Abstract
Human bone marrow expresses a pseudogene that encodes an antimicrobial peptide homologous to rhesus monkey circular minidefensins (θ-defensins). We prepared the putative ancestral human peptide by solid-phase synthesis and named it “retrocyclin.” Retrocyclin did not cause direct inactivation of HIV-1, and its modest antibacterial properties resembled those of its rhesus homologs. Nevertheless, retrocyclin had a remarkable ability to inhibit proviral DNA formation and to protect immortalized and primary human CD4+ lymphocytes from in vitro infection by both T-tropic and M-tropic strains of HIV-1. Confocal fluorescent microscopy studies performed with BODIPY-FL-labeled RC-101, a close analog of retrocyclin, showed that the peptide formed patch-like aggregates on the surface of CD4+ cells. These findings suggest that retrocyclin interferes with an early stage of HIV-1 infection and that retrocyclin-like agents might be useful topical agents to prevent sexually acquired HIV-1 infections.
Defensins are cysteine-rich, cationic antimicrobial peptides expressed by the leukocytes and epithelial cells of birds and mammals (1–4). Three defensin subfamilies exist in vertebrates: α-defensins, β-defensins, and circular (θ) minidefensins (5). All derive from an ancestral gene that existed before reptiles and birds diverged (6), all contain six cysteines, and all have largely β-sheet structures that are stabilized by three intramolecular disulfide bonds. Although α- and β-defensins differ in the spacing and connectivity of their cysteines, they have similar topology (7). The human α− and β-defensin genes cluster on chromosome 8p23 (8), a locus that also contains the pseudogene for retrocyclin (GenBank accession nos. U10267 and AF238378). RTD-1, a type of defensin (5), was recently detected in bone marrow from the rhesus monkey, Macacca mulatta. It had 18 residues and was circular—having been formed by the fusion of two truncated α−defensin precursors (called “demidefensins” in this report), each of which contributed 3 cysteines to the mature peptide. Subsequently, two additional rhesus circular minidefensins, RTD-2 and RTD-3, were described (9, 10). The cellular machinery responsible for processing RTDs is capable of generating molecular diversity posttranslationally (9, 10), and remains operational in human leukocytes (5).
Here, we report that human bone marrow expresses mRNA that is homologous to the precursors of rhesus monkey circular minidefensins. Although a stop codon within its signal sequence suggests that the human transcript now represents an expressed pseudogene, we used its sequence and information derived from studies on rhesus monkeys to synthesize “retrocyclin,” the putative ancestral human circular minidefensin. Retrocyclin dramatically protected human CD4+ cells from infection by HIV-1 in vitro, was noncytotoxic, and killed certain bacteria effectively in physiologic saline.
Methods
Cloning.
The term “demidefensin” denotes an α-defensin transcript that contains a stop codon between the third and fourth cysteine of its defensin domain. We prepared two primers 5′-TCAGGCAAGAGCTGATGAAGCT-3′ (P1) and 5′-ACTCTCAAAGTAACTTCTGAGC-3′ (P2) based on the monkey demidefensin cDNA sequences (GenBank accession nos. AF 184156, 184157, and 184158) and performed PCR on Marathon-Ready human bone marrow cDNA (CLONTECH), which resulted in an ≈264-bp amplified product. To obtain its 3′ side sequence, we amplified Marathon-Ready human bone marrow cDNA with P1 and Abridged Universal Amplification Primer (AUAP) using a 3′-RACE (rapid amplification of cDNA ends) kit (GIBCO/BRL). After an ≈352-bp PCR product was cloned and sequenced, we obtained 5′ side sequence with the 5′-RACE kit from Boehringer Mannheim by amplifying Marathon-Ready human bone marrow cDNA with oligo(dT) anchor primer and antisense primer (5′-ACTCTCAAAGTAAATTCTGAGC-3′). The ≈407-bp PCR product was cloned and sequenced. As expected, 264-bp nucleotides were overlapping in the sequences of our two PCR products.
Peptides.
