Summary
The tumor microenvironment (TME) plays a critical role in both tumor progression and therapeutic efficacy. Here, we present a protocol to perform transcription factor and surface marker staining for a comprehensive analysis of the TME of mouse intestinal tumors by combining multiplex immunofluorescence imaging with the “Swiss roll” technique. We describe steps for sample preparation, sectioning, staining, and mounting. We then detail procedures for imaging and data processing. This protocol supports quantitative assessments such as calculating cell-to-cell distances.
For complete details on the use and execution of this protocol, please refer to You et al.1
Subject areas: Cancer, Immunology, Microscopy
Graphical abstract

Highlights
-
•
Preparation of mouse small intestine and colon using the “Swiss roll” technique
-
•
Combination of transcription factor and surface marker staining
-
•
Multiplex immunofluorescence with up to seven-color staining
-
•
Quantitative analysis of cellular spatial distribution
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
The tumor microenvironment (TME) plays a critical role in both tumor progression and therapeutic efficacy. Here, we present a protocol to perform transcription factor and surface marker staining for a comprehensive analysis of the TME of mouse intestinal tumors by combining multiplex immunofluorescence imaging with the “Swiss roll” technique. We describe steps for sample preparation, sectioning, staining, and mounting. We then detail procedures for imaging and data processing. This protocol supports quantitative assessments such as calculating cell-to-cell distances.
Before you begin
The following protocol outlines the detailed steps for imaging mouse spontaneous intestinal tumors, along with the subsequent quantification of fluorescence images.2 It encompasses the entire workflow, including sample preparation, staining, imaging, data processing and quantitative analysis. This protocol has been successfully applied to studying the tumor micro-environment in the ApcMin/+ mouse tumor model,3 using a combination of cell surface markers and transcription factors. Additionally, this protocol can also be adapted for imaging non-tumorous intestines as well as other organs, such as the lungs, lymph nodes, spleen, and liver.
A comprehensive list of materials and equipment is provided in the key resources table, with step-by-step instructions for solution preparation and storage in the materials and equipment section.
Institutional permissions
All mice were maintained under specific pathogen-free condition at the Laboratory Animal Center, Xiamen University. ApcMin/+ mice spontaneously develop tumors in both small intestine and colon at 4-month-old. All animal experiments described in this protocol were approved by the Institutional Animal Care and Use Committee and were in strict accordance with good veterinary practice as defined by the Xiamen University Laboratory Animal Center.
Prepare buffers
Timing: 60 min
-
1.Prepare fixation buffer.
 CRITICAL: Prepare before use.
CRITICAL: Prepare before use.-
a.BD fixation and permeabilization solution were diluted with 1×PBS at a ratio of 1:2.Note: Total 3 mL fixation buffer per intestine.
-
b.Mix well and place it on ice until use.
 CRITICAL: This fixation buffer is effective for transcription factor staining and does not require antigen retrieval.
CRITICAL: This fixation buffer is effective for transcription factor staining and does not require antigen retrieval. CRITICAL: This fixation buffer must be handled under a chemical hood.
CRITICAL: This fixation buffer must be handled under a chemical hood.
-
a.
-
2.Prepare 30% sucrose, Blocking buffer and Antibody Dilution Buffer as described in materials and equipment.
-
a.Sterilize the buffers with 0.22 μm filter into a 50 mL centrifuge tube. Store at 4°C for up to 12 months.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| eFluor 450 anti-mouse CD326 (EpCAM) monoclonal antibody (clone G8.8) | eBioscience | Cat# 48-5791-82, RRID:AB_10717090 |
