Abstract
Biological water at the interface of proteins is critical to their equilibrium structures and enzyme function and to phenomena such as molecular recognition and protein–protein interactions. To actually probe the dynamics of water structure at the surface, we must examine the protein itself, without disrupting the native structure, and the ultrafast elementary processes of hydration. Here we report direct study, with femtosecond resolution, of the dynamics of hydration at the surface of the enzyme protein Subtilisin Carlsberg, whose single Trp residue (Trp-113) was used as an intrinsic biological fluorescent probe. For the protein, we observed two well separated dynamical solvation times, 0.8 ps and 38 ps, whereas in bulk water, we obtained 180 fs and 1.1 ps. We also studied a covalently bonded probe at a separation of ≈7 Å and observed the near disappearance of the 38-ps component, with solvation being practically complete in (time constant) 1.5 ps. The degree of rigidity of the probe (anisotropy decay) and of the water environment (protein vs. micelle) was also studied. These results show that hydration at the surface is a dynamical process with two general types of trajectories, those that result from weak interactions with the selected surface site, giving rise to bulk-type solvation (≈1 ps), and those that have a stronger interaction, enough to define a rigid water structure, with a solvation time of 38 ps, much slower than that of the bulk. At a distance of ≈7 Å from the surface, essentially all trajectories are bulk-type. The theoretical framework for these observations is discussed.
Water is essential for the stability and function of biological macromolecules, proteins and DNA. Hydration plays a major role in the assembly of a protein's structure and dynamics. For example, water molecules around hydrophobic and hydrophilic sites are important to the understanding of the activity of enzyme proteins (see, e.g., refs. 1–3) and are part of the recognition process by other molecules or proteins. The water molecules that make up the hydration shell in the immediate vicinity of the surface are particularly relevant to the function and, in that sense, are termed biological water; this distinction has been discussed clearly by Nandi and Bagchi (4) in relation to dielectric relaxations. The nature of this shell “layer” has been the focus of numerous studies both theoretically and experimentally (see refs. 5–12), yet there is no generalized picture of the dynamics at the local molecular level.
X-ray crystallography, neutron diffraction, and molecular dynamics studies have shown (5–10) that at protein surfaces, water molecules are site-selective and can be restricted in their motion, even existing in the form of clusters in some cases. For example, neutron diffraction experiments (9) followed by molecular dynamics simulations on carboxymyoglobin (10) revealed that among the 89 water molecules associated with the protein, 4 remain bound during the entire length of the molecular dynamics simulation (50 ps), whereas the rest undergo continuous exchange between bound and free states on a variety of time scales. On the basis of dielectric measurements (11) and NMR studies (12), it has been shown that the relaxation times for water associated with biomolecules occur on different time scales: from a few tens of picoseconds to the nanosecond time regime (see below).
Recently, these time-dependent relaxations were described in a model of a dynamical equilibrium between the free and bound water molecules (4). The so-called moment–moment correlation function shows a bimodal behavior in the time range from picoseconds to nanoseconds. Solvation dynamics in protein environments using dye molecules as extrinsic probes (13–14) have been studied and show a nonexponential behavior. Molecular dynamics simulations have provided a range of time scales, with distinct difference for the residence time for different sites at the surface (7, 8, 10). To understand the dynamics at the local molecular scale, several questions need to be addressed: What are the time scales for water relaxation in the immediate vicinity of a protein surface? Are there local “rigid” structures of water at the surface, and how uniform is the protein surface? How does solvation at the surface differ from that of bulk water, and what are the entropic contributions?
In this study, we use the single Trp residue (Trp-113) in the enzyme (serine endopeptidase) Subtilisin Carlsberg (SC) as an intrinsic fluorescent probe. The single indole side chain of SC at Trp-113 is on the surface of the protein and is significantly exposed to the water environment (see Fig. 1 Top for the crystal structure of SC). The main advantage of using Trp to study the local molecular dynamics of the protein surface is that its intrinsic nature rules out any ambiguity about the location of the probe and about the heterogeneity at different sites. Although the ground state of Trp has a very small dipole moment, a hydrophobic residue, the electronically excited 1La state, has a large static dipole, and it is the emitting state in water. The energy gap to the electronic ground state is very sensitive to the local environment, and the spectral shifts follow the polarity of the medium, including proteins (15); in polar media, the spectrum shifts to the red. In the SC protein, the Trp residue resides in a site that is significantly exposed to water. Thus, we are able, with UV excitation, to follow its solvation in the dynamic Stokes shift, caused by water relaxation on the femtosecond to picosecond time scale. We then compare with the results obtained for free Trp in bulk water (16, 17).