Peptides were synthesized at a 0.25-mmol scale with a Perkin–Elmer Applied Biosystems 431 A Synthesizer, using prederivatized polyethylene glycol polystyrene arginine resin (PerSeptive Biosystems, Framingham, MA), FastMoc chemistry, and double coupling for all residues. The crude peptide was reduced under nitrogen for 15 h at 50°C with excess DTT in 6 M guanidine⋅HCl, 0.2 M Tris⋅HCl, and 0.2 mM EDTA (pH 8.2). The reaction was stopped with glacial acetic acid (final concentration, 5%), and the reduced peptide was stored under nitrogen until purified by RP-HPLC. After this step, the peptide appeared homogeneous, and its mass [1942.5, by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) MS] agreed well with its theoretical mass. The reduced peptide (0.1 mg/ml) was oxidized, cyclized, and purified essentially as described by Tang and colleagues (5). The MALDI-TOF MS mass of retrocyclin (1918.5 Da) agreed well with its expected mass. CD spectra were obtained at 25°C from an AVIV 62DS spectropolarimeter (Aviv Associates, Lakewood, NJ) as described previously (11).
Antimicrobial Activity.
Two stage radial diffusion assays were done as previously described (12). Clear zone diameters were measured to the nearest 0.1 mm after overnight incubation. The x-intercept of the relationship of zone diameter to log10 peptide concentration was determined by least mean squares regression, and equated to the MEC (minimal effective concentration).
Cytotoxicity Assay.
Cytotoxicity determinations were made with a Cell Proliferation Kit purchased from Boehringer Mannheim according to the manufacturer's instructions. The procedure measures the reduction of the yellowish MTT molecule (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) to a dark blue formazan.
Antiviral Activity.
Cells.
HIV-1 strain IIIB uses the CXCR4 chemokine coreceptor for entry. Immortalized CD4+ H9 cells, which are permissive for infection with this strain, were maintained in RPMI 1640 supplemented with 10% heat-inactivated FCS, 10 mM Hepes, 2 mM glutamine, 100 units of penicillin/ml, and 10 μg of streptomycin/ml. In addition, primary CD4+ lymphocytes from HIV-1-seronegative donors were generated from freshly purified peripheral blood mononuclear cells stimulated with a CD3-CD8 bispecific monoclonal antibody (13–15). After ≈7 days, when 98% of these cells coexpressed CD3 and CD4 (data not shown), they were infected with HIV-1. These cells were maintained in RPMI 1640 containing 10% FCS supplemented with 2 mM glutamine, 100 units of penicillin/ml, 10 μg of streptomycin/ml, and 50 units of IL-2/ml.
HIV-1 stocks.
HIV-1 IIIB (T-tropic) was produced by fresh infection of T1 cells, and supernatant was harvested 4 to 5 days after infection (16). HIV-1 JR-CSF (M-tropic) was produced by fresh infection of peripheral blood mononuclear cells (PBMC) after 3 days of phytohemagglutinin (PHA) stimulation (PHA blasts), followed by supernatant harvest 7 or 8 days after infection (17). Aliquots of virus stocks were cryopreserved at −80°C and thawed immediately before use. Viral titers were determined as previously described (18).
Antiviral assays.
Approximately 106 CD4+ cells/ml were resuspended at 37°C in fresh RPMI 1640–10% FCS medium that contained 50 units of IL-2 when primary CD4+ PBL cells were tested. Retrocyclin (final concentration, 20 μg/ml) was added, and 3 h later the cells were challenged with an inoculum of HIV-1 that gave a multiplicity of infection (moi) of 10−2 TCID50 per cell (TCID50 signifies “50% tissue culture infectious dose”). After incubation for 3 h at 37°C, the cells were washed twice and added to 24-well plates at 5 × 105 cells per well in 2 ml of medium containing retrocyclin at the indicated final concentrations. At 3-day intervals, 1 ml of supernatant was removed from each well for HIV-1 p24 antigen quantitation by ELISA (ELISA; DuPont) and replaced with 1 ml of fresh medium supplemented with retrocyclin at the final concentrations.
HIV-1 Provirus.