| BV480 rat anti-mouse CD4 (clone RM4-5) | BD Biosciences | Cat# 565634, RRID:AB_2739312 |
| Alexa Fluor 488 rat anti-mouse CD4 (clone RM4-5) | BD Biosciences | Cat# 557667, RRID:AB_396779 |
| Alexa Fluor 647 rat anti-mouse CD4 (clone RM4-5) | BD Biosciences | Cat# 557681, RRID:AB_396791 |
| Alexa Fluor 594 anti-mouse CD326 (Ep-CAM) antibody (clone G8.8) | BioLegend | Cat# 118222, RRID:AB_2563322 |
| Alexa Fluor 488 anti-mouse CD3 antibody (clone 17A2) | BioLegend | Cat# 100210, RRID:AB_389301 |
| Alexa Fluor 647 anti-mouse CD31 antibody (clone MEC13.3) | BioLegend | Cat# 102516, RRID:AB_2161029 |
| Alexa Fluor 488 anti-mouse F4/80 monoclonal antibody (clone BM8) | eBioscience | Cat# 53-4801-82, RRID:AB_469915 |
| BV480 rat anti-mouse CD45R/B220 (clone RA3-6B2) | BD Biosciences | Cat# 565631, RRID:AB_2739311 |
| CD45R (B220) monoclonal antibody (RA3-6B2), eFluor 450 | eBioscience | Cat# 48-0452-82, RRID: AB_1548761 |
| Alexa Fluor 647 anti-mouse CD8a antibody (clone 53–6.7) | BioLegend | Cat# 100724, RRID: AB_389326 |
| CD4 monoclonal antibody (RM4-5), Alexa Fluor 700 | eBioscience | Cat# 56-0042-82, RRID:AB_494000 |
| Alexa Fluor 488 anti-mouse LYVE1 monoclonal antibody (clone ALY7) | eBioscience | Cat# 53-0443-82, RRID:AB_1633415 |
| LYVE1 monoclonal antibody (ALY7), eFluor 450 | eBioscience | Cat# 48-0443-80, RRID: AB_2784722 |
| Alexa Fluor 488 anti-GFP antibody | BioLegend | Cat# 338008, RRID: AB_2563287 |
| eFluor 570 anti-mouse Foxp3 monoclonal antibody (clone FJK-16s) | eBioscience | Cat# 41-5773-82, RRID:AB_11219073 |
| Alexa Fluor 647 mouse anti-Ki-67 (clone B56) | BD Biosciences | Cat# 561126, RRID:AB_10611874 |
| Alexa Fluor 700 mouse anti-Ki-67 (clone B56) | BD Biosciences | Cat# 561277, RRID:AB_10611571 |
| BV480 rat anti-mouse CD8a (clone ALY7) | BD Biosciences | Cat# 566096, RRID:AB_2739500 |
| Goat anti-rabbit IgG (H + L) cross-adsorbed secondary antibody, Alexa Fluor 647 | Invitrogen | Cat# A-21244, RRID:AB_2535812 |
| Chemicals, peptides, and recombinant proteins | ||
| Fixation and permeabilization solution | BD Biosciences | Cat# 554722 |
| Normal goat serum | Jackson ImmunoResearch Laboratories, Inc. | Cat# 005-000-121, RRID: AB_2336990 |
| Normal mouse serum | Jackson ImmunoResearch Laboratories, Inc. | Cat# 015-000-120, RRID: AB_2337194 |
| Normal rabbit serum | Jackson ImmunoResearch Laboratories, Inc. | Cat# 011-000-120, RRID: AB_2337123 |
| Normal rat serum | Jackson ImmunoResearch Laboratories, Inc. | Cat# 012-000-120, RRID: AB_2337141 |
| PBS | HyClone | Cat# SH30256.01 |
| Triton X-100 | Sangon Biotech | Cat#A600198-0500 |
| Tissue-Tek O.C.T. compound | Sakura Finetek | Cat# 4583 |
| Sucrose | Sigma-Aldrich | Cat# v900116 |
| Fluoromount-G | SouthernBiotech | Cat# 0100-01 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| Bovine serum albumin | Sigma-Aldrich | Cat# B2064-100G |
| Experimental models: Organisms/strains | ||
| Mouse: ApcMin/+ | The Jackson Laboratory | JAX: 002020 |
| Mouse: C57BL/6 | The Jackson Laboratory | JAX: 000664 |
| Mouse: Foxp3DTR | The Jackson Laboratory | JAX:016958 |
| Software and algorithms | ||
| Prism 8 | GraphPad | https://www.graphpad.com/ |
| Imaris 10.0 | Bitplane | https://imaris.oxinst.com/ |
| R 4.3.2 | N/A | https://www.r-project.org/ |
| Python 2.7 | N/A | https://www.python.org/ |
| Leica LAS X Suite software | Leica | N/A |
| Other | ||
| Charged slides | Premiere | Cat# 9308W |
| Standard NorGlas microscope cover glass | MeVid | Cat# CS01-2450 |
| Reagent reservoirs, 7 mL, 8 channels | Sangon Biotech | Cat#F619713-0001 |
| Hydrophobic Barrier PAP Pen | Vector | Cat# H-4000 |
| Leica CM1950 cryostat | Leica | CM1950 |
| Leica TCS SP8 | Leica | N/A |
| Leica STELLARIS 8 STED | Leica | N/A |
Materials and equipment
Fixation buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Fixation and Permeabilization Solution | N/A | 1 mL |
| PBS | N/A | 2 mL |
| Total | N/A | 3 mL |
Total 3 mL fixation buffer per intestine. Prepare before use.