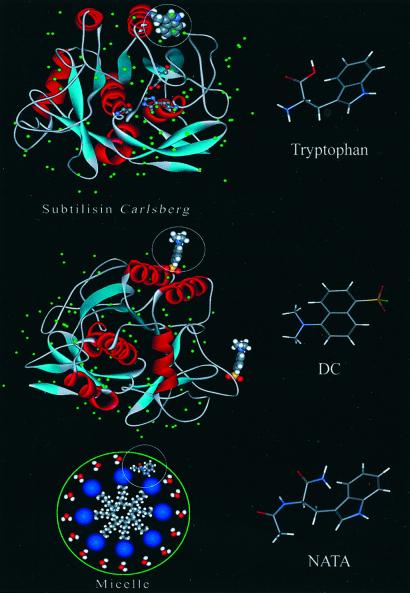
Figure 1.
High-resolution x-ray structure of the protein SC. This structure was downloaded from the Protein Data Bank and processed with WEBLAB-VIEWERLITE, Accelrys, San Diego, CA. (Top) We depict the position of the single Trp residue of the protein. Note the bound water molecules around this residue. (Middle) Two of the nine potential binding sites for DC labeling are shown. (Bottom) Illustration of a micelle with a NATA molecule included. Molecular structures of the probes are presented at the right of each illustration.
To examine the distance dependence, we made another set of femtosecond studies of hydration for a covalently attached dansyl chromophore. This labeling is well known (18) to occur at the ɛ-amino groups of the lysine and arginine residues, which are water-exposed. There are nine such sites in SC. Two of these binding sites are depicted in Fig. 1 Middle. On the basis of bond lengths, we estimate that the location (≈7 Å) of the dansyl probe when bound to SC is such that it sees relatively smaller interaction with the protein backbone and side chains in comparison with bulk water. The dansyl chromophore undergoes a twisted intramolecular charge transfer in the excited state to generate the fluorescing state (19), and the process of solvation leads to continuous red shift of the emission spectrum. These dynamics are also determined by solvent relaxation (20).
For all systems, we examined the time-dependent anisotropy of the emission, and the results elucidate the degree of rigidity of the probe. To “mimic” the surface effect of the protein, we also studied the solvation and local rigidity dynamics in the palisade layer of a neutral micelle formed with reduced triton X-100 (TX-100). In this case, we used the indole chromophore of N-acetyl-tryptophan amide (NATA), which is again a Trp analogue.
Experimental
Up-Conversion.
Our experimental setup uses a Ti-sapphire, 1-kHz regeneratively amplified femtosecond laser. Excitation pulses were formed in a two-stage 800-nm pumped optical parametric amplifier. The resulting near-IR pulses were either sum-frequency mixed with the fundamental and then frequency doubled (for 288- and 296-nm excitation) or frequency doubled twice in βBBO crystals (for 325-nm excitation). The energy of the excitation pulses was kept at approximately 200 nJ. The samples were placed in a rotating circular cell with a 1-mm path length. The emission from the sample was collected by a pair of parabolic focus mirrors and crossed with the gate beam (800 nm) in a 0.3-mm βBBO crystal (θcut = 44°). The polarization axis of the pump beam was adjusted to be at the magic angle (54.7°) with respect to the acceptance axis of the up-conversion nonlinear crystal and the polarization axis of the gate beam. The resulting UV signal was dispersed in a double-grating monochromator and detected by a photomultiplier tube. More details about the setup can be found elsewhere (20, 21).
Sample Preparation.
The protein SC, l-tryptophan, NATA, and TX-100 were purchased from Sigma and used without further purification. The SC protein was in the lyophilized powder form (highest grade). Dansyl chloride (DC) was acquired from Molecular Probes. Aqueous biological solutions were prepared in a phosphate buffer (0.1 M, pH 7) in water from a Nanopure purification system. The concentrations of Trp, NATA, and protein SC in the buffer solution are 5 mM, 5 mM, and 400 μM, respectively.
The covalent attachment of the dansyl probe to SC (adduct formation) was achieved following the procedure from Molecular Probes. Briefly, DC was first dissolved in a small amount of dimethyl formamide and then added drop by drop into a sodium bicarbonate solution (0.1 M) of SC (pH 8.3) with continuous stirring. At this pH, the solvent-exposed lysine and arginine residues of SC react with DC to form dansyl-labeled sites. From the known x-ray structure (22), there are a total of nine potential binding sites (two of them are shown in Fig. 1 Middle). The steady-state fluorescence spectrum of DC has a maximum at 510 nm in methanol and shifts to 500 nm for the dansyl–SC adduct in water. This shift is similar to that of Trp, from 356 nm in bulk water to 351 nm in the SC protein. Proper stoichiometry was maintained to obtain 1:1 dansyl-labeled SC adducts (D-SC). The reaction was terminated by adding a small amount of freshly prepared hydroxylamine (1.5 M, pH 8.5) after incubating it for 1 h at 4°C.
