Abstract
A mass difference between the measured molecular weight of a protein and that of its DNA-predicted sequence indicates sequence errors and/or posttranslational modifications. In the top-down mass spectrometry approach, the measured molecular ion is dissociated, and these fragment masses are matched against those predicted from the protein sequence to restrict the locations of the errors/modifications. The proportion of the ion's interresidue bonds that are cleaved determines the specificity of such locations; previously, ubiquitin (76 residues) was the largest for which all such bonds were dissociated. Now, cleavages are achieved for carbonic anhydrase at 250 of the 258 interresidue locations. Cleavages of three spectra would define posttranslational modifications at 235 residues to within one residue. For 24 of the 34 possible phosphorylation sites, the cleavages of one spectrum would delineate exactly all −PO3H substitutions. This result has been achieved with electron-capture dissociation by minimizing the further cleavage of primary product ions and by denaturing the tertiary noncovalent bonding of the molecular ions under a variety of conditions.
The explosive growth in the applications of mass spectrometry (MS) to proteomics can be traced back to the discovery four decades ago by Biemann, who showed that the mass spectrum of a peptide can directly indicate its sequence of amino acids (1). This approach was extended to larger peptides (2), to mixtures using MS/MS (3), and to proteins (4–10), even at the 10−17 mol level (11, 12). For “top down” protein MS/MS (7–10), masses of 7601.5 and 7865.7 Da for two fragment ions from carbonic anhydrase indicate the presence of Tyr and Thr (Tyr, 163.06 Da; Thr, 101.05 Da), matching the DNA-predicted Tyr-192 and Thr-191 at this mass distance from the C terminus (7). For the more difficult structural proteomics problem of characterizing posttranslational modifications, a molecular ion mass 80 Da higher than predicted could indicate phosphorylation (HPO3, 79.97 Da; ref. 13); if the fragment mass ∼7601.5 is present, but 7865.7 is replaced by ∼7946, the +80-Da modification is at Tyr-192 or Thr-191. An intermediate cleavage is necessary to differentiate these; a 7764.6-Da peak would show that Thr-191 is modified. This limitation in the proportion of interresidue bonds cleaved has been far more severe for such larger proteins. Top-down Fourier transfrom MS of the 8.6-kDa ubiquitin gave masses representing all interresidue cleavages, making possible its complete sequencing de novo (14). However, MS/MS of β-casein (3 variants, ≈24 kDa) identified the phosphorylation site at Ser-15, limited three more sites to five locations, but only restricted the last site to four possibilities (the protein contains 27 possible sites; ref. 13). Many MS/MS spectra of carbonic anhydrase (259 residues) combined gave cleavages between 135 different residue pairs, by far the most reported for any protein ion (15). Improvements here to electron capture dissociation (ECD; refs. 15–19) result in the cleavage of 250 of the 258 interresidue bonds of carbonic anhydrase molecular ions.
Common MS methods for protein characterization require separate steps of protein purification and degradation (e.g., proteolysis) before MS analyses (4–6). In the top down MS/MS approach (7–10), partial purification (e.g., capillary liquid chromatography; ref. 12) precedes electrospray ionization to produce multiple-charged protein ions, from which the desired molecular ions are purified by mass separation (MS-I) and dissociated (MS-II). The mass values of the resulting fragment ions can be measured with ppm accuracy by using Fourier transform MS (20–22), which also has unique capabilities for ECD (15–19). Conventional energetic MS-II methods include nozzle-skimmer (NS; ref. 23), collisionally activated dissociation (CAD; refs. 23–25), infrared multiphoton dissociation (26), and blackbody infrared dissociation (27, 28). Dissociation of the weakest amide bonds is favored for these, so that all were only able to cleave a combined total of 91 of the 258 interresidue bonds of carbonic anhydrase (Fig. 1 top spectrum, E). However, the new technique ECD itself gave 116 cleavages (15). For ECD, the electron neutralization releases sufficient energy (≈6 eV), so that backbone dissociation is essentially nonselective, independent of the bond dissociation energy, and occurs before this energy can be randomized. This nonergodic dissociation results in backbone fragmentation without cleavage of posttranslational modifications or, counterintuitively, of the noncovalent bonds of the protein's tertiary structure. These bonds must first be dissociated [activated ion (AI) ECD; ref. 15], such as by “in-beam” collisions of the ions with a gas pulse as they enter the MS cell (18), a process already used for trapping the ions in the cell. Without such activation, ECD gave no cleavage products from carbonic anhydrase (15). Even with AI, ECD effected no cleavages between residues 94 to 188, or in the central portions of the other larger proteins studied. This lack of larger fragment ions was postulated (15) to result from (i) secondary e− capture that is favored for these larger primary fragment ions, and thus converts them to smaller products, and (ii) incomplete prior AI dissociation of the tertiary noncovalent bonds. Conditions are described here that ameliorate these problems, producing cleavages between nearly all of the amino acids of carbonic anhydrase.