30% sucrose
| Reagent | Final concentration | Amount |
|---|---|---|
| Sucrose | 30% | 30 g |
| PBS | N/A | add to 100 mL |
| Total | N/A | 100 mL |
Store at 4°C for up to 12 months.
Blocking buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| BSA | 1% | 1 g |
| Triton X-100 | 0.2% | 0.2 mL |
| PBS | N/A | add to 100 mL |
| Total | N/A | 100 mL |
Store at 4°C for up to 12 months.
Antibody Dilution Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| BSA | 1% | 1 g |
| PBS | N/A | add to 100 mL |
| Total | N/A | 100 mL |
Store at 4°C for up to 12 months.
Step-by-step method details
Swiss roll technique for sample preparation
Timing: 3 days (1 h for sample preparation, 1 day for fixation, 1 day for dehydration, 1 day for embedding)
This step outlines the use of the “Swiss roll”4 technique to process intestinal tissues, either with or without tumors, into a compact structure. Following fixation, dehydration, and embedding, the tissue block is prepared for frozen sectioning. Once prepared, the tissue block can be stored at −80°C for 3–5 years.
-
1.Euthanasia:
-
a.Sacrifice mice via CO2 asphyxiation in a chamber until unconsciousness and cessation of all movement are confirmed.
-
b.Perform cervical dislocation to ensure euthanasia.
-
a.
-
2.Tissue Harvesting:
-
a.Harvest the colon and distal small intestine (approximately 12 cm from the cecum).
-
b.Using a 20 mL syringe filled with pre-cooled 1×PBS, gently flush the intestinal lumen to remove feces, taking care not to damage the intestinal villi.
-
a.
Note: A blunt needle is used to prevent tissue damage.
-
3.Fixation:
-
a.Aspirate the fixative solution (see the materials and equipment setup, fixation buffer) into a 5 mL syringe and inject it into the intestinal tissue.
-
b.Place the tissue in an 8-channel polystyrene reservoir at room temperature (16°C–25°C) for fixation.
-
a.
Note: Fix for 15 min for the colon and 1 h for the small intestine.
-
4.Tissue Preparation:
-
a.Place the fixed intestinal tissue on PBS-moistened filter paper.
-
b.Using fine forceps, carefully remove any fat from the surface of the intestine.
-
c.Cut the tissue open longitudinally and gently flatten it.
-
a.
-
5.
Rolling: Trim the delicate ends of the intestine using a scalpel. Roll the tissue into a “Swiss roll” configuration and secure it by passing a 30G needle through the center. See Methods video S1 for steps 1–5.
-
6.Final Fixation:
-
a.Place the rolled intestine into a 7 mL round-bottom centrifuge tube containing 1 mL fixative solution.
-
b.Incubate overnight at 4°C (generally 16–24 h).
-
a.
-
7.Dehydration:
-
a.Remove the fixative solution and rinse tissue with 1 mL PBS.
-
b.Wash the sample on a shaker at approximately 80 rpm for 10 min.
-
c.Incubate the tissue with 1 mL of 30% sucrose overnight at 4°C with gentle shaking (generally 16–24 h).
-
a.
-
8.OCT Equilibration:
-
a.Remove all the sucrose solution from the sample and gently dab the tissue dry with a paper towel.
-
b.Transfer the tissue to a 7 mL round-bottom tube containing 1 mL of OCT.
-
c.Shake at room temperature (16°C–25°C) for 4 h to ensure the OCT fully penetrates the “Swiss roll” (recommended shaking speed, 80 rpm).
-
a.
-
9.Embedding and Freezing:
-
a.Remove the needle from the tissue and embed the intestinal tissue vertically in an embedding mold filled with OCT.
-
b.Freeze the mold on dry ice and store it at −80°C.
-
a.
Note: Ensure there are no bubbles in the OCT, particularly around the tissue, as they can make cryosectioning difficult.
Pause Point: After this step is completed, tissue block can be stored at −80°C for 3–5 years.
Cryosectioning and immunofluorescence staining
Timing: 2–3 days
This step outlines the use of a cryostat to prepare slides, followed by immunofluorescence staining procedures. Standard staining involves the use of unconjugated primary antibodies paired with fluorescent-labeled secondary antibodies, as well as directly fluorescent-labeled antibodies (Figure 1). After staining, the slides can be stored in a light-protected environment at 4°C for up to one month.
-
10.Cryosectioning:
-
a.Prior to sectioning, equilibrate the tissue at −20°C for 1–2 h.
-
b.Set the chamber and sample holder temperatures to −20°C and −18°C, respectively.
-
c.Using a cryostat, cut 15 μm thick sections and collect them on charged slides.
-
a.