To verify whether retrocyclin blocked the formation of proviral DNA in HIV-1JR-CSF-inoculated CD4+-selected PBMC, quantitative real time PCR was performed as described in refs. 19 and 20. In brief, DNA was obtained from frozen cell pellets after exposure to urea lysis buffer and subsequent phenol/chloroform extraction as previously described (21). Amplification and detection was performed on an Applied Biosystems PRISM 7700 Sequence Detection System using the Taqman Reagent Kit. Oligonucleotides that were used for detection of early HIV DNA [R-U5 regions of the long-term repeat region (LTR)] have been described previously [SR1 and AA55 (19–21)]. The fluorescent probe ZXF (200 nM) corresponds to nucleotide numbers 584 to 616 in the JR-CSF LTR. The oligonucleotides that were used to detect full-length (LTR-gag) HIV reverse transcripts included primers that bind sequences in the beginning of gag, and have been previously described [SR1 and M667 (19–21)]. The probe for this condition is identical to that used for total HIV reverse transcripts. The standard curve that was used to determine HIV DNA levels ranged from 10 to 20,000 copies of cloned HIV DNA (19–21). The standard curve used to determine levels of beta-globin gene sequences consisted of DNA derived from 10 to 100,000 normal human peripheral blood lymphocytes. Quantitation of HIV sequences was achieved by extrapolation from these standard curves. Thus, the combination of primers SR1 and AA55 amplified sequences in R/U5 of the LTR, and detected early events in reverse transcription. The combination of SR1 and M667 amplified the LTR/gag junction, formed near the completion of the reverse transcription process.
Reverse Transcription–PCR.
Human total placental RNA (0.5 μg) was reverse transcribed with 1 unit of rHIV reverse transcriptase (U.S. Biological, Swampscott, MA) by using an Advantage RT-for-PCR kit (CLONTECH) in the presence or absence of 5 μM retrocyclin, T140, or 3′-azido-3′-deoxythymidine (AZT). PCR was performed for 25 cycles according to the kit's protocol. Amplified products were electrophoresed in a 1% agarose gel and visualized by using ethidium bromide.
Confocal Microscopy.
RC-101 was joined to the aminereactive fluorescent dye, BODIPY-FL (Molecular Probes), according to the manufacturer's protocol, and the conjugate (RC-101BODIPY-FL) was purified by reverse-phase HPLC and resuspended in 0.01% acetic acid at concentrations up to 240 μg/ml. RC-101BODIPY-FL (20 μg/ml) was incubated with 2.5 × 105 CD4+-selected PBMC cells for 15 min at room temperature and washed once in fresh R10–50 media. Specimens were imaged on a Leica (Heidelberg, Germany) TCS-SP Confocal Microscope equipped with an argon laser for 488 nm blue excitation of BODIPY-FL. Images were collected by using Leica Confocal Software. Confocal microscopy was performed in the Carol Moss Spivak Cell Imaging Facility at the University of California at Los Angeles Brain Research Institute.
Results and Discussion
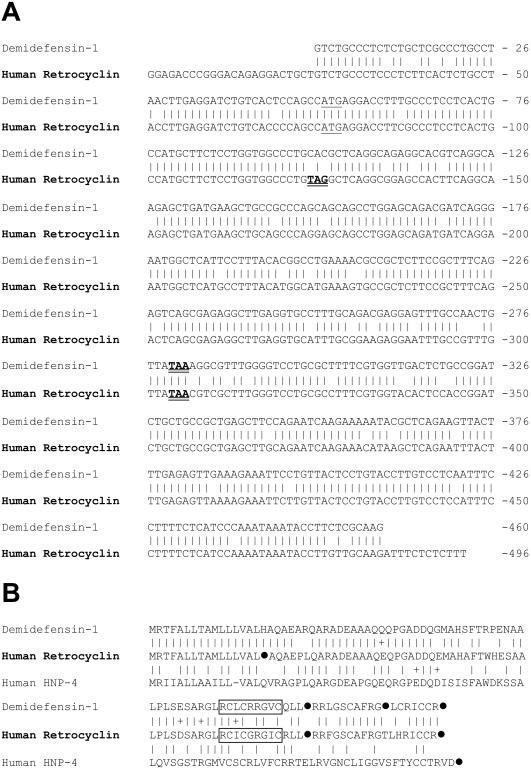
Fig. 1A compares the cDNA sequence of the human retrocyclin transcript to that of rhesus demidefensin 1. At the nucleotide level, retrocyclin is 88.9% identical to rhesus demidefensin 1, and shows similar identity to rhesus RTD-1 precursors 1a and 1b (5). Although a stop codon (TAG) within the human transcript's signal sequence should abort translation, the otherwise high conservation of rhesus and human mRNAs suggests that humans acquired this mutation relatively recently in evolution.
Figure 1.
