Abstract
The mechanism of stop codon recognition during translation has long been a puzzle. Only recently has it been established that a tripeptide in the bacterial release factors (RFs) 1 and 2 serves as the “anticodon” in deciphering stop codons in mRNA. However, the molecular basis of the accuracy of stop codon recognition is unknown. Although specific tripeptides in the RFs are primarily responsible for selective reading of cognate stop codons, charge-flip variant RF proteins, altered at conserved Glu residues adjacent to the tripeptide-anticodon, are shown here to be crucial to codon recognition. Changes of these Glu residues are capable of triggering polypeptide release at noncognate stop codons and also at sense codons. These changes also reverse the growth inhibition by RFs containing “harmful” tripeptide-anticodon changes. These findings suggest that electrostatic interactions involving negative charges in domain C of the RFs mediate their accurate docking in the ribosome. Our results also establish that the charge flipping creates a phenotype/translation termination by “codon bypassing” via relaxed positioning of the RF tripeptide-anticodon in the decoding pocket of the ribosome.
Since the deciphering of the genetic code in the early 1960s, it has remained uncertain how the three stop codons, UAG, UGA, and UAA, are recognized and how the ribosome terminates translation. The termination of protein synthesis takes place on the ribosomes in response to a stop codon rather than a sense codon in the “decoding” site (A site). Polypeptide release factors (RFs) are essential to this process and are speculated to possess a tRNA-like property, because they are likely to decipher the genetic code instead of tRNA (1, 2). Prokaryotes generally have two codon-specific factors, RF1 (for UAG/UAA) and RF2 (for UGA/UAA), whereas eukaryotes have only one factor, eRF1, which normally recognizes all three stop codons (3).
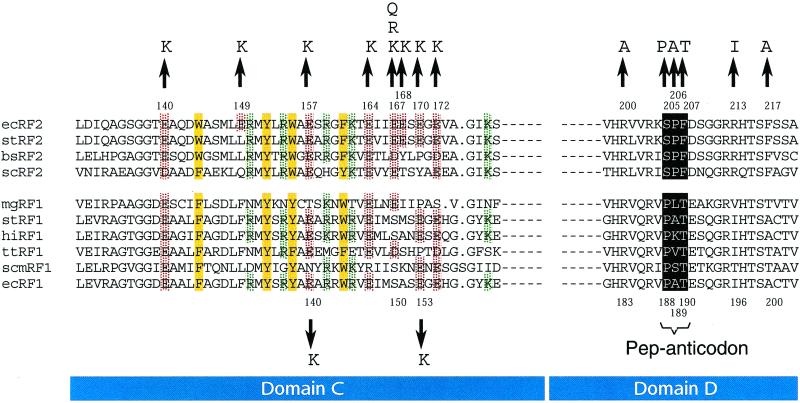
The discovery that two bacterial RFs share conserved regions led to a path that may reveal how RFs are involved in decoding stop codons (1). This insight led to a seven-domain model of RF in which the domains are designated A–G and to a hypothesis of “molecular mimicry” between RF and tRNA (4). Domain D of RFs share (partial) sequence homology with the C-terminal portion, domain IV, of elongation factor G, the crystal structure of which mimics the shape of the anticodon helix of tRNA (5). It therefore would seem that domain D of RF constitutes a “tRNA-mimicry” domain necessary for RF binding to the ribosomal A site, leading to the proposal that RFs contain an anticodon-mimicry element. This prediction was confirmed recently by the discovery of a tripeptide determinant responsible for stop codon recognition (6): RF1 and RF2 are distinguished functionally by the tripeptides Pro-(Ala)-Thr and Ser-Pro-Phe, respectively. These tripeptides discriminate between the second and third bases of the mRNA stop codon (Fig. 1). Thus, they functionally represent a tripeptide “anticodon” that deciphers stop codons in mRNA (hereafter referred to as a pep-anticodon; ref. 7). The site on the terminating ribosome for binding to this functionally assigned tripeptide-anticodon of RF1 has been mapped at the decoding site of the 30S subunit by directed hydroxyl radical probing (8).
Figure 1.