Note: The recommended thickness for intestinal tissue sections is 15 μm, but this can be adjusted as needed.
CRITICAL: Ambient humidity and temperature are crucial for preserving the sections. Excessive humidity can reduce the storage duration and negatively affect the subsequent staining quality.
Pause Point: After this step is completed, these sections can be stored at −20°C for 3–6 months.
-
11.Slide Preparation:
-
a.Allow the slides to equilibrate at room temperature for 30 min.
-
b.Use a hydrophobic pen to draw a circle around the tissue to define the staining area.
-
c.Let the wax dry completely for 30 min.
-
a.
-
12.
Washing: Place the slides in a humidified chamber and wash the tissue with PBS twice, for 5 min each wash.
Note: Prepare a slide without staining, wash the OCT with PBS and seal it directly as a negative control. In theory, this slide should not fluoresce upon excitation, except for autofluorescence and any fluorescent proteins present.
-
13.Blocking:
-
a.To minimize non-specific binding of primary and secondary antibodies, prepare 50 μL of blocking buffer per sample containing 1% normal serum from both mouse and the species in which the secondary antibodies were raised.
-
b.Block the sections at room temperature (16°C–25°C) for 0.5–1 h.
-
a.
-
14.
Washing: Wash the slides with PBS 3 times at room temperature (16°C–25°C), for 5 min per wash.
Note: The volume used is based on the size of the hydrophobic zone, approximately 1–2 mL. Allow it to stand for 5 min without shaking.
-
15.
Primary antibody staining: Incubate each section with 50 μL of antibody dilution buffer containing unconjugated primary antibodies. Add 1% normal serum from both mouse and the species in which the secondary antibodies were raised to the prepared buffer.
-
16.
Incubation: Incubate the slides in a dark, humidified chamber at room temperature (16°C–25°C) for 16–18 h.
CRITICAL: If the staining includes transcription factors, the incubation time can be extended to 24 h. Ensure the chamber is closed to retain moisture and prevent evaporation of the buffer.
-
17.
Washing: Remove the staining solution and wash the slides with PBS 5 times at room temperature, for 5 min per wash.
-
18.
Secondary antibody staining: Prepare 50 μL of antibody dilution buffer containing fluorescent-labeled secondary antibodies. Add 1% serum from both mouse and the species in which the secondary antibodies were raised to the buffer. Incubate the slides in dark at room temperature (16°C–25°C) for 3 h.
-
19.
Washing: Wash the slides with PBS 5 times at room temperature (16°C–25°C), for 5 min per wash.
Note: If all primary antibodies are fluorescent-conjugated, steps 13–19 can be skipped.
-
20.
Blocking: Prepare 50 μL of blocking buffer per sample, adding 1% serum from both mouse and the species in which the primary antibodies were raised. Incubate the slides at room temperature (16°C–25°C) for 1 h to block non-specific binding.
-
21.
Washing: Wash the slides with PBS 3 times at room temperature (16°C–25°C), for 5 min per wash.
-
22.
Fluorescent-labeled antibody staining: Incubate each section with 50 μL of antibody dilution buffer containing fluorescent-labeled primary antibodies in dark at room temperature (16°C–25°C) for 3 h. Add 1% normal serum from both mouse and the species in which the secondary antibodies were raised to the prepared buffer.
-
23.
Washing: Wash the slides with PBS 5 times at room temperature, for 5 min per wash.
-
24.
Optional step: DAPI Staining: Prepare the DAPI solution according to the manufacturer’s instructions and stain the tissues at room temperature (16°C–25°C) for 5–10 min. If DAPI staining is not required, this step can be skipped.
Note: Due to the broad emission spectrum of DAPI, consider reducing the concentration of the DAPI solution or shortening the staining time to minimize fluorescence spillover.
-
25.Mounting:
-
a.Carefully remove any liquid from the slide surface.
-
b.Apply 75 μL of mounting medium to each slide and gently place a cover slip on top, ensuring no bubbles are trapped.
-
a.
Note: It is recommended to use an anti-fade mounting medium that does not contain DAPI.
-
26.
Drying and Storage: Allow the mounting medium to dry at room temperature (16°C–25°C), protected from light.
Pause Point: After this step is completed, these slides can be stored at 4°C for up to 1 month.
Figure 1.
The staining process flowchart
The gray section represents both direct and indirect staining. The orange section highlights the subsequent steps for direct staining, while the blue section outlines the steps following indirect staining.
Imaging and data processing
Timing: 4–6 h
This step describes how to use a confocal microscope to image stained slides and process the images using software like Imaris.
-
27.
