bears strong similarity to streptococcal group antigens. In the late 1950s, Elliot (30, 31) recognized that type antigens within the group D streptococci were similar in structure and composition to the group-specific substances of streptococcal groups A, B, C, E, F, and G. Pazur and coworkers (32, 33) demonstrated several unique carbohydrates in the cell wall of E. faecalis strain N, including a diheteroglycan and tetraheteroglycan. The tetraheteroglycan of Pazur and coworkers (32, 33), composed of galactose, rhamnose, N-acetylgalactosamine, and β-d-glucose-1-phosphate, bears similarity to the composition of the common E. faecalis cell wall carbohydrate. Minor differences in composition between the carbohydrate of Pazur and coworkers and the current study may be explained by the fact that we found Streptococcus faecalis strain N, by current classification schemes [API20S, and PCR analysis (34)], to be an E. faecium isolate. The observation that the rhamnopolysaccharide biosynthetic pathway is invariant among E. faecalis strains, and that antiserum to this carbohydrate fails to detect it on the surface of E. faecalis (27, 28), are consistent with a model where this carbohydrate is localized to deeper layers of the cell wall. As a result of inaccessibility, there may be little immune pressure to vary.
Variable serotype-specific carbohydrates appear to represent capsules, as demonstrated by electron microscopy (2) and agglutination in the current study. The composition of the capsular carbohydrate studied by Huebner et al. (2) and the type 2 carbohydrate examined in this study is similar. Huebner et al. (2) reported the structure of their capsular polysaccharide to be a repeating glucose-glycerolphosphate polymer. Determination of the nature of linkages and structure of the type 2 antigen studied here will be necessary to resolve the basis of serological discrimination of these two carbohydrates.
The model also recognizes the existence of a small (15 kDa) molecular mass polysaccharide, which consists of glycerol, phosphate, and glucose, and depicts it as being an integral cell wall teichoic acid. Its small size corresponds to that of other essential cell wall teichoic acids (35, 36). At present, however, there is no other direct evidence for this assignment.
The group D antigen, or lipoteichoic acid, is shown anchored to the cell membrane buried within the cell wall. Studies dating back to Lancefield (37) have described the difficulty in raising antiserum to this antigen. Further, studies by Watson et al. (38) found that digestion of the enterococcal cell wall with lysozyme enhanced detection of the group D antigen with antiserum, suggesting that this antigen may be hidden within the cell wall.
This model for the organization of E. faecalis cell wall carbohydrate polymers is coherent with the results of all studies published to date and should provide a rational basis for the design of immunotherapeutics and anti-infectives for treating life-threatening infections caused by an organism increasingly refractory to conventional antibiotic therapies. The current findings suggest that the specific targeting of antibody to capsular polysaccharides should prove beneficial for treating multidrug-resistant enterococcal infections.
Supplementary Material
Acknowledgments
We acknowledge Shizue Maekawa, Yasuyoshi Ike, and Koichi Tanimoto for providing the typing serum and bacterial typing strains used in this study. Preliminary compositional analysis of partially purified material was kindly provided by Dr. Ying Wang, Channing Laboratory, and confirmed by Dr. Parastoo Azadi, Complex Carbohydrate Research Center. We also gratefully acknowledge Phil Coburn, Brett Shepard, Keeta Gilmore, and Wolfgang Haas for helpful discussions. This research was supported by the U.S. Public Health Service Grants AI41108 and DE13244 and an unrestricted grant from Research to Prevent Blindness.
Abbreviation
- cfu
colony-forming units
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Hancock, L. E. & Gilmore, M. S. 97th Annual Meeting of the American Society for Microbiology, May 4–8, 1997, Miami Beach, FL.