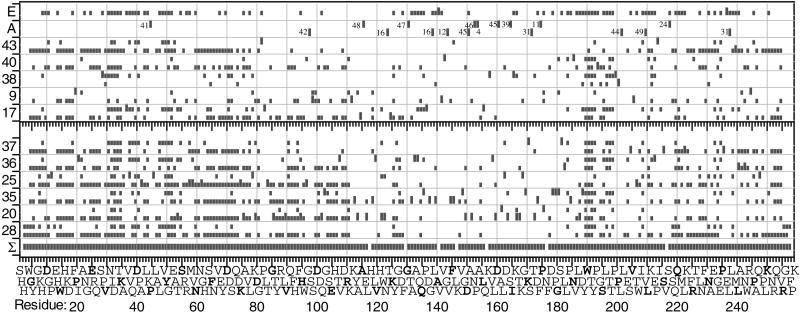
Figure 1.
Unique cleavages in ECD spectra of carbonic anhydrase (taller vertical bars). Each spectrum shows two rows of vertical bars indicating cleavages from a⋅, b, or y ions (Upper) and those from c or z⋅ ions (Lower). (Upper, E) Energetic spectrum, all cleavages reported in NS, CAD, infrared multiphoton dissociation, and blackbody infrared dissociation spectra. (Upper, second from top) Unique cleavages from other numbered spectra; see Table 1. (Lower, Σ spectrum) Cleavages from all spectra in Fig. 1.
Materials and Methods
N-terminal acetylated bovine carbonic anhydrase B, obtained from Sigma, was electrosprayed in a 20 μM solution with a nanospray emitter into a 6-T Finnigan Fourier transform ion cyclotron resonance mass spectrometer adapted for ECD, as described (15–18). A molecular ion distribution in which ≈80% of the charge states were 19–35+ was achieved with a 49:49:2 solution of MeOH/H2O/CH3COOH; 17–26+ from a 10:90:0.02 solution; 23–30+ from 30:69:1; 26–35+ from 5:95:0.1; 29–34+ from 30:70:0.2; and 32–38+ from a 50:43:5:2 solution of MeOH/H2O/glycerol/CH3COOH (the last solution produced less reliable electrospray conditions and instrument contamination).
Positive ions and electrons are added to the Fourier transform mass spectrometry ion cell from opposite ends of the magnetic axis. Activation of ions entering the ion cell is effected by in-beam collisions (18) with nitrogen (or Xe or Kr, as stated) introduced through a pulsed valve. After an ≈5 ms N2 gas pulse [cell pressure ≅ 10−7 torr (1 torr = 133 Pa)], the second and third quadrupoles of the ion introduction system are turned on to allow the electrospray ionization ion beam (typically, 300 pA for 2 s) to be transmitted to the ion cell. The outer trapping plate electrodes, conventionally used for electron containment (16–18), are used instead for ion trapping, with +5 eV at the ion introduction “front” end and +6 eV at the electron introduction end. At the same time, the electron filament heating grid is biased to admit electrons to the ion cell for 2 s, varying their current and energy (usually < +0.2 eV, relative to the closer outer trapping electrode), as noted. Spectra are measured with +1 eV on the normal (cylindrical) trapping electrodes and reported as averages of multiple scans (Table 1).
Table 1.
Effect of experimental conditions on ECD spectra
| No. | Experiment | Scans | Mn+, % | b | y | a⋅ | c | z⋅ | Σ |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.028 μA | 292 | 79 | 5 | 8 | 6 | 63 | 23 | 94 |
| 2 | 0.03 μA | 390 | 84 | 3 | 7 | 1 | 61 | 20 | 90 |
| 3 | 0.04 μA | 150 | 59 | 3 | 12 | 6 | 48 | 18 | 82 |
| 4 | 0.05 μA | 135 | 54 | 0 | 12 | 3 | 44 | 17 | 71 |
| 5 | 0.06 μA | 163 | 34 | 3 | 9 | 1 | 39 | 17 | 66 |
| 6 | 0.10 μA | 60 | 35 | 7 | 4 | 2 | 38 | 17 | 60 |
| 7 | 0.15 μA | 65 | 25 | 0 | 9 | 3 | 43 | 22 | 73 |