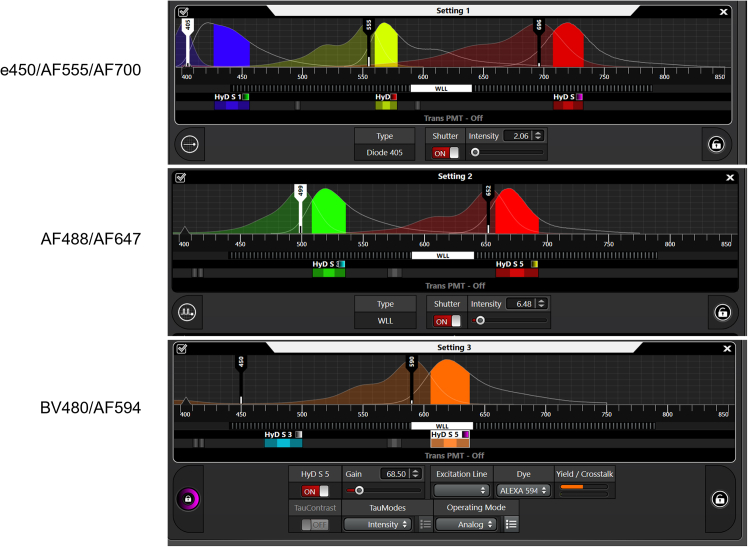
Imaging: Acquire images using the Leica STELLARIS 8 confocal microscope following the manufacturer’s instructions. Use the recommended channel configuration (Figure 2) and corresponding parameters (Table 1) for the seven-color panel.
Note: Select an appropriate objective lens based on the experimental goals and staining panel. To achieve high-quality images while minimizing imaging time, begin with a low-magnification objective (10x, 20x……) to scan the entire section. Then, use a high-magnification objective (40x, 63x, 100x……) to examine regions of interest (ROI) in greater detail.
CRITICAL: Before starting the experiment, it is essential to verify the excitation and emission spectra of the fluorescent dyes used in your panel by a fluorescence spectral viewer. Ensure minimal overlap between the emission spectra of the fluorophores. To reduce fluorescent spillover, apply sequential scanning for dyes with closely overlapping spectra. Optimize laser power and detector gain to maximizing signal-to-noise ratio and minimize photobleaching.
-
28.Image Processing and Quantification: Use the surface reconstruction feature in Imaris 10.0 to generate statistical data tailored to your experimental objectives. This data can be further analyzed or visualized using software such as R or Prism.
-
a.Use Imaris File Converter to transform the “.lif” file obtained from the confocal microscope into a “.ims” file compatible with Imaris. Then perform surface reconstruction for the channels requiring quantitative analysis (Figure 3, step 1, red arrow).
-
b.Once the surface reconstruction is completed, revies the “object-object statistics” for all required surfaces (Figure 3, step 2, red box). Next, extract the statistical data in the software (Figure 3, step 3, red arrow). You can choose to view the data as “overall”, “Spatial”, “Detailed” and “Selection”, according to your specific needs (Figure 3, step 4, red box).
-
c.For example, to analyze a single surface, select the “Detailed” module and choose “specific value” from the drop-down list, and review the statistical information related to distance in the statistics section (Figure 3, step 5, red box). Then you can save the extracted statistical data such as distance and fluorescence intensity of the channel, as a “.csv” or “.xlsx” file (Figure 3, step 6, red box). This data can be further analyzed and plotted by software such as R, Excel, Prism, etc.
-
a.
Note: The surface distance calculation feature is available only in Imaris 9.5 and later versions. Earlier versions of Imaris do not support this functionality. If you are using an earlier version, you can export the position information of each surface (i.e. xyz coordinates) and manually calculate distances using Python or R.
Figure 2.
Recommended settings for the seven-color program for a confocal microscope with continuously adjustable pulsed white lasers (Leica STELLARIS 8 STED as an example)
This configuration minimizes cross-talk effects.
Table 1.
Recommended imaging parameters for the seven-color program
| Scan unit manufacturer and model | Leica STELLARIS 8; Leica SP8 |
Details |
|---|---|---|
| Acquisition Model | stage | Using the stage model |
| Sequential Scan | Between stacks, Sequential setting provided in Figure 2 | |
| xy | Format | 512∗512 for 20X; 1024∗1024 for 40X |
| Speed | 400 Hz | |
| Scan directionality | bidirectional | |
| Zoom factor | 1 | |
| Line average | 2 for 20X; 3 for 40X | |
| Pinhole | 1 A.U. | |
| z-stack | z-stack size | 1.5 μm |
| Stage | Filed size | According to experimental requirements |
| Tile Scan Stitching Overlap | x: 10%, y: 10% | |
| Accessories | laser | 405 nm; WLL2 (440 nm–790 nm) |
| Objective | 20X (NA 0.75, Air); 40X (NA 1.3, oil) | |
| Detector | Power HyD S |
Figure 3.