Sequence comparison of human and rhesus demidefensins. (A) The sequences of rhesus demidefensin-1 (GenBank accession no. AF184156) and human pro-retrocyclin (GenBank accession no. AF 355799) were both obtained in our laboratory. A vertical line connects identical bases, start codons are underlined, and stop codons are bolded and doubly underlined. The sequences are 89.4% identical. The demidefensin-1 sequence is 92% (422/457) identical to RTD-1 precursor 1a and 97% identical to RTD-1 precursor (445/457) 1b–both as described by Tang and colleagues (5). (B) The sequences of rhesus demidefensin-1, human retrocyclin, and human defensin 4 (HNP-4) are shown. Solid circles (●) indicate a stop codon in the corresponding cDNA. Vertical bars connect identical residues, and + signs connect similar residues. Residues represented in mature retrocyclin and RTD-molecules are boxed. The demidefensin-1 sequence (GenBank accession no. AF184156) was derived from the monkey mRNA shown in A.
Rhesus RTD-1 is the fused and trimmed product of two different truncated α-defensin precursors (5), designated 1a and 1b. Recently, we purified and sequenced two additional circular minidefensins from rhesus bone marrow (9, 10). Their sequences indicated that these peptides arose after homologous pairing of the aforementioned 1a and 1b precursors. Fig. 1B shows the peptide sequences of demidefensin 1 [equivalent to RTD precursor 1b (5)], prepro-retrocyclin, and prepro-defensin 4. Residues incorporated into the circular minidefensins are boxed, and stop codons are represented by solid circles.
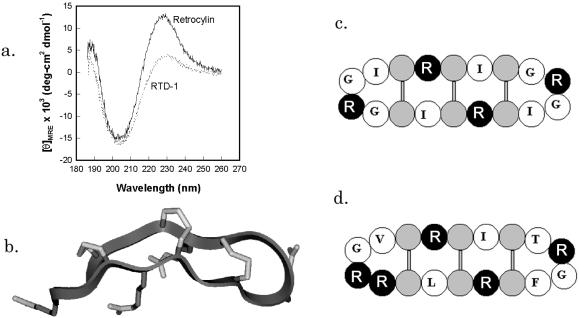
Our synthetic retrocyclin represents the circular minidefensin that would have formed (i) if the signal sequence mutation were absent or inoperative, and (ii) if the precursor underwent homologous pairing so that its boxed residues formed both halves of the circular molecule. RTD-1 and retrocyclin have very similar CD spectra, with largely β-sheet structures stabilized by disulfide linkages and connected by turns (Fig. 2A; ref. 5). Antimicrobial peptides with similar spectra include tachyplesins (22), protegrins (11), and circularized defensins (23). Fig. 2B, a backbone ribbon model of retrocyclin, was made by templating its sequence on the structure of protegrin PG-1 (11) and cyclizing it. The resulting structure was annealed by molecular dynamics and energy minimized. Fig. 2C is a cartoon version of Fig. 2B, designed primarily to show the placement of the cysteine and arginine molecules. Fig. 2D is a similar cartoon of rhesus RTD-1.
Figure 2.
Structural characterization of retrocyclin. (a) CD spectrum demonstrating the similarity in structure between retrocyclin and RTD-1, both at 0.5 mg/ml in a 1:1 mixture of trifluoroethanol in PBS at pH 7.4. (b) A hypothetical model of retrocyclin made by templating its sequence on the backbone of a similar peptide from porcine neutrophils, protegrin-1 (PDB accession code: 1PG1). (c) A cartoon version of b, wherein arginines are black, cysteines are gray, and the other residues are identified by single letter code. (d) A similar cartoon of rhesus RTD-1, indicating the similarity in structure with retrocyclin.
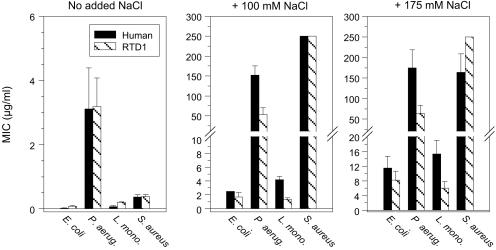
The effects of NaCl on the antimicrobial activity of retrocyclin and RTD-1, from two-stage radial diffusion assays, are compared in Fig. 3. The peptides showed very similar behavior. Neither was hemolytic for human red cells, and retrocyclin showed minimal cytotoxicity for two human cell lines, ME-180 cervical epithelial cells and H9 CD4+ T lymphocytes, even when tested at 100–200 μg/ml (data not shown). Under low salt conditions, both peptides were highly effective (minimal effective concentration < 3 μg/ml) against all four test organisms: Pseudomonas aeruginosa, Escherichia coli, Listeria monocytogenes, and Staphylococcus aureus (Fig. 3). Their strong activity against E. coli and L. monocytogenes persisted in physiological (100 mM) NaCl, and even hypersalinity (175 mM NaCl) was only modestly inhibitory. In contrast, neither peptide was effective (minimal effective concentration > 50 μg/ml) against S. aureus or P. aeruginosa in physiological or high salt concentrations.