| 8 | Xe, 0.03 μA | 133 | 95 | 0 | 6 | 0 | 18 | 5 | 28 |
| 9 | Xe, 0.06 μA | 112 | 83 | 3 | 5 | 6 | 28 | 17 | 56 |
| 10 | Xe, 0.10 μA | 120 | 90 | 2 | 5 | 1 | 37 | 14 | 56 |
| 11 | Xe, 0.15 μA | 112 | 67 | 5 | 12 | 6 | 54 | 21 | 91 |
| 12 | Xe, 0.20 μA | 80 | 37 | 7 | 10 | 8 | 67 | 29 | 108 |
| 13 | Xe, 0.30 μA | 150 | 22 | 9 | 11 | 3 | 47 | 33 | 96 |
| 14 | 55 V | 350 | 90 | 19 | 14 | 7 | 64 | 15 | 98 |
| 15 | 60 V | 240 | 84 | 14 | 18 | 9 | 69 | 15 | 106 |
| 16 | 65 V | 300 | 80 | 9 | 18 | 1 | 60 | 7 | 88 |
| 17 | 75 V | 100 | 9 | 25 | 21 | 7 | 53 | 23 | 109 |
| 18 | 100 V | 100 | 10 | 18 | 22 | 4 | 15 | 10 | 61 |
| 19 | 120 V | 150 | 7 | 23 | 28 | 5 | 9 | 14 | 69 |
| 20 | i50°C | 375 | 90 | 4 | 8 | 5 | 78 | 33 | 113 |
| 21 | i56°C | 150 | 59 | 0 | 12 | 3 | 44 | 17 | 71 |
| 22 | i62°C | 150 | 48 | 8 | 13 | 3 | 55 | 28 | 96 |
| 23 | 17–26+, 0.06 μA | 200 | 5 | 6 | 8 | 2 | 36 | 13 | 62 |
| 24 | 32–38+, 0.06 μA | 100 | 13 | 5 | 16 | 6 | 50 | 27 | 95 |
| 25 | 29–34+ | 105 | 17 | 10 | 13 | 10 | 86 | 30 | 133 |
| 26 | 29–34+, c40°C | 120 | 47 | 11 | 14 | 8 | 74 | 37 | 124 |
| 27 | 29–34+, c50°C | 80 | 54 | 13 | 15 | 6 | 67 | 37 | 117 |
| 28 | 29–34+, c60°C | 130 | 42 | 19 | 18 | 14 | 81 | 45 | 138 |
| 29 | 29–34+, c80°C | 120 | 27 | 14 | 13 | 5 | 69 | 34 | 114 |
| 30 | 29–34+, c100°C | 120 | 30 | 19 | 14 | 7 | 72 | 35 | 123 |
| 31 | 29–34+, 2.5 W | 120 | 60 | 9 | 11 | 4 | 63 | 31 | 109 |
| 32 | 29–34+, 5 W | 120 | 21 | 13 | 11 | 2 | 56 | 32 | 100 |
| 33 | 29–34+, 7.5 W | 120 | 20 | 22 | 19 | 3 | 62 | 41 | 123 |
| 34 | 29–34+, 25 W | 120 | 18 | 12 | 12 | 7 | 8 | 13 | 41 |
| 35 | Plasma | 300 | 46 | 16 | 18 | 8 | 71 | 31 | 123 |
| 36 | 75V, 0.06 μA | 175 | 16 | 25 | 17 | 8 | 53 | 26 | 113 |
| 37 | 75V, 0.10 μA | 80 | 4 | 24 | 14 | 3 | 45 | 30 | 99 |
| 38 | CAD | 90 | 50 | 16 | 14 | 5 | 4 | 7 | 40 |
| 39 | e−0.4 eV | 280 | 59 | 6 | 15 | 2 | 34 | 15 | 66 |
| 40 | e−0.6 eV | 295 | 34 | 1 | 13 | 5 | 47 | 22 | 83 |
| 41 | e−11 eV, 0.20 μA | 150 | 27 | 17 | 8 | 8 | 32 | 22 | 74 |
| 42 | 23–30+, 0.06 μA | 150 | 6 | 5 | 6 | 6 | 68 | 26 | 102 |
| 43 | 23–30+, 0.10 μA | 80 | 8 | 10 | 6 | 4 | 57 | 31 | 98 |
| 44 | 29–34+, 0.10 μA | 100 | 35 | 2 | 16 | 7 | 48 | 27 | 91 |
| 45 | 26–35+ | 150 | 80 | 7 | 5 | 3 | 49 | 11 | 68 |
| 46 | 26–35+, 0.06 μA | 150 | 41 | 12 | 5 | 5 | 62 | 23 | 94 |
| 47 | Kr, 0.15 μA | 150 | 51 | 2 | 7 | 1 | 27 | 9 | 46 |
| 48 | Kr, 0.20 μA | 150 | 45 | 4 | 1 | 3 | 35 | 13 | 52 |
| 49 | Kr, 0.25 μA | 110 | 29 | 3 | 3 | 0 | 28 | 14 | 48 |
Experimental: 0.10 μA, e− current (none, 0.03 μA); Xe or Kr, collision gas (none, N2); c60°C, ion cell temperature (none, 25°C); i50°C, capillary inlet temperature (none, 56°C); 2.5 W, IR laser power (none, zero); 31–35+, ion charge states (none, 19–35+); 120 V, nozzle skimmer voltage (none, 60 V); e−0.6 eV, electron energy (none, <0.2 eV); CAD, excitation 46 dB after ECD; Plasma ECD, 0.3 μA e− into collision gas.