The solution was then dialyzed exhaustively against phosphate buffer (0.1 M) to separate the D-SC adducts from any unreacted DC and its hydrolysis product. It should be noted that D-SC complexes are quantitatively formed because of covalent synthesis (18). The labeling was checked by static fluorescence and time-resolved fluorescence anisotropy measurements by using a FluoTime200 time-correlated single photon-counting instrument with 400-nm picosecond excitation (PicoQuant, Berlin). The temporal behavior of the fluorescence anisotropy of the D-SC system in water is consistent with the probe being bound to the protein (see Results).
NATA–micelle complexes were prepared by mixing NATA (1 mM) with the micellar solution of Triton X-100 (TX-100; 100 mM). Assuming the aggregation number of the TX-100 micelle to be 100, and based on the concentration ratio of NATA/micelle, the 1:1 complexes must be the dominant species with little multioccupancy at these concentrations.
Steady-State Optical Studies.
Trp in bulk water shows a fluorescence maximum at 356 nm, with 288 or 296 nm excitation. In the SC protein, it shifts to 351 nm (for 296-nm excitation). The fluorescence maximum and the shape of the spectrum on ≈296-nm excitation are consistent with previous studies of this protein (23, 24). NATA in neat water solution exhibits a fluorescence spectrum similar to Trp, but on inclusion in the TX-100 micelles, its peak shifts to the blue by 15 nm. DC is sparingly soluble in water but dissolves in methanol. The emission maximum in methanol is at 510 nm. When the dansyl chromophore is covalently attached to SC, the fluorescence emission increases by about a factor of two, and the spectrum is 10 nm blue-shifted with respect to the spectrum of DC in methanol at the same optical density. Static absorption spectra were obtained in a Cary 500 spectrometer (Varian). Steady-state fluorescence spectra were taken in a Fluormax2 spectrofluorometer (Instruments SA, Edison, NJ).
Results
Tryptophan in Bulk Water.
Fluorescence up-conversion scans of aqueous Trp solutions are presented in Fig. 2 Upper. The excitation wavelength for this experiment was 288 nm. As indicated in a previous communication from this and another group (16, 17), the ultrafast decays, observed on the blue side of the spectrum, and the corresponding rises, in the red edge, reflect a dynamic spectral shift that is a signature of relaxation of the solvent around the large dipole of the fluorescing state of Trp: 1La. The transients were fitted to the sum of exponentials convoluted with a Gaussian instrument response function with a full width half maximum of 350 fs. The time constant of one of these exponentials was fixed at 3 ns to represent the slow decay component from the fluorescent state at equilibrium. Using a total of 10 of these transients at a series of wavelengths, we obtained the time-resolved emission spectra; the transients were normalized to the fluorescence spectrum of Trp at 20 ps. The fluorescence spectrum used to normalize these data was constructed from the known lifetimes and fluorescence spectra of the two rotamers of Trp, as described in a study by Shen and Knutson (16).
Figure 2.
Femtosecond-resolved fluorescence (Upper) and the solvent response function (Lower) obtained from up-conversion measurements of Trp in aqueous solutions. (Inset) Normalized spectral evolution at five delay times from the t = 0 point (see text). The excitation wavelength was 288 nm.
The values for fluorescence maximum, νmax(t) in cm−1, were obtained from fits of the time-resolved spectra to logarithm-normal functions at each delay point. With our time resolution, we observed that the fluorescence spectrum has a maximum at ≈320 nm at t = 0; this value is 12 nm red-shifted from the fluorescence maximum observed in nonpolar environments (15). The corresponding solvent response function, which is a correlation function describing the influence of solvent fluctuations on the emitting dipoles, C(t), is shown in Fig. 2 Lower and is defined as C(t) = [νmax(t)-νmax(∞)]/[νmax (0)-νmax(∞)] (for a review, see ref. 25). We have used the spectrum at 20 ps to define the νmax(∞) value, because the spectral evolution related to solvation is completed by this time (16, 17), whereas the nanosecond spectral change occurs because of the different fluorescence lifetimes of the two rotamers of Trp (26). In this and the rest of our measurements, we did not include wavelengths where a significant contribution from Raman scattering by water was evident (about ± 5 nm, ≈3,400 cm−1 OH band from the respective excitation wavelength).