Figure 3.
Effect of salt on antibacterial activity of circular minidefensins. Human retrocyclin and monkey RTD-1 were tested against our standard lab stains: E. coli ML-35p, P. aeruginosa MR 3007, L. monocytogenes, EGD, and S. aureus 930918. The bars show minimal effective concentration values ± SE values that resulted from three to six radial diffusion assays per organism and assay condition.
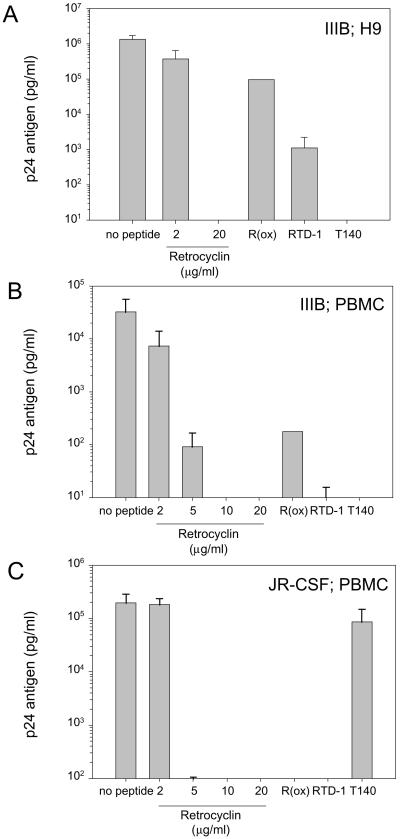
The anti-HIV-1 properties of retrocyclin are shown in Fig. 4. Retrocyclin (10–20 μg/ml) afforded complete protection to CD4+-selected human PBMCs challenged with two different strains of HIV-1: IIIB (T-cell tropic) and JR-CSF (M-tropic), or H9 human T cells challenged with IIIB. The rhesus circular minidefensins RTD-1 (Fig. 4) and RTD-2 and RTD-3 (data not shown) were also protective, but to a lesser extent than retrocyclin. Additionally, the anti-HIV-1 activities of T140 (Fig. 4) and T22 (not shown), analogs of polyphemusins from horseshoe crabs that were previously shown to protect against T-tropic, but not M-tropic, infections (24, 25), were confirmed in the present study. In similar assays, we also determined that the minimal concentration of T140 that affords complete protection against T-tropic HIV-1 was 64-fold less than retrocyclin (day 9 of incubation; n = 3).
Figure 4.
Anti-HIV-1 activity of retrocyclin. Two strains of HIV-1 and two types of human target cells were used. The IIIB strain is T cell-tropic and utilizes the CXCR4 coreceptor for entry; the JR-CSF strain is M-tropic and uses CCR5 for entry. PBMC signifies CD4+-selected peripheral blood mononuclear cells. Results indicate p24 antigen concentration in pg/ml, as determined by quantitative ELISA assay. (A) Two concentrations of retrocyclin (2 μg/ml, 20 μg/ml), 20 μg/ml of the rhesus circular defensin “RTD-1,” 20 μg/ml of an oxidized and linearized variant of retrocyclin [“R(ox)”], and 20 μg/ml of a horseshoe crab-derived peptide “T140,” which previously was reported to prevent only T cell-tropic infections (25), were tested in antiviral assays of against strain IIIB in H9 cells (n = 2–6 per peptide; error bars indicate SEM). (B) To confirm our results with primary human cells, similar assays were performed by using IIIB virus and CD4+ PBMC or (C) JR-CSF virus and CD4+ PBMC (n = 2–4 per peptide; error bars indicate SEM). Assay sensitivity = 10 pg/ml.