Reconstruct signal in Imaris and get statistics information such as distance
Expected outcomes
In this protocol, we use a commercial fixation and permeabilization solution (BD Biosciences, Cat# 554722) instead of conventional formaldehyde (PFA) solution. This approach enables the staining of transcription factors, such as Foxp3, RORγt,5 GATA36 and Ki67, in conjunction with cell surface markers. Using Foxp3DTR-EGFP mice, in which a human diphtheria toxin receptor (DTR) and an enhanced green fluorescent protein (EGFP) were knocked into the Foxp3 locus, we observed precise co-localization of Foxp3 expression with GFP+ Treg cells (Figure 4). These results demonstrate the high accuracy and efficiency of transcription factor staining achieved with our protocol.
Figure 4.
Verification of Foxp3 staining with Foxp3DTR-EGFP mice
Immunofluorescence staining of Tregs in lymph nodes from Foxp3DTR-EGFP mice. Scale bar, 20 μm.
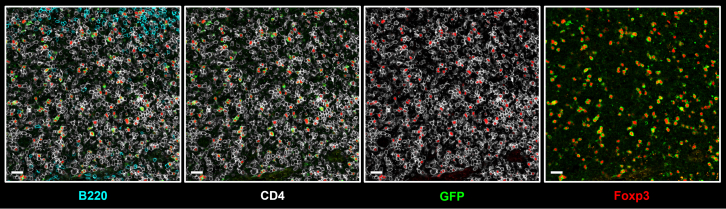
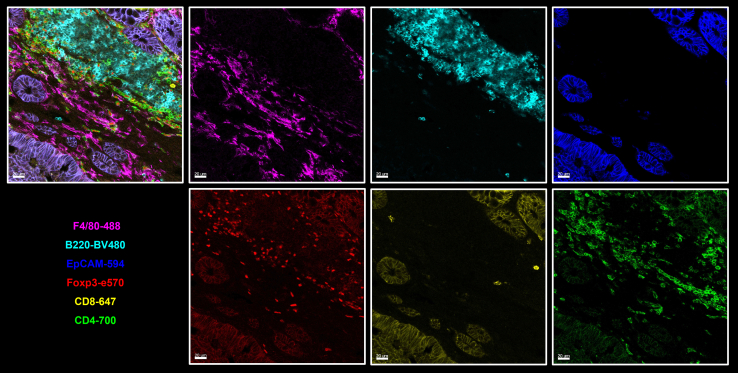
Additionally, this protocol supports up to a seven-color staining panel across multiple tissue types. For example, it enables simultaneous visualization of CD4+ and CD8+ T cells along with tissue vasculature in the small intestine. EPCAM marks epithelial cells and can substitute nuclear staining to outline intestinal tissue morphology. Lyve-1 and CD31 highlight lymphatic vessels and blood vessels, respectively. Ki67+ cells represent intestinal epithelial stem cells and transient-amplifying cells in intestinal crypts. CD8, CD4 and Foxp3 staining further reveal the spatial distribution of cytotoxic, helper, and regulatory T cells within the lamina propria of the small intestine (Figure 5).
Figure 5.
Immunofluorescence staining of T cell distribution in the lamina propria of the small intestine
The image illustrates the epithelium, immune cell populations and vascular structures of the small intestine. Imaging acquired by Leica STELLARIS 8 STED. Scale bar, 50 μm.