For some spectra, simultaneous activation of the ions during their introduction to the ion cell is effected with a continuous CO2 IR laser (25 W) at the same end of the magnetic axis as the slightly off-axis electron gun (26). Spectra are run at different ion cell temperatures, which should heat the ions by blackbody infrared radiation (27, 28). Assignment of fragment masses and compositions from the spectral data were made with the computer program THRASH (29). Most spectra were measured only above m/z 500 so that their terminal fragment ions were not determined. The low-mass ions were measured in only one spectrum, and for this spectrum the two terminal bond cleavages were not observed.
Prediction of the monoisotopic mass for an individual isotopic cluster involves matching its peak intensities against those predicted theoretically, so that the probability of a prediction in error by ±1 Da increases with decreasing signal/noise ratio (8, 18). The possibility of such mass errors leading to sequence assignment errors is made less probable by mass measurement of the same product ion in other charge states and by comparing the ≈115 ECD spectra of carbonic anhydrase measured in this study.
Results and Discussion
Previous studies have established that the primary products (≈90%) of electron capture dissociation are c and z⋅ ions Eq. 1 plus the minor products a⋅ and y Eq. 2 (16–18).
The collisional cooling by the gas pulse to trap the introduced ion beam can cause CAD (23–25) of the ions to produce b and y ions, with these y ions indistinguishable from those from ECD; however, without ECD, the normal ion introduction conditions with collisional cooling give minimum amounts of b and y ions. Accompanying CAD can be advantageous, as it is favored on the N-terminal side of Pro, where ECD does not form c,z⋅ ions, and the unique 16.02-Da mass difference between y and z⋅ ions shows that these represent C-terminal fragments (18).
ECD Spectral Reproducibility.
As will be shown, many experimental factors affect the cleavages achieved for the carbonic anhydrase protein ions studied here. Spectra measured weeks or months apart under ostensibly the same conditions can vary substantially (e.g., spectra 2, 15, and 21). With 258 interresidue bonds, many hundreds of product ions are possible from Eq. 1 and Eq. 2, and detection of individual ions depends on signal/noise levels that are affected by ion quantity (charge repulsion), number of scans, etc. Furthermore, the complex mixture of more open conformers produced by the denaturing conditions is undergoing diverse refolding during electron capture, as indicated by the recent kinetic studies of the unfolding and refolding of gaseous cytochrome c (30) and ubiquitin (31) ions. The spectra of Table 1, Fig. 1, and Fig. 2 represent the widest range of bond cleavages achieved of the ≈115 ECD spectra measured.
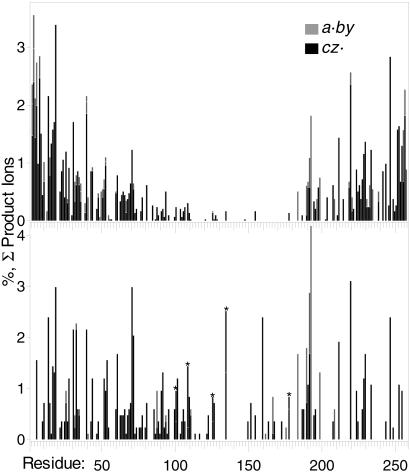
Figure 2.
(Upper) ECD spectra 28, 29–34 + carbonic anhydrase ions, increased (25 to 60°C) ion cell temperature. (Lower) ECD spectra 20, 19–35 + ions, lowered (56 to 50°C) capillary inlet temperature. *, both c (Upper) and z⋅ complementary ions observed from this cleavage.
The previous AI ECD study of such large proteins (15) pointed out that cleavages could not be effected in central regions of the backbone, yielding no c,z⋅ ions between residues 94 and 188. Approaches to this major problem were to decrease multiple electron reductions and to increase denaturation of the tertiary noncovalent structure.
Minimization of Secondary ECD Products.