References
- 1.Hancock L E, Gilmore M S. In: Gram-Positive Pathogens. Fischetti V, Novick R, Ferretti J, Portnoy D, Rood J, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 251–258. [Google Scholar]
- 2.Huebner J, Wang Y, Krueger W A, Madoff L C, Martirosian G, Boisot S, Goldmann D A, Kasper D L, Tzianabos A O, Pier G B. Infect Immun. 1999;67:1213–1219. doi: 10.1128/iai.67.3.1213-1219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharpe M E. J Gen Microbiol. 1964;36:151–160. doi: 10.1099/00221287-36-1-151. [DOI] [PubMed] [Google Scholar]
- 4.Maekawa S, Yoshioka M, Kumamoto Y. Microbiol Immunol. 1992;36:671–681. doi: 10.1111/j.1348-0421.1992.tb02070.x. [DOI] [PubMed] [Google Scholar]
- 5.Clewell D B, Tomich P K, Gawron-Burke M C, Franke A E, Yagi Y, An F Y. J Bacteriol. 1982;152:1220–1230. doi: 10.1128/jb.152.3.1220-1230.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callegan M C, Jett B D, Hancock L E, Gilmore M S. Infect Immun. 1999;67:3357–3366. doi: 10.1128/iai.67.7.3357-3366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 8.Pospiech A, Neumann B. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 9.Huycke M M, Spiegel C A, Gilmore M S. Antimicrob Agents Chemother. 1991;35:1626–1634. doi: 10.1128/aac.35.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahm D F, Kissinger J, Gilmore M S, Murray P R, Mulder R, Solliday J, Clarke B. Antimicrob Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gish W, States D J. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 12.Cruz-Rodz A L, Gilmore M S. Mol Gen Genet. 1990;224:152–154. doi: 10.1007/BF00259462. [DOI] [PubMed] [Google Scholar]
- 13.Eichenbaum Z, Federle M J, Marra D, de Vos W M, Kuipers O P, Kleerebezem M, Scott J R. Appl Environ Microbiol. 1998;64:2763–2769. doi: 10.1128/aem.64.8.2763-2769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H G, Cowman M K. Anal Biochem. 1994;219:278–287. doi: 10.1006/abio.1994.1267. [DOI] [PubMed] [Google Scholar]
- 15.DeAngelis P L. Anal Biochem. 2000;284:167–169. doi: 10.1006/abio.2000.4699. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y C. Anal Biochem. 1990;189:151–162. doi: 10.1016/0003-2697(90)90099-u. [DOI] [PubMed] [Google Scholar]
- 17.Rouser G, Fleischer S, Yamamoto A. Lipids. 1970;5:494–496. doi: 10.1007/BF02531316. [DOI] [PubMed] [Google Scholar]
- 18.Jett B D, Hatter K L, Huycke M M, Gilmore M S. BioTechniques. 1997;23:648–650. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- 19.Glantz S A. Primer of Biostatistics. New York: McGraw–Hill; 1987. [Google Scholar]
- 20.Rest R F, Speert D P. Methods Enzymol. 1994;236:91–108. doi: 10.1016/0076-6879(94)36010-3. [DOI] [PubMed] [Google Scholar]
- 21.Llull D, Lopez R, Garcia E, Munoz R. Biochim Biophys Acta. 1998;1443:217–224. doi: 10.1016/s0167-4781(98)00213-9. [DOI] [PubMed] [Google Scholar]
- 22.Stevenson G, Neal B, Liu D, Hobbs M, Packer N H, Batley M, Redmond J W, Lindquist L, Reeves P. J Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koplin R, Brisson J, Whitfield C. J Biol Chem. 1997;272:4121–4128. doi: 10.1074/jbc.272.7.4121. [DOI] [PubMed] [Google Scholar]
- 24.Maekawa S, Habadera S. Kansenshogaku Zasshi. 1996;70:168–174. doi: 10.11150/kansenshogakuzasshi1970.70.168. [DOI] [PubMed] [Google Scholar]
- 25.Huycke M M, Gilmore M S. Adv Exp Med Biol. 1997;418:781–784. doi: 10.1007/978-1-4899-1825-3_184. [DOI] [PubMed] [Google Scholar]
- 26.Richards M J, Edwards J R, Culver D H, Gaynes R P. Infect Control Hosp Epidemiol. 2000;21:510–515. doi: 10.1086/501795. [DOI] [PubMed] [Google Scholar]
- 27.Xu Y, Murray B E, Weinstock G M. Infect Immun. 1998;66:4313–4323. doi: 10.1128/iai.66.9.4313-4323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, Singh K V, Qin X, Murray B E, Weinstock G M. Infect Immun. 2000;68:815–823. doi: 10.1128/iai.68.2.815-823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hancock L E, Gilmore M S. Adv Exp Med Biol. 1997;418:1049–1050. doi: 10.1007/978-1-4899-1825-3_247. [DOI] [PubMed] [Google Scholar]
- 30.Elliot S D. Nature (London) 1959;184:1342. [Google Scholar]
- 31.Elliot S D. J Exp Med. 1960;111:621–630. doi: 10.1084/jem.111.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pazur J H, Andeson J S, Karakawa W W. J Biol Chem. 1971;246:1793–1798. [PubMed] [Google Scholar]
- 33.Pazur J H. J Biol Chem. 1982;257:589–591. [PubMed] [Google Scholar]
- 34.Dutka-Malen S, Evers S, Courvalin P. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joseph R, Shockman G D. Infect Immun. 1975;12:333–338. doi: 10.1128/iai.12.2.333-338.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chorpenning F W, Lynch J J, Jr, Cooper H R, Oldfather J W. Infect Immun. 1979;26:262–269. doi: 10.1128/iai.26.1.262-269.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lancefield R C. J Exp Med. 1933;57:571–595. doi: 10.1084/jem.57.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watson B K, Moellering R C, Kunz L J. Am J Clin Pathol. 1976;66:73–79. doi: 10.1093/ajcp/66.1.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.