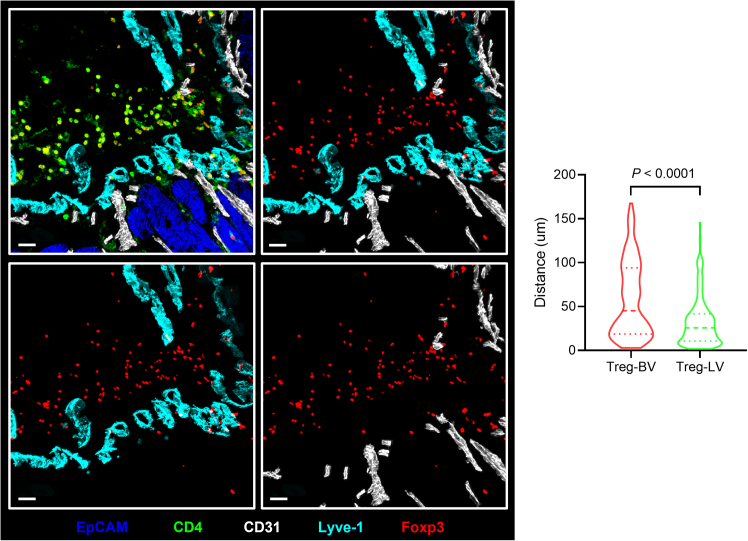
Next, we analyzed the distribution of regulatory T cells (Tregs) in colonic tumors of ApcMin/+ mice. Consistent with our previous findings,1 Tregs were observed to accumulate along the vasculature within the tumor stroma. To perform a quantitative analysis, we measured the distance of Tregs to lymphatic and blood vessels combined the Histo-Cytometry method.7 First, we used the surface function in Imaris software to reconstruct the channels for analysis (Foxp3, Lyve1 and CD31). Cells were then segmented based on marker expression: Foxp3+CD4+ cells were defined as Tregs, Lyve-1+ and Lyve-1−CD31+ structures were identified as lymphatic vessels and blood vessels, respectively. For each Treg cell, the shortest distance to the surfaces of blood and lymphatic vessels was calculated in Imaris. The statistical output was exported as a “.csv” file, which was then processed in Excel, Prism, or R for further analysis. Our results showed that in the stromal regions of colonic tumors in ApcMin/+ mice, despite the presence of a dense vascular network comprising both blood and lymphatic vessels, Tregs were preferentially co-localized with lymphatic vessels (Figure 6).
Figure 6.
Immunofluorescence staining and quantification of the association between Tregs and lymphatic or blood vessels in colon tumors from ApcMin/+ mice
Left: immunofluorescence staining; right: quantification. Scale bar, 30 μm.
Quantification and statistical analysis
Immunofluorescence images are analyzed using Imaris software. Statistical significance was calculated by paired two-tailed Student’s t test.
Limitations
A significant challenge in multiplex immunofluorescence imaging, especially with panels involving up to seven colors, is managing fluorophore spillover. The Leica STELLARIS 8 confocal microscopy, equipped with a white light laser, helps address this issue by enabling precise selection of excitation wavelengths and facilitating appropriate sequential scanning. As a result, the quality of imaging largely depends on the instrument and its optimal settings.
In comparison to fluorescent in situ hybridization (FISH), multiplex immunofluorescence imaging relies heavily on the antibodies used. Antibody specificity is crucial, as it directly influences the signal-to-noise ratio during image acquisition and the overall quality of image processing. While many antibodies perform well in flow cytometry, factors such as fixation and antigen preservation can affect their compatibility with immunofluorescence imaging. Therefore, thorough validation of antibodies is essential before use in this technique.
Troubleshooting
Problem 1
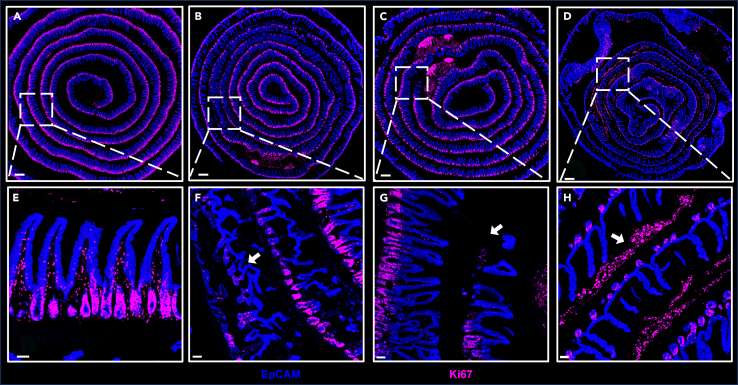
Destruction of tissue structure (Figures 7B and 7C).
Figure 7.
Representative images of properly and improperly processed tissues
(A) Properly processed tissues.
(B–D) Improperly processed tissues.
(E)–(H) show images of the enlarged region highlighted by the dashed white square in (A)–(D). Scale bars, 500 μm (A–D) and 50 μm (E–H).
Potential solution
-
•
Avoid using forceps to directly touch the villi of the small intestine.
-
•
When flushing the intestinal contents with a syringe, use a blunt needle. Handle it gently to prevent scratching the intestinal wall.
-
•
When cutting the intestine longitudinally with scissors, take care not to touch or damage the lower intestinal wall.
Problem 2
Strong nonspecific signal outside the intestinal muscle layer (Figure 7D).
Potential solution
This issue could be attributed to residual fat tissue surrounding the sample, as fluorescent antibodies tend to bind to fat. To mitigate this, ensure the tissue is thoroughly cleaned prior to rolling.
Problem 3
Low signal-to-noise ratio or high background.