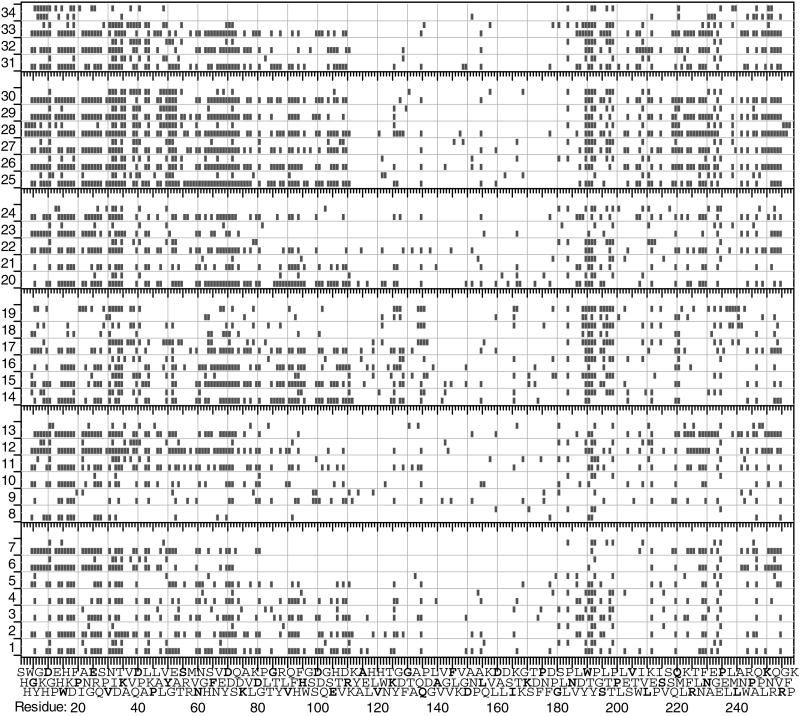
The larger of the primary c,z⋅ product pair has the larger charge and, thus, a far larger electron capture cross section (proportional to the square of the charge; ref. 16) for secondary capture to produce smaller products. This primary product dissociation could account for an abundance distribution skewed to smaller ions (Fig. 2 Upper). For AI ECD, the lowest electron beam current used previously was 0.1 μA, for which the summation of 50 or more scans was necessary to obtain spectra with good signal/noise (15). For this heated filament electron gun, the lowest useful beam current that can be achieved is 0.028 μA, which requires more scans (Fig. 3, spectrum 1; Table 1, Expt. 1, 292 scans). For the central third of the backbone, bonds 87 to 172, the number of cleavages (Fig. 3, spectra 7 to 1) for e− beam was only admitted to the trapped ions during the last third of the standard 2 s exposure (1.33–2 s); a very poor ECD spectrum (not shown) was obtained. Exposing the ions to the 0.03 μA e− currents of 0.15, 0.10, 0.06, 0.05, 0.04, 0.03, and 0.028 μA, were 0, 0, 9, 19, 17, 23, and 25, respectively. The combined spectra gave 39 cleavages in bonds 87 to 172.
Figure 3.
Effect on ECD spectra (see Fig. 1) of experimental conditions (see Table 1).
To lower further the number of electrons, the 0.03 μA e− beam was only admitted to the trapped ions during the last third of the standard 2s exposure (1.33–2s); a very poor ECD spectrum (not shown) was obtained. Exposing the ions to the 0.03 μA e− current during the first third (0–0.67 s) gave a far better spectrum; repeating this with a 0.3 μA electron beam yielded 21 cleavages in the bonds 87 to 172. Extending this, the 0.3 μA e− beam of <0.2 eV was admitted for 1 s starting with the N2 gas pulse, and then the ion beam was admitted to produce (Fig. 1, spectrum 35) 35 cleavages in bonds 87 to 172 in a single spectrum. In addition to decreased secondary electron capture, this increase in larger products also could result from the thermalization of the electrons in the bath gas to make their capture more efficient at the instant of incoming ion denaturation. This “plasma ECD” technique requires further investigation.
In an attempt to denature the ions further by heating them as they enter the mass spectrometer, it was found that increasing the normal inlet capillary temperature from 56 to 62°C (spectra 21, 22) actually reduced the cleavages between residues 87 and 172 from 18 to 14. However, lowering the inlet temperature from 56 to 50°C (Fig. 2 Lower, spectrum 20), the lowest value giving useful signal/noise, produced a surprising increase to 34 cleavages by using a much larger number of scans. Five of these cleavages result in a complementary pair of c,z⋅ ions. This lower inlet temperature produced more stable molecular ions (90% recovered vs. 59% for spectrum 21, Table 1), which is consistent with increased noncovalent bonding. Apparently, this additional bonding decreases the chance that an e− capture results in separated products, which also decreases the probability that a separated product will undergo secondary electron capture. Reflecting this, the product distribution (Fig. 2 Lower) shows far less skewing to smaller ions.
Effect of Charge Density.
Changing the electrospray conditions changed the average charge state in three sets of experiments from 21.2 to 27.8 to 34.5 (the last was achieved by adding glycerol to the electrospray solution; ref. 32). For 0.06 μA electron current spectra, the number of cleavages in the residue 87–172 region increased from 2 to 9 to 10 (spectra 23, 5, and 24), and the total number increased from 62 to 66 to 95 (Table 1). For the 0.03 μA e− current, the 19–35+ and 29–34+ charge states (spectra 2 and 25) gave 23 and 27 cleavages from 87–176, respectively, and 90 and 133 overall. The higher electrostatic repulsion mainly increases cleavages nearer the termini, but without a concomitant decrease of central cleavages, as found for the use of higher electron currents.
Increased Ion Denaturation.