Amino acid changes in the tripeptide-anticodon and Glu residues conserved in domain C of prokaryotic RFs. The similarity alignments of RFs were performed as described (4). Conserved acidic and basic residues in domain C are marked in red and green, respectively, and conserved aromatic residues are marked in yellow. The tripeptide-anticodons are shown by outline characters. The numbers refer to the amino acid position counting from the N-terminal Met of Escherichia coli RF1 (Lower) and RF2 (Upper). The arrows show relevant substitutions used in this study. The species abbreviations are: ec, E. coli; st, Salmonella typhimurium; bs, Bacillus subtilis; sc, Streptomyces coelicolor; mg, Mycoplasma genitalium; hi, Haemophilus influenzae; tt, Thermus thermophilus; and scm, Saccharomyces cerevisiae mitochondria.
The fidelity of stop codon recognition by RF1 and RF2 during translation is extremely high (i.e., error rate <10−5). In addition to the recognition of two stop codons each, RF1 and RF2 must discriminate against the 61 sense codons of the genetic code. Unlike the elongation process, GTP energy-driven proofreading is not required for termination (9), suggesting that the high fidelity of translation termination is governed by precise protein-RNA interactions. This remarkable specificity of RFs must be based on a highly complex A-site design that holds both the protein and mRNA in sterically well defined positions (10). In other words, the accurate recognition of stop codons by the pep-anticodon would not be possible without specific positional interactions between RF domain(s) around the pep-anticodon and docking site(s) of the ribosome.
We found previously that variant RF2 proteins containing tripeptide-anticodon changes cause growth inhibition (6). To identify RF domain(s) other than the pep-anticodon that contribute to the accuracy of stop codon recognition, we isolated additional mutations in bacterial RF2 that reverse the growth inhibition exhibited by these variant RF2 proteins. These studies revealed that multiple Glu residues conserved in domain C are crucial for the accurate reading of stop codons by RFs and that their charge-flip (i.e., negative-to-positive) variants trigger polypeptide termination at noncognate stop codons and also at sense codons. This phenotype, which leads to polypeptide termination without the cognate-codon recognition, is designated here as “codon bypassing.”
Materials and Methods
Strains and Manipulations.
The E. coli K-12 strains used were W3110 and its derivatives, RM789A (prfA∷KmR; refs. 6 and 11), RM789B (prfB∷CmR; ref. 11), and RM718 [prfB286 (temperature-sensitive) recA∷Tn10; ref. 12]. RM789A and RM789B were RF1 and RF2 knockout strains, respectively, and carried plasmid pSUIQT-RF2* (11) as a maintainer plasmid. pSUIQT-RF2* is a lac promoter-controlled expression plasmid of an RF2 variant, RF2*, that is able to recognize UAA/UGA/UAG; hence it complements the single or double RF1/RF2 knockout alleles in the presence of isopropyl-1-thio-β-d-galactoside (IPTG).
Plasmids.
The parental plasmid, pSUIQ-RF2, bearing the E. coli RF2 gene under the control of the lac promoter was described previously (13). Most of the RF2 constructs except for those used for the in vivo interference test and suppressor selection substituted Ala for Thr-246. Pep-anticodon variants of pSUIQ-RF2 were constructed by site-directed mutagenesis of the pep-anticodon sequence using designed primers containing the substitutions (see Fig. 1) according to standard procedures (14). Throughout this work, plasmid variants were named with the variant alleles following the parental plasmid names (e.g., pSUIQ-RF2-F207T or pBR322-RF2-E167K). Plasmids pEF-UGA and pEF-UCA5 (used for an in vitro S-30 translation assay) are pET30a(+) (Novagen) derivatives carrying the 3A′ gene sequence, the NcoI–ClaI segment from pAB94 (15), and additional UGA- and UCA-pentatriplet sequences inserted into a linker between the segments coding for the second and third A domains. The first A domain is fused in its N terminus with the 26-kDa protein glutathione S-transferase for the better resolution of products by SDS/PAGE.
Mutagenesis of RF2 and Selection of Suppressors.
The pSUIQ-RF2-F207T DNA was mutagenized with hydroxylamine as described (11). A temperature-sensitive lethal RF2 mutant of E. coli (RM718) was transformed with the mutagenized DNA, and temperature-resistant chloramphenicol-resistant (CmR) colonies were selected at 42°C on YT agar plates containing 1 mM IPTG. Plasmid DNAs were recovered from these survivors and retransformed into the same parental strain, and those that gave a reproducible phenotype were characterized further.