Potential solution
First, ensure the antibody used has high specificity and is not contaminated. Optimize the confocal settings to achieve ideal parameters. Adjust the antibody concentration, staining duration, and staining conditions as needed. For detailed scenarios and corresponding solutions, refer to Table 2.
Table 2.
Evaluating of staining effectiveness and adjustment recommendations based on signal-to-noise ratio
| Target protein | Background | Potential solutions |
|---|---|---|
| Strong signal | Weak signal | Good performance |
| Strong signal | Strong signal |
|
| Weak signal | Strong signal |
|
| Weak signal | Weak signal | This antibody may not be suitable for immunofluorescence |
Problem 4
Fluorescent spillover between channels (Figure 8).
Figure 8.
An example of fluorescent spillover
Imaging acquired by Leica SP8. Scale bar, 20 μm.
Potential solution
To minimize fluorescent spillover, choose an appropriate combination of antibodies with fluorescent dyes. For antigens with high expression levels, use dimmer dyes, while for low-expression antigens, opt for brighter dyes. When two antigens, such as CD4 and Foxp3, are expressed in the same cell population, choose fluorophores with minimal spectral overlap. During imaging acquisition, configure these channels to be captured sequentially. If fluorescent spillover persists, utilize the “dye separation” function in the Leica software with single-color staining for fluorescence compensation.
Generally, the fluorescence intensity of short-wavelength fluorescent dyes (e.g., e450, BV421) is weaker, while dyes with longer wavelengths (e.g., AF594, AF647) exhibit stronger fluorescence intensity. However, when the wavelength extends into the near-infrared region (e.g., AF700, AF750), the fluorescence intensity detectable by the detector decreases due to its limited sensitivity to near-infrared light.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kairui Mao (maok@xmu.edu.cn).
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by the technical contact, Kairui Mao (maok@xmu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The code generated during this study is available at https://github.com/SSforestLL/StarProtocols.
Acknowledgments
We thank Qingfeng Liu in the Core Facility of Biomedical Sciences of Xiamen University for confocal technical support and the Laboratory Animal Center of Xiamen University for experimental animal management. This work was supported by grants from the National Key R&D Program of China (2020YFA0803500 to K.M.), the National Natural Science Foundation of China (32270953 and 81971557 to K.M.), the National Natural Science Foundation of China Major Research Program Integration Project (92042305 to K.M.), the Natural Science Foundation of Fujian Province of China (2021J06004 to K.M.), and the Fundamental Research Funds for the Central Universities of China-Xiamen University (20720210001 to K.M., 20720220003 to K.M., and 20720210111 to K.M.). The graphical abstract was created using BioRender.com.
Author contributions
K.M. conceived this study and designed the experiments. S.L. performed most experiments and analyzed the data. S.Y. provided mice used in this experiment. K.M., S.L., and S.Y. wrote the manuscript. K.M. supervised the project.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2025.103776.
References
- 1.You S., Li S., Zeng L., Song J., Li Z., Li W., Ni H., Xiao X., Deng W., Li H., et al. Lymphatic-localized Treg-mregDC crosstalk limits antigen trafficking and restrains anti-tumor immunity. Cancer Cell. 2024;42:1415–1433.e12. doi: 10.1016/j.ccell.2024.06.014. [DOI] [PubMed] [Google Scholar]
- 2.Piyadasa H., Angelo M., Bendall S.C. Spatial proteomics of tumor microenvironments reveal why location matters. Nat. Immunol. 2023;24:565–566. doi: 10.1038/s41590-023-01471-8. [DOI] [PubMed] [Google Scholar]
- 3.Moser A.R., Pitot H.C., Dove W.F. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 4.Moolenbeek C., Ruitenberg E.J. The "Swiss roll": a simple technique for histological studies of the rodent intestine. Lab. Anim. 1981;15:57–59. doi: 10.1258/002367781780958577. [DOI] [PubMed] [Google Scholar]
- 5.Mao K., Baptista A.P., Tamoutounour S., Zhuang L., Bouladoux N., Martins A.J., Huang Y., Gerner M.Y., Belkaid Y., Germain R.N. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature. 2018;554:255–259. doi: 10.1038/nature25437. [DOI] [PubMed] [Google Scholar]
- 6.Huang Y., Mao K., Chen X., Sun M.A., Kawabe T., Li W., Usher N., Zhu J., Urban J.F., Jr., Paul W.E., Germain R.N. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science. 2018;359:114–119. doi: 10.1126/science.aam5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerner M.Y., Kastenmuller W., Ifrim I., Kabat J., Germain R.N. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37:364–376. doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code generated during this study is available at https://github.com/SSforestLL/StarProtocols.