Although spectra 35 and 20 together provide cleavage information on 56% of the interresidue bonds in the central third of the backbone, residual noncovalent bonding seems to be a problem at the remaining sites. For all these ECD spectra, collisions of the ions entering the cell with a bath gas is the primary method for ion activation. As a first additional method (29–34+ ions, 80 to 130 scans), the ion cyclotron resonance cell temperature was raised to heat the ions by blackbody infrared radiation (27, 28), spectra 25–30. For cell temperatures of 25, 40, 50, 60, 80, and 100°C, the number of cleavages between bonds 87 and 172 was surprisingly similar (27, 21, 20, 24, 11, 20, respectively), with total cleavages of 133, 124, 117, 138, 114, and 123 providing the highest value observed in this study (Fig. 2 Upper, spectrum 28), although the spectrum is skewed to smaller products. The different temperatures did, however, activate somewhat different bonds; for the regions 1–86, 87–172, and 173–258, the total cleavages of spectra 25–30 were 78, 45, 57, respectively (sum = 180), whereas just the 25 and 60°C spectra (spectra 29 and 40) together gave 78, 31, and 52 cleavages (sum = 161).
Heating the carbonic anhydrase ions during their 2 s introduction with an IR laser (ref. 26; 25 W, 0.03 μA e− beam, spectra 25 and 31–34) with 0%, 10%, 20%, 30%, and 100% of the duty cycle gave 27, 16, 10, 13, and 1 cleavages, respectively, in the 87–172 bond region, with the IR excitation only providing a total of 7 new cleavages. Intermediate laser power of 7.5 W did double the number of b,y ions, presumably by infrared multiphoton dissociation (26), but the full 25 W removed most c,z⋅ ions. In NS activation, the incoming ions are accelerated in high pressure (≈10−3 torr) region under a variable voltage difference, with ≈60 V stripping off adducted solvent molecules, etc., but causing minimal CAD (25). For NS potentials of 55, 60, 65, 75, 100, and 120 V (spectra 14–19), the number of 87–172 interresidue cleavages are 30, 33, 27, 27, 8, and 13, respectively, although intermediate NS values of 65 and 75 V (spectra 16 and 17) did produce 12 central-region cleavages not in the 60-V spectrum. For both NS and laser IR activation, high excitation leads to dissociation before the c,z⋅ product ions can be formed and also produces spectra of poor quality (signal/noise, resolution), possibly from ions trapped off-axis.
Higher energy collisional interactions were tried with xenon (spectra 8–13) and krypton (spectra 47–49) in place of nitrogen, but the spectra were of low signal/noise with far fewer total cleavages. For e− currents of 0.30, 0.20, 0.15, 0.10, 0.06, and 0.03 μA, the Xe spectra show 6, 11, 16, 8, 18, and 3 cleavages, respectively, in the 87–172 region. Although these represent 39 different cleavages, only one is unique vs. other ECD spectra using N2 collision gas. The use of Xe seems to lower the efficiency of electron capture; the 0.03 μA e− current dissociates only 5% of the molecular ions, whereas higher currents increase central cleavages. Similarly, with Kr collision gas, 0.05 and 0.10 μA e− currents give only 7 and 16 total cleavages, respectively (spectra not shown). However, the three higher current Kr (spectra 47–49) each show a unique cleavage despite their low total number of cleavages.
For higher energy electrons, those of 0.6 eV (spectrum 40) gave a reduced number of cleavages, but several were unique. Zubarev and coworkers (personal communication) have recently shown that ECD of peptides can be achieved with “hot” electrons (HECD; 3–13 eV), adding sufficient additional energy to cause side chain cleavage (e.g., to distinguish Leu and Ile). ECD of carbonic anhydrase (spectrum 41; others not listed) using 0.2 μA electrons of 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, and 12 eV gave 4, 5, 26, 81, 95, 78, 64, 77, 91, 74, and 57 total cleavages, respectively, with 0, 0, 0, 4, 5, 3, 1, 0, 1, 2, 0 in the 87 to 172 region. HECD of ubiquitin (8.6 kDa) gave nearly complete sequence coverage. HECD of these proteins without in-beam activation required a 100-fold higher current (surface emitter source; ref. 33), as found necessary for HECD of peptides (R. A. Zubarev, personal communication). In fact, such high current, high energy (≈10 eV) electron bombardment is a surprisingly efficient alternative to in-beam collisional activation; when followed by 0.15 μA e− capture, an ECD spectrum similar to spectrum 7 (Fig. 3) is obtained.
For the effect of kinetic energy of the ions, the minimum value is determined by the potential on the front trapping electrode that admits ions into the grounded cell. A value of +5 V was used in most experiments, whereas +4 V (with +5 V on the rear electrode) gave poor ECD spectra. Even higher voltages seem to deposit more energy on ECD, but a detailed study was not made. For the rear trapping electrode, a value +1 V higher than the front electrode was optimum; ions that have higher energy after passing through the collision gas then exit the trap without being reflected back. Setting the back electrode +2 V higher than the front electrode gave much poorer ECD spectra, possibly by affecting the collision process.