In bulk water, the Trp solvation C(t) curve can be fitted to the sum of two exponential decays: τ1 = 180 fs (20%) and τ2 = 1.1 ps (80%). This dynamic Stokes shift behavior is typical of solvation of a molecular probe in bulk water, and the time scale is consistent with our previous study of Trp at pH 2 with excitation at 266 nm (17). The ultrafast behavior of the C(t) in this case corresponds to relaxation of the water molecules involving both inertial (sub-100 fs) and diffusive (overdamped) rotational motions (up to ≈1 ps) (27). Rotational motions of the solvent molecules dominate the relaxation trajectories, and the contribution from the translational motion is small. The inertial part of the response is because of pair-wise solute–solvent interactions, whereas for the diffusive part, more collective motions of the solvent molecules are present (25).
Tryptophan of the Protein.
Fig. 3 Top shows the hydration correlation function obtained from up-conversion experiments on aqueous solutions of the SC protein. We used an excitation wavelength of 296 nm to minimize the excitation of the tyrosine residues. In this case, C(t) was obtained by using the same procedure outlined above; the long time-decay component was fixed at 2.8 ns, which is the weighted average of the two longer (2.42 and 4.64 ns) decay components from a previous nanosecond time-resolved fluorescence study of SC (23). The freely varying preexponential factors and decay times are not very sensitive to the specific value of this long time component, because the range of our studies is concerned only with the first ≈200 ps.
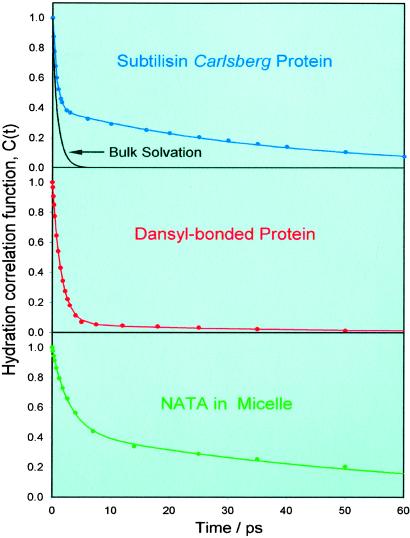
Figure 3.
Hydration correlation function obtained from femtosecond fluorescence up-conversion measurements of aqueous solutions of the SC protein (Top), dansyl-labeled SC (Middle), and the NATA–TX-100 micelle system (Bottom). For comparison with Trp in bulk water, we show the C(t) obtained in Fig. 2 as a decay curve in the top section.
To construct the time-resolved spectra of the SC, we performed a direct normalization of the up-conversion data by multiplying the multiexponential decays by the factor: Fss(ν)/Σai⋅τi, where Fss(ν) is the measured steady-state fluorescence spectrum, and ai and τi are the preexponential factors and decay times from the fits. The value of νmax(∞) was taken from the spectrum at 200 ps. In all other respects, the calculation of the C(t) function follows the procedure of Trp indicated above.
The hydration dynamics of the SC protein show important differences when compared with the results of Trp in bulk water. In particular, the C(t) decay shows a considerably slower decay component and is described by the sum of two exponentials, with τ1 = 800 fs (61%) and τ2 = 38 ps (39%), much different from the observation made in bulk water. The overall spectral shift we observed is 1,440 cm−1; νmax (0) = 30,710 cm−1, νmax(200 ps) = 29,270 cm−1. Clearly, the local protein environment of the indole chromophore at the surface of SC changes the dynamics of hydration.
To measure the local rigidity of the indole rings in SC and also to verify that the spectral-shift dynamics we observed are not complicated by internal conversion between the 1La and 1Lb states, we studied the fluorescence polarization anisotropy. Up-conversion scans were acquired at 370 nm with parallel and perpendicular relative polarizations of the pump and gate beams to obtain the anisotropy decay: r(t) = [I∥(t) − I⊥(t)]/[I∥(t) + 2⋅I⊥(t)]. The anisotropy (not displayed) shows a slow decay that, together with the t = 0 anisotropy value of ≈0.2, indicates that any 1Lb→1La internal conversion must occur within our time resolution (17). The t = 0 value of the anisotropy of emission from the indole chromophore depends on the specific wavelength of excitation (16).
The anisotropy decay of free aqueous Trp (excitation, 288 nm; fluorescence, 340 nm; data not shown) was studied carefully to compare with the results from the protein and the micelle. We observed a rapid change of the anisotropy from a value of about 0.13 to a value of 0.07, well within our instrument response time (this is observed in the raw anisotropy data). After this decay, the anisotropy continues to decrease from 0.07 to a value of zero with a time constant of 35 ps. This decay describes the rotational relaxation of the free molecule in bulk solution. These observations are consistent with other femtosecond time-resolved anisotropy studies of free aqueous Trp (16, 17).