Because sexually transmitted HIV-1 stains are typically M-tropic, genetic mutations in humans that eliminate the CCR5 coreceptor afford strong resistance to HIV infection (26). A consequence of the strong selectivity of T140 for the CXCR4 coreceptor is that it affords little or no protection against M-tropic strains of HIV-1. In contrast, retrocyclin is effective against M-tropic as well as T-tropic HIV-1 strains, lending credence to the potential utility of retrocyclin-like agents for prophylaxis.
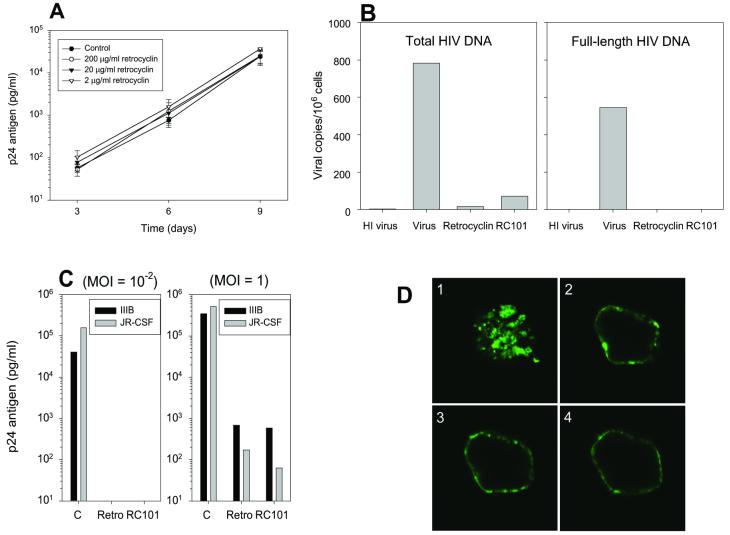
To ascertain whether retrocyclin directly inactivated HIV-1 virions, we incubated HIV-IIIB (moi 10−2) or JR-CSF (data not shown) with 2 μg/ml, 20 μg/ml, or 200 μg/ml retrocyclin in R10–50 media. The mixture was diluted in R10–50 media well below its empirically determined antiviral concentrations, and used to infect CD4+-selected PBMC. Viral replication was measured by collecting supernatant for 9 days at 3-day intervals to quantify HIV-1 p24 antigen by ELISA (Fig. 5A). HIV titer was not reduced even with the highest concentration (200 μg/ml) of retrocyclin, suggesting that retrocyclin does not directly target the virion.
Figure 5.
Mechanism of action of retrocyclin. (A) Retrocyclin (2–200 μg/ml) was incubated with HIV-IIIB (moi = 10−2) diluted in R10–50 media before infecting CD4+-selected PBMC. p24 antigen release was measured by ELISA (n = 3). Adding retrocyclin directly with HIV-1-IIIB does not reduce infection. Limit of detection = 10 pg/ml. (B) Retrocyclin (“Retro”) and RC-101 inhibited the formation of DNA from both early events (total HIV DNA) and later events (full-length HIV DNA) of reverse transcription. Data are an average of two experiments, except for RC-101 (one experiment). “HI virus” is a heat-inactivated virus control for background levels of viral DNA. (C) The p24 titers from day 9 CD4+ PBMC infected with IIIB or JR-CSF at the indicated moi, with or without 20 μg/ml retrocyclin or RC-101, are shown. RC-101 and retrocyclin were comparably active in inhibiting HIV-1 replication at low moi (Left) and higher moi (Right). (D) RC-101BODIPY-FL was detected by confocal fluorescence microscopy in patches on the surface of CD4+-selected PBMC. The focal plane of D1 is at the surface of a representative cell, whereas D2–D4 are representative sections from the middle of the same cell. Note that RC-101 aggregates in patches on the surface of PBMC, suggesting that it inhibits viral fusion or entry. Cell diameter = 15 μm.
Retrocyclin, a circular peptide, lacks free N-terminal or side-chain amine groups, rendering radiolabeling or fluorescent-conjugation somewhat problematic. To overcome this limitation, we synthesized RC-101 (GICRCICGKGICRCICGR). This peptide contained an Arg9→Lys9 substitution (bolded) that preserves the net cationic charge of the peptide while adding the free amino group of lysine's side chain. Circular dichroism confirmed that the CD spectra of RC-101 and retrocyclin were nearly identical (data not shown).