ECD does not yield cleavages on the N-terminal side of Pro (13–15), but such cleavages were identified for all 19 Pro residues of carbonic anhydrase ions by the presence of b,y product ions (none from c,z⋅ ions). The energetic conditions used to enhance noncovalent bond cleavage produce more b,y product ions with electron capture than without.
Such improved ECD conditions are advantageous for smaller, less folded protein ions. For ubiquitin, 8.6 kDa, 0.1 μA electrons with in-beam activation cleaved 70 of 75 interresidue bonds, whereas plasma ECD cleaved 73 of 75 bonds, each in a single spectrum.
Conclusions
Of the 258 interresidue bonds in carbonic anhydrase, ECD has cleaved 138 bonds in spectrum 28; 201 bonds in the five spectra 28, 20, 35, 25, and 36; 230 bonds including spectra 37, 17, 9, 38, 40, 43; 248 in 26 ECD spectra; and 250 including the energetic MS/MS spectra (6, 18, 22). The cleavages of spectra 20, 28, and 35 would define posttranslational modifications at 235 of the 259 residues to within one residue. The two terminal cleavages were not found, but little effort was expended to detect these low-mass ions. For the first third of the backbone from the N terminus, all cleavages 2–86 were identified, with all but 6 from the five spectra 28, 20, 35, 25, and 36. For the 173–257 C-terminal cleavages, all but two were identified, with all but 17 identified by the first five spectra (Fig. 1 Bottom). Of the central 86 interresidue cleavages, four were not identified, with all but 24 identified in the first five spectra.
For a real problem in protein characterization, such as a protein molecular weight from MS-I that is higher than that of the DNA predicted sequence, the experimental conditions for the first ECD spectrum should be chosen to maximize the number of cleavages; electrospray should yield highly charged ions (here 29–34 +), whereas ECD should use low (here 0.03 μA) electron current (or plasma) conditions, such as spectra 25, 28, or 35. For spectrum 35, the largest information gap results from the absence of products from cleavages 155–162; if cleavages 154 and 163 indicated a mass error between these, its location would be narrowed by cleavages 159 and 161 in spectrum 20 and by cleavages 159 and 162 in spectrum 25, with specific identification possibly requiring cleavages 155, 157, 158, or 160 from spectra 17, 38, 36, or 31, respectively. Such specificity is not required for many cases, such as site-specific posttranslational modifications. Thus, phosphorylations are restricted to Ser, Thr, or Tyr, of which carbonic anhydrase contains 34; for 24 of these, the cleavages of spectrum 35 alone would define exactly the position of phosphorylation (13). A reduction (or increase) in the measured molecular weight vs. that predicted could result from a mutation, such as the 28-Da lower value for carbonic anhydrase A; the cleavages of spectrum 25 would pinpoint this mutation as Arg-56 (156.10 Da) to Gln-56 (128.06 Da), although 5 ppm accuracy (20–22) for the mass difference of c55 vs. c56 would be necessary to exclude Lys-56 (Δ0.04 Da).
The fact that small changes in experimental conditions, especially those that should affect the conformational structure, produce almost random changes in bond cleavages is consistent with the large variety of rapidly interchanging conformers postulated previously for the unfolding/folding studies of gaseous cytochrome c (30) and ubiquitin (31) ions, and for the negligible effect of bond dissociation energy on the nonergodic ECD reactions (16–18). Minimizing secondary ECD products and achieving near-complete ion denaturation have been possible for this 29-kDa ion. Extension to larger proteins with plasma ECD deserves further study.
Acknowledgments
We thank Kathrin Breuker, Barry Carpenter, and Roman Zubarev for valuable discussions and the National Institutes of Health (Grant GM16609) for generous financial support.