For the protein, the value of r (0) we observed is in excellent agreement with that obtained from the study by Shen and Knutson, where the excitation wavelength was systematically varied in a fluorescence up-conversion study of aqueous Trp (16). The anisotropy decays to a value of ≈0.1 in 200 ps. The r(t) decay can be fitted to a sum of two terms: a 55-ps decay (50%) and a constant term (50%). This constant anisotropy level, not observed for free Trp in bulk water, suggests that the rotational reorientation of the probe (and the protein) occurs on a much longer time scale; the 55-ps decay is accordingly a reflection of the fluctuations of the indole chromophore orientation when anchored in the protein backbone.
Protein–DC Adduct.
The results of the femtosecond up-conversion experiments on the D-SC were obtained following the methodology used in the study of the protein and free Trp systems. The static fluorescence spectrum of D-SC is centered at 500 nm. At the wavelength used, 325 nm, excitation of the native protein chromophores is negligible, and only fluorescence from the extrinsic dansyl probe can be observed. The up-conversion scans were again fitted to the sum of exponentials. In this case, we fixed the long time component to 4.8 ns, a value that was obtained from a lifetime measurement by time-correlated single photon counting, as mentioned before. We then obtained C(t) by the same procedure.
The anisotropy measurement for D-SC is consistent with the binding to the protein. The results of the anisotropy measurement from time-correlated single-photon counting gives r (0) ≈0.4 and shows two decays, one of ≈100 ps (46%) and another of ≈1 ns (28%), after which the r(t) at ≈26% persists for at least 4 ns. This long time persistence is consistent with the probe being attached to the protein, which has a very long rotational relaxation time. Note that the experiments were performed on dialyzed samples with which free DC were excluded.
As shown in Fig. 3 Middle, the hydration correlation function C(t) decays much differently from that of Trp in the native SC. Here, the C(t) function has an ultrafast decay with a time constant of 1.5 ps (94%), and only a very small amount of spectral shift occurs with a time constant of 40 ps (6%). The total shift is of 1,180 cm−1; νmax (0) = 21,430 cm−1, νmax (200 ps) = 20,250 cm−1. The near absence of the ≈40-ps decay indicates that at the separation of ≈7 Å from the surface, the solvent dynamics resemble, to a large extent, those seen in bulk water.
Micelle–NATA Complex.
Femtosecond time-resolved fluorescence study of the probe NATA in a micelle formed by TX-100 amphiphilic molecules gave results that are different from that of the protein and DC-adduct decays in time and form. This micellar system can be considered a chemical model to study the solvation phenomenon for the probe-exposed site at a surface in contact with water, similar to the protein situation. In this case, the indole moiety faces to some degree the aqueous environment, whereas the rest of the NATA molecule is embedded in the micelle. We have verified that up-conversion scans of aqueous solutions of NATA produced very similar results to that of Trp in bulk water.
The C(t) curve for the NATA–TX-100 system was obtained following the procedure outlined above (see Fig. 3 Lower). It also displays a biexponential decay with τ1 = 2.9 ps (45%) and τ2 = 58 ps (55%), and the total spectral shift in this case is: 670 cm−1; νmax (0) = 30,420 cm−1, νmax (250 ps) = 29,750 cm−1. We measured the anisotropy decay of the fluorescence (340 nm) from the NATA–micelle system and observed that r(t) has a value of ≈0.2 at time 0. It can be fitted to a 63-ps decay (36%) and a constant level term (64%). The behavior of the r(t) decay confirms that the NATA probe is included in the micellar structure; the persistent anisotropy reflects the slow rotational reorientation of the probe (and micelle), and the 63-ps decay describes the local motion of the probe.
Discussion
Summarizing the above results, we can make the following points: (i) Solvation dynamics of intrinsic protein Trp are very different from those in bulk water. We note that the excitation of Trp carries with it negligible excess vibrational energy and switches a dipole that is not present in the ground state hydrophobic residue. (ii) For the protein, a “bimodal” solvation with two time scales, 1 and 40 ps, was observed, almost with equal contribution, with our time resolution. (iii) For the indole chromophore (NATA) in the micelle water structure, we recovered the protein-type solvation behavior but with longer decay times (≈3 and 60 ps). (iv) The covalently bonded dansyl chromophore at a distance of ≈7 Å from the protein surface exhibited solvation behavior similar to bulk solvation with a small contribution ≈5% of a 40-ps component. (v) In bulk water, the anisotropy of Trp decays in less than 100 fs and then from the value of 0.07 (excitation at 288 nm) to zero in 35 ps; the 35-ps decay is the orientation relaxation time, whereas the femtosecond component is because of the intrinsic states mixing (1La, 1Lb) of Trp, as discussed in the text. (vi) The picosecond anisotropy of Trp distinctly changes in the protein, from 35 ps in bulk water to a decaying anisotropy, by ≈60 ps, but remains constant at the value of ≈0.1 (296-nm excitation); for the micelle–NATA and protein–dansyl systems, similar behavior was observed.