Quantitative, real-time PCR was performed to compare the ability of retrocyclin and RC-101 to block formation of proviral DNA in HIV-JR-CSF-inoculated CD4+-selected PBMC. This method (20) is even more sensitive than measuring p24 release and can detect infection even when p24 values may be affected by virus carried over from the original inoculum. CD4+-selected PBMC were incubated with either HIV-1 strain JR-CSF (moi = 0.1), heat-inactivated virus (background control), JR-CSF + 20 μg/ml retrocyclin, or JR-CSF + 20 μg RC-101. Retrocyclin and RC-101 inhibited the formation of total and full-length HIV-1 proviral DNA (Fig. 5B). Consequently, retrocyclin and RC-101 act very early in the infection process, either by inhibiting reverse transcription or events (e.g., binding, virion uptake, etc) that must precede it. The complete lack of proviral DNA formation with the R-U5 primers suggested that inhibition by retrocyclin occurred at steps before reverse transcription, consistent with in vitro tests with purified HIV-1 reverse transcriptase in a cell-free system (data not shown).
The anti-HIV-1 activity of RC-101 was compared with retrocyclin. Primary CD4+-selected PBMC lymphocytes from HIV-1-seronegative donors were generated from freshly purified peripheral blood mononuclear cells and were infected with HIV-1 in the presence or absence of test peptide. The cells were maintained in R10–50 media for 9 days in the presence of 20 μg/ml test peptide. Similar to retrocyclin, RC-101 afforded complete protection to CD4+-selected PBMC challenged with a low inoculum of HIV-IIIB and HIV-JR-CSF and partial protection with higher inocula (Fig. 5C).
To explore how retrocyclins interacted with cells, we used fluorescence confocal microscopy to examine CD4+-selected PBMC cells that had been incubated with 20 μg/ml RC-101BODIPY-FL (Fig. 5D). RC-101BODIPY-FL bound to the cell membrane, mostly concentrated in patches, and was not internalized. Patching, or “microaggregation,” has been reported to occur with hormone-occupied epidermal growth factor receptors (27), and “rafts” are involved in signaling through the selective confinement of chemokine receptors to discrete regions of the cell membrane (reviewed in ref. 28). Studies are underway to identify the precise molecular target(s) of retrocyclin.
In summary, we used a forensic approach to recreate a “dead” human peptide (retrocyclin) from its remains, an expressed pseudogene. That the antimicrobial properties of retrocyclin closely resembled those of rhesus circular minidefensins (θ defensins) was not very surprising. However, the ability of retrocyclin to prevent infection by HIV-1 was both remarkable and tinged with irony. Although it is not possible to know whether the evolutionary loss of retrocyclin contributed to the susceptibility of humans to HIV-1 infection, retrocyclin is a promising lead for designing agents (e.g., topical microbicides) that can prevent human HIV infections.
Acknowledgments
A.M.C. thanks Dr. Tomas Ganz for his continued guidance and support. We thank Mark Sherman (Beckman Research Institute, City of Hope, Duarte, CA) for help with molecular modeling; Dr. Matthew Schibler for assistance with confocal imaging; and Hwee Ng for her expert technical assistance. This work was supported by grants AI 22839 and AI 37945 (to R.I.L.) and AI 43202 (to O.O.Y.) from the National Institutes of Health, by a grant from the University of California Universitywide AIDS Research Program (to A.M.C. and O.O.Y.), and by a postdoctoral training grant from the American Lung Association (to A.M.C).
Abbreviations
- moi
multiplicity of infection
- PBMC
peripheral blood mononuclear cell
- LTR
long-term repeat region
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF355799).