Abbreviations
- MS
mass spectrometry
- ECD
electron capture dissociation
- NS
nozzle skimmer
- CAD
collisionally activated dissociation
- AI
activated ion
References
- 1.Biemann K. Mass Spectrometry. Organic Chemical Applications. New York: McGraw–Hill; 1962. pp. 260–296. [Google Scholar]
- 2.Barber M L, Jolles P, Vilkas E, Lederer E. Biochem Biophys Res Commun. 1965;18:469–473. doi: 10.1016/0006-291x(65)90775-8. [DOI] [PubMed] [Google Scholar]
- 3.McLafferty F W, Venkataraghavan R, Irving P. Biochem Biophys Res Commun. 1970;39:274–278. doi: 10.1016/0006-291x(70)90789-8. [DOI] [PubMed] [Google Scholar]
- 4.Pandey A, Mann M. Nature (London) 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 5.Griffin T J, Han D K M, Gygi S P, Rist B, Lee H, Aebersold R, Parker K C. J Am Soc Mass Spectrom. 2001;12:1238–1246. doi: 10.1016/S1044-0305(01)00316-6. [DOI] [PubMed] [Google Scholar]
- 6.Service R F. Science. 2001;294:2074–2077. doi: 10.1126/science.294.5549.2074. [DOI] [PubMed] [Google Scholar]
- 7.Kelleher N L, Lin H Y, Valaskovic G A, Aaserud D J, Fridriksson E K, McLafferty F W. J Am Chem Soc. 1999;21:806–812. [Google Scholar]
- 8.McLafferty F W, Fridriksson E K, Horn D M, Lewis M A, Zubarev R A. Science. 1999;284:1289–1290. doi: 10.1126/science.284.5418.1289. [DOI] [PubMed] [Google Scholar]
- 9.Forbes A J, Mazur M, Patel H M, Walsh C T, Kelleher N L. Proteomics. 2001;1:927–933. doi: 10.1002/1615-9861(200108)1:8<927::AID-PROT927>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 10.Ge Y, Lawhorn B G, ElNaggar M, Strauss E, Park J-H, Begley T P, McLafferty F W. J Am Chem Soc. 2002;124:672–678. doi: 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- 11.Valaskovic G A, Kelleher N L, McLafferty F W. Science. 1996;273:1199–1202. doi: 10.1126/science.273.5279.1199. [DOI] [PubMed] [Google Scholar]
- 12.Settlage R E, Russo P S, Shabanowitz J, Hunt D F. J Microcolumn Sep. 1998;10:281–285. [Google Scholar]
- 13.Shi S D-H, Hemling M E, Carr S A, Horn D M, Lindh I, McLafferty F W. Anal Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- 14.Horn D M, Zubarev R A, McLafferty F W. Proc Natl Acad Sci USA. 2000;97:10313–10317. doi: 10.1073/pnas.97.19.10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horn D M, Ge Y, McLafferty F W. Anal Chem. 2000;72:4778–4784. doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- 16.Zubarev R A, Kelleher N L, McLafferty F W. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 17.Zubarev R A, Kruger N A, Fridriksson E K, Lewis M A, Horn D M, Carpenter B K, McLafferty F W. J Am Chem Soc. 1999;121:2857–2862. [Google Scholar]
- 18.Zubarev R A, Horn D M, Fridriksson E K, Kelleher N L, Kruger N A, Lewis M A, Carpenter B K, McLafferty F W. Anal Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 19.Haselmann K F, Budnik B A, Olsen J V, Nielsen M I, Reis C A, Clausen H, Johnson A H, Zubarev R. Anal Chem. 2000;73:2998–3005. doi: 10.1021/ac0015523. [DOI] [PubMed] [Google Scholar]
- 20.Beu S C, Senko M W, Quinn J P, McLafferty F W. J Am Soc Mass Spectrom. 1993;4:190–192. doi: 10.1016/1044-0305(93)85077-B. [DOI] [PubMed] [Google Scholar]
- 21.Bruce J E, Anderson G A, Wen J, Harkewicz R, Smith R D. Anal Chem. 1999;71:2595–2599. doi: 10.1021/ac990231s. [DOI] [PubMed] [Google Scholar]
- 22.Hughey C A, Hendrickson C L, Rodgers R P, Marshall A G, Qian K. Anal Chem. 2001;73:4676–4681. doi: 10.1021/ac010560w. [DOI] [PubMed] [Google Scholar]
- 23.Loo J A, Udseth H R, Smith R D. Rapid Commun Mass Spectrom. 1988;2:207–210. [Google Scholar]
- 24.Gauthier J W, Trautman T R, Jacobsen D B. Anal Chim Acta. 1991;246:211–225. [Google Scholar]
- 25.Senko M W, Speir J P, McLafferty F W. Anal Chem. 1994;66:2801–2808. doi: 10.1021/ac00090a003. [DOI] [PubMed] [Google Scholar]
- 26.Little D P, Speir J P, Senko M W, O'Connor P B, McLafferty F W. Anal Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 27.Price W D, Schnier P D, Williams E R. Anal Chem. 1996;68:859–866. doi: 10.1021/ac951038a. [DOI] [PubMed] [Google Scholar]
- 28.Ge Y, Horn D M, McLafferty F W. Int J Mass Spectrom. 2001;210/211:203–214. [Google Scholar]
- 29.Horn D M, Zubarev R A, McLafferty F W. J Am Soc Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 30.Horn D M, Breuker K, Frank A J, McLafferty F W. J Am Chem Soc. 2001;123:9792–9799. doi: 10.1021/ja003143u. [DOI] [PubMed] [Google Scholar]
- 31.Breuker, K., Oh, H.-B., Horn, D. M., Cerda, B. A. & McLafferty, F. W. (2002) J. Am. Chem. Soc., in press. [DOI] [PubMed]
- 32.Iavarone A T, Jurchen J C, Williams E R. J Am Soc Mass Spectrom. 2000;11:976–985. doi: 10.1016/S1044-0305(00)00169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsybin Y O, Hakansson P, Budnik B A, Haselmann K F, Kjeldsen F, Gorshkov M, Zubarev R A. Rapid Commun Mass Spectrom. 2001;15:1849–1854. doi: 10.1002/rcm.448. [DOI] [PubMed] [Google Scholar]