These results are significant in view of the common picture for protein hydration as one in which the protein involves a water layer that is “static” and different from bulk water. A recent high-resolution x-ray crystallographic study of the protein SC (28) found that a layer of about 100 water molecules is associated with the protein; the occupancy is near unity, and there are a number of water molecules that occupy the vicinity of the Trp residue. This number of ≈100 is dictated by the equilibrium thermodynamic properties of hydrogen and van der Waals bonding and by the averaging in the crystal over space and time. Accordingly, the use of “ordered water” should be defined in this context. In solution, however, the dynamics and kinetics of water at the surface of the protein are determined by the time scale of solvation and by exchange with the bulk.
Our results indicate that the apparent “bimodality” reflects two types of solvation. The subpicosecond decay of the solvation correlation function, C(t), in SC has the similar characteristic of bulk solvation, primarily because of rotational inertial and diffusive relaxation of water around the chromophore. In contrast, the 38-ps component, which is orders of magnitude slower, reflects hydration by water molecules on the surface hindered for a long enough time under the influence of the protein potential of interaction. As such, this water structure, formed by dynamical ordering, is “rigid” but for a finite time. Molecular dynamics studies of the protein carboxymyoglobin (see, for example, ref. 10) have shown that the interaction of water molecules at the surface has a distribution of interaction energies, which gives rise to a distribution of water residence times, ranging from subpicoseconds to 100 ps or so, at the different hydration sites. Moreover, recent work by Clary's group on trajectories of a single water molecule attacking a protein shows distinct time scales for the trajectories (D. Clary, personal communication).
Molecular dynamics simulations by Rocchi et al. (29) on the protein plastocyanin have indicated that the dynamics of water molecules are significantly altered by their proximity to the surface of the protein. In particular, the study showed that the average rotational reorientation time for water molecules within a 4-Å distance shell from the protein's surface is significantly longer than that of a 14-Å shell. Although the study did not focus on specific molecular details at the protein's surface, it gives a clear indication that interactions with the protein's exposed sites, on average, tend to slow down the rotational dynamics of water molecules.
In a simple kinetic model, one can provide an order-of-magnitude estimate of the time scale for the exchange of surface water molecules with those of the bulk. The equilibrium between free and bound water is determined by the barrier and the free energy difference of the two forms. As suggested by Bagchi and Nandi, taking into consideration the number of hydrogen-bonding sites and their energies (typically −1.4 to −4.0 kcal/mol), the free-to-bound rate can be as small as 1.5 × 109 s−1 and as large as 1.3 × 1011 s−1 for a barrier of ΔG* = 1.5 kT; the bound-to-free rate can range from 4 × 107 s−1 to 3 × 109 s−1 (4). We note that these rates are the result of exchange between free and bound water, but these results represent a spatial averaging and depend on barrier height. One must also take into account the relative motion of the probe; however, in our case, the probe Trp itself is actually restricted in motion, not like in bulk water, as evidenced by our results of the anisotropy.
In contrast to the case of the native Trp in SC, when the chromophore is the dansyl moiety bound to the surface of the protein, almost bulk-like solvation behavior is recovered, and the long decay component (40 ps) corresponds to only about ≈5% of the total C(t) decay. Because in the D-SC system the chromophore resides at a distance of a few bond lengths from the protein surface (see Fig. 1 Middle), solvation is significantly influenced by bulk free water molecules.
For the micelle–NATA system, the observed solvation dynamics resemble that of the native SC protein, but the time scales are different: the fast decay component has a time constant of 2.9 ps, compared with the 0.8 ps seen in SC, and the long decay component becomes 60 ps instead of 40 ps in SC. The surface of the TX-100 micelle is considerably polar, and the hydrogen-bonding network of water molecules at the surface defines an extended solvation shell (30). Hence, the 60-ps decay is indicative of relaxation in this type of rigid water structure. The comparison with the protein is not direct, but the important point is that in both cases the dynamical nature of the water layer is important.
Finally, it should be mentioned that the above results of solvation dynamics in all systems studied, except Trp in bulk water, were obtained by using the time window from 0 to 200 ps, the value used for νmax(∞). However, longer-time Stokes shifts may be present, reflecting the influences of more rigid structures, either of water or protein itself. Several studies have provided evidence of such Stokes shift in proteins on time scales of hundreds of picoseconds to nanoseconds (refs. 31 and 32); some of these studies have used single-photon counting techniques to cover such a range, without the femtosecond resolution provided in this work.