References
- 1.Lehrer R I, Lichtenstein A K, Ganz T. Annu Rev Immunol. 1993;11:105–128. doi: 10.1146/annurev.iy.11.040193.000541. [DOI] [PubMed] [Google Scholar]
- 2.Schonwetter B S, Stolzenberg E D, Zasloff M A. Science. 1995;267:1645–1648. doi: 10.1126/science.7886453. [DOI] [PubMed] [Google Scholar]
- 3.Schroder J M. Cell Mol Life Sci. 1999;56:32–46. doi: 10.1007/s000180050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harwig S S, Swiderek K M, Kokryakov V N, Tan L, Lee T D, Panyutich E A, Aleshina G M, Shamova O V, Lehrer R I. FEBS Lett. 1994;342:281–285. doi: 10.1016/0014-5793(94)80517-2. [DOI] [PubMed] [Google Scholar]
- 5.Tang Y Q, Yuan J, Osapay G, Osapay K, Tran D, Miller C J, Ouellette A J, Selsted M E. Science. 1999;286:498–502. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- 6.Zhao C, Nguyen T, Liu L, Sacco R E, Brogden K A, Lehrer R I. Infect Immun. 2001;69:2684–2691. doi: 10.1128/IAI.69.4.2684-2691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann G R, Legault P, Selsted M E, Pardi A. Biochemistry. 1995;34:13663–13671. doi: 10.1021/bi00041a048. [DOI] [PubMed] [Google Scholar]
- 8.Liu L, Zhao C, Heng H H Q, Ganz T. Genomics. 1997;43:316–320. doi: 10.1006/geno.1997.4801. [DOI] [PubMed] [Google Scholar]
- 9.Leonova L, Kokryakov V N, Aleshina G M, Hong T, Nguyen T, Zhao C, Waring A J, Lehrer R I. J Leukocyte Biol. 2001;70:461–464. [PubMed] [Google Scholar]
- 10.Tran, D., Tran, P. A., Tang, Y. Q., Yuan, J., Cole, T. & Selsted, M. E. (October 23, 2001) J. Biol. Chem., 10.1074/jbc.M109117200.
- 11.Harwig S S, Waring A, Yang H J, Cho Y, Tan L, Lehrer R I. Eur J Biochem. 1996;240:352–357. doi: 10.1111/j.1432-1033.1996.0352h.x. [DOI] [PubMed] [Google Scholar]
- 12.Steinberg D A, Lehrer R I. Methods Mol Biol. 1997;78:169–186. doi: 10.1385/0-89603-408-9:169. [DOI] [PubMed] [Google Scholar]
- 13.Wilson C C, Wong J T, Girard D D, Merrill D P, Dynan M, An D D, Kalams S A, Johnson R P, Hirsch M S, D'Aquila R T. J Infect Dis. 1995;172:88–96. doi: 10.1093/infdis/172.1.88. [DOI] [PubMed] [Google Scholar]
- 14.Wong J K, Hezareh M, Gunthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 15.Yang O O, Tran A C, Kalams S A, Johnson R P, Roberts M R, Walker B D. Proc Natl Acad Sci USA. 1997;94:11478–11483. doi: 10.1073/pnas.94.21.11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salter R D, Cresswell P. EMBO J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 18.Yang O O, Swanberg S L, Lu Z, Dziejman M, McCoy J, Luster A D, Walker B D, Herrmann S H. J Virol. 1999;73:4582–4589. doi: 10.1128/jvi.73.6.4582-4589.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole S W, Naliboff B D, Kemeny M E, Griswold M P, Fahey J L, Zack J A. Proc Natl Acad Sci USA. 2001;98:12695–12700. doi: 10.1073/pnas.221134198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorry P, Bristol G, Zack J, Ritola K, Swanstrom R, Birch C, Bell J, Bannert N, Crawford K, Wang H, et al. J Virol. 2001;75:10073–10089. doi: 10.1128/JVI.75.21.10073-10089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 22.Oishi O, Yamashita S, Nishimoto E, Lee S, Sugihara G, Ohno M. Biochemistry. 1997;36:4352–4359. doi: 10.1021/bi962171f. [DOI] [PubMed] [Google Scholar]
- 23.Tam J P, Lu Y A, Yang J L. Biochemistry. 2000;39:7159–7169. doi: 10.1021/bi0003487. [DOI] [PubMed] [Google Scholar]
- 24.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, et al. J Exp Med. 1997;186:1389–1393. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamamura H, Xu Y, Hattori T, Zhang X, Arakaki R, Kanbara K, Omagari A, Otaka A, Ibuka T, Yamamoto N, et al. Biochem Biophys Res Commun. 1998;253:877–882. doi: 10.1006/bbrc.1998.9871. [DOI] [PubMed] [Google Scholar]
- 26.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 27.Zidovetzki R, Yarden Y, Schlessinger J, Jovin T M. EMBO J. 1986;5:247–250. doi: 10.1002/j.1460-2075.1986.tb04205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manes S, Lacalle R A, Gomez-Mouton C, del Real G, Mira E, Martinez A. Semin Immunol. 2001;13:147–157. doi: 10.1006/smim.2000.0306. [DOI] [PubMed] [Google Scholar]