Conclusion
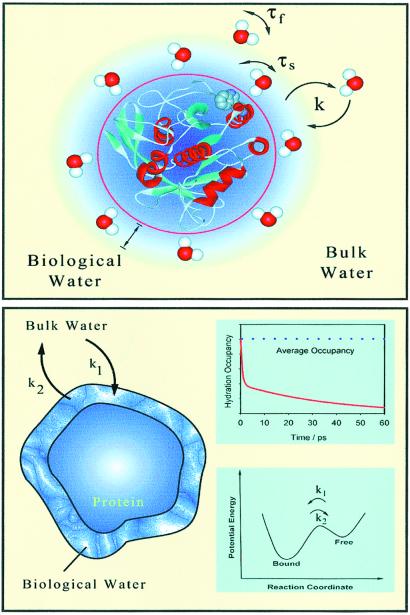
The study we present here is, to our knowledge, the first characterization, with femtosecond resolution, of the dynamics of solvation at the surface of a protein and using a single native Trp residue. Thus, the probing of biological water at the interface without spatial averaging or the position inhomogeneity of an extrinsic probe is achieved. Two types of trajectories of solvation have been observed, bulk type and protein layer type; both are dynamically involved and reflect the distribution in the residence times at the local surface site of the protein. The time scales observed in the apparent “bimodal” behavior of the hydration correlation function determine local order and rigidity. The probe itself is relatively rigid, restricted in motion in the protein, as evidenced by its anisotropy decay, but the water reorientation in the network is what determines the rigidity and order of the layer. By about 7 Å, essentially all water is bulk type. The model of our picture is shown in Fig. 4, and we used it as a schematic of what we term “dynamically ordered water” of proteins.
Figure 4.
A representation of the dynamic model discussed in the text for the solvation of proteins. (Upper) A water structure is shown, but it should not be viewed as a static layer. Instead, the distribution in residence time at the surface site defines two types of water motions: those that are as fast as bulk water, τf (weak interactions), and those much slower, τs (strong interactions). The exchange of surface and bulk water is displayed with two rate constants, k = k1 or k2. (Lower) We correlate the solvation behavior to a hydration occupancy, but the time scale shown is only for illustration. The free energy change is shown (Inset).
NMR studies of protein hydration in aqueous solution give a range of residence times, 10−2–10−8, for interior water and subnanosecond for surface water, as shown by the groups of Wüthrich (12) and Forsén (33) and others. Because of the femtosecond resolution achieved here, we decided to reexamine the same protein, but when denatured. Our preliminary results show again nonexponential behavior: the ultrafast type behavior (≈1 ps) and multiple solvation times (10–100 ps), reflecting the induced heterogeneity with some random coiling. We note that the average lifetime of tryptophan changes from 2.1 to 0.49 ns in the native and denatured forms (23), respectively, but both are longer than the reported solvation times.
The significance of these findings to the biological function of the enzyme protein SC may now be considered. The enzymatic activity of the protein, which is a surface function, depends on the efficiency of recognition of negatively charged and polar amino acids of the substrate peptide (34). The bulk-like environment in the close vicinity of the surface would enhance the interaction with the substrate. On the other hand, the structured water molecules are needed around the protein surface to be part of an efficient chemistry and possibly maintain a three-dimensional structure. Thus, the time scale for hydration and exchange dynamics is crucial to the tradeoff between the structure and its enzymatic function, and it is possible that the bimodal behavior reported here is of fundamental importance for such function.
Acknowledgments
We thank Dongping Zhong for continued interest and discussion and Spencer Baskin for helpful discussion about the anisotropy studies. This work was supported by the National Science Foundation.
Abbreviations
- NATA
N-acetyl-l-tryptophan amide
- SC
Subtilisin Carlsberg
- DC
dansyl chloride
- D-SC
dansyl-labeled SC
- TX-100, reduced Triton X-100.
References
- 1.Gregory R B, editor. Protein-Solvent Interactions. New York: Dekker; 1995. [Google Scholar]
- 2.Colombo M F, Rau D C, Parsegian V A. Science. 1992;256:655–659. doi: 10.1126/science.1585178. [DOI] [PubMed] [Google Scholar]
- 3.Launnas V, Pettit B M. Proteins Struct Funct Genet. 1994;18:148–160. doi: 10.1002/prot.340180207. [DOI] [PubMed] [Google Scholar]
- 4.Nandi N, Bagchi B. J Phys Chem B. 1997;101:10954–10961. [Google Scholar]
- 5.Burling F T, Weis W I, Flaherty K M, Brunger A T. Science. 1996;271:72–77. doi: 10.1126/science.271.5245.72. [DOI] [PubMed] [Google Scholar]
- 6.Svergun D I, Richard S, Koch M H J, Sayers Z, Kuprin S, Zaccai G. Proc Natl Acad Sci USA. 1998;95:2267–2272. doi: 10.1073/pnas.95.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerstein M, Chothia C. Proc Natl Acad Sci USA. 1996;93:10167–10172. doi: 10.1073/pnas.93.19.10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makarov V A, Feig M, Andrews B K, Pettitt M. Biophys J. 1998;75:150–158. doi: 10.1016/S0006-3495(98)77502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng X, Schoenborn B P. J Mol Biol. 1991;220:381–399. doi: 10.1016/0022-2836(91)90020-7. [DOI] [PubMed] [Google Scholar]
- 10.Gu W, Schoenborn B P. Proteins Struct Funct Genet. 1995;22:20–26. doi: 10.1002/prot.340220104. [DOI] [PubMed] [Google Scholar]
- 11.Grant E H, McClean V E R, Nightingale N R V, Sheppard R J, Chapman M J. Bioelectromagnetics. 1986;7:151–162. doi: 10.1002/bem.2250070206. [DOI] [PubMed] [Google Scholar]
- 12.Gottfried O, Edvards L, Wüthrich K. Science. 1991;254:974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- 13.Barret P C, Choma C T, Gooding E F, DeGrado W F, Hochstrasser R M. J Phys Chem B. 2000;104:9322–9329. [Google Scholar]
- 14.Jordanides X, Lang M, Song X, Fleming G R. J Phys Chem B. 1999;103:7995–8005. [Google Scholar]
- 15.Vivian J T, Callis P R. Biophys J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen X, Knutson J R. J Phys Chem B. 2001;105:6260–6265. [Google Scholar]
- 17.Zhong D, Pal S K, Zhang D, Chan S I, Zewail A H. Proc Natl Acad Sci USA. 2002;99:13–18. doi: 10.1073/pnas.012582399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haugland R P. Handbook of Fluorescent Probes and Research Chemicals. 6th Ed. Eugene, OR: Molecular Probes; 1996. pp. 8–13. [Google Scholar]
- 19.Ren B, Gao F, Tong Z, Yan Y. Chem Phys Lett. 1999;307:55–61. [Google Scholar]
- 20.Zhong D, Pal S K, Zewail A H. ChemPhysChem. 2001;2:219–227. doi: 10.1002/1439-7641(20010417)2:4<219::AID-CPHC219>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 21.Peon J, Zewail A H. Chem Phys Lett. 2001;348:255–262. [Google Scholar]
- 22.Schmitke J L, Stern L J, Klibanov A M. Proc Natl Acad Sci USA. 1998;95:12918–12923. doi: 10.1073/pnas.95.22.12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swaminathan R, Krishnamoorthy G, Periasamy N. Biophys J. 1994;67:2013–2023. doi: 10.1016/S0006-3495(94)80685-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lakshmikanth G S, Krishnamoorthy G. Biophys J. 1999;77:1100–1106. doi: 10.1016/S0006-3495(99)76960-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stratt R M, Maroncelli M. J Phys Chem. 1996;100:12981–12996. [Google Scholar]
- 26.Szabo A G, Rayner D M. J Am Chem Soc. 1980;102:554–563. [Google Scholar]
- 27.Jimenez R, Fleming G R, Kumar P V, Maroncelli M. Nature (London) 1994;369:471–473. [Google Scholar]
- 28.Fitzpatrick P A, Ringe D, Klibanov A M. Biochem Biophys Res Commun. 1994;198:675–681. doi: 10.1006/bbrc.1994.1098. [DOI] [PubMed] [Google Scholar]
- 29.Rocchi C, Bizzarri A R, Cannistraro S. Phys Rev E. 1998;57:3315–3325. [Google Scholar]
- 30.Molina-Bolivar J A, Aguiar J, Carnero Ruiz C. Mol Phys. 2001;99:1729–1741. [Google Scholar]
- 31.Vincent M, Gilles A-M, Li de la Sierra I M, Briozzo P, Bârzu O, Gallay J. J Phys Chem B. 2000;104:11286–11295. [Google Scholar]
- 32.Pal S K, Mandal D, Sukul D, Sen S, Bhattacharyya K. J Phys Chem B. 2001;105:1438–1441. [Google Scholar]
- 33.Halle B, Andersson T, Forsén S, Lindman B. J Am Chem Soc. 1981;103:500–508. [Google Scholar]
- 34.Voet D, Voet J G. Biochemistry. 2nd Ed. Somerset, NJ: Wiley; 1995. pp. 398–400. [Google Scholar]