Protein Purification and Polypeptide Release Assay.
Histidine-tagged RF genes were cloned downstream of a T7 RNA polymerase promoter in plasmid pET30a (Novagen), and proteins were purified after overexpression in E. coli through affinity (nickel-nitrilotriacetic acid agarose, Qiagen, Chatsworth, CA) and Mono Q column (ÄKTA, Amersham Pharmacia) as described (11, 13). The ability of purified RF proteins (note that RF2 substituted Ala for Thr-246) to terminate protein synthesis was monitored by the rate of N-formylmethionine (fMet) release at the stop codon from the in vitro termination complex composed of f[3H]Met-tRNAfMet, a 9-mer minimessenger, and the ribosome isolated from E. coli strain MRE600 essentially as described (11). The relative specificity value was calculated according to the following formula: relative specificity = (fMet-release activity of 1 unit of RF at a noncognate codon)/(fMet-release activity of 1 unit of RF at a cognate codon).
S-30 Termination Assay Using the 3A′ Gene.
Template DNAs containing the 3A′ gene and its 3′ and 5′ flanking sequences were amplified from pEF-UGA and pEF-UCA5 by PCR using the primers 5′-GGTGCATGCAAGGAGATGG-3′ and 5′-CAGCGATCGCGTATTTCG-3′. The 3A′ transcripts were synthesized in an in vitro transcription reaction by using the amplified DNA templates and T7 RNA polymerase (TaKaRa) according to the manufacturer's instructions. These 3A′ transcripts were added to the in vitro translation reaction containing E. coli S-30 extract system for circular DNA (Promega) and redivue ProMix l-[35S] in vitro cell-labeling mix (Amersham Pharmacia), and the reaction mix was incubated at 37°C for 30 min according to the manufacturer's instructions. The 35S-labeled 3A′ and 2A′ proteins were subjected to SDS/PAGE, and the read-through (RT) value was calculated as described (13).
Results
Growth Interference by Pep-Anticodon Variant RF2.
We have shown previously that swapping of the tripeptide-anticodon residues between native RF1 and RF2 sequences leads to inactivation of the RF activity (6). Here we examined systematically how single amino acid substitutions between the pep-anticodons affect the RF activity. Three residues of the pep-anticodon of RF2 were changed individually to the counterpart residues of RF1 (i.e., S205P, P206A, and F207T; see Fig. 1). These variant RF2 genes were cloned into a lac promoter-controlled expression plasmid pSUIQ (13) and transformed to wild-type E. coli cells (W3110). The resulting transformants grew well in the absence of IPTG but failed to grow in the presence of IPTG (Table 1), showing that the pep-anticodon variant RF2 interfered with cell growth. The same growth interference also was observed after expression of variant RF2 proteins containing substitutions of amino acids conserved in RF1 for those in RF2 such as R200A, R213I, and F217A (see Fig. 1) in the vicinity of the pep-anticodon (Table 1). These results suggest that amino acids surrounding the pep-anticodon are required also for the termination fidelity.
Table 1.
Growth interference and suppression by mutations in the tripeptide anticodon and domain C of E. coli RF2
| Pep-anticodon change | IPTG | Effect on cell growth of domain C change
|
||||
|---|---|---|---|---|---|---|
| Wild type | E149K | E157K | E167K | E170K | ||
| Wild type | − | ++ | ++ | ++ | ++ | ++ |
| + | ++ | ++ | ++ | ++ | ++ | |
| R200A | − | ++ | ++ | ++ | ++ | ++ |
| + | − | ++ | + | + | + | |
| S205P | − | ++ | ++ | ++ | ++ | ++ |
| + | − | ++ | ++ | ++ | ++ | |
| P206A | − | ++ | ++ | ++ | ++ | ++ |
| + | − | ++ | ++ | ++ | ++ | |
| F207T | − | ++ | ++ | ++ | ++ | ++ |
| + | − | ++ | ++ | ++ | ++ | |
| R213I | − | ++ | ++ | ++ | ++ | ++ |
| + | − | ++ | ++ | ++ | ++ | |
| F217A | − | ++ | ++ | ++ | ++ | ++ |
| + | − | ++ | ++ | ++ | ++ | |
The variant RF2 genes containing the indicated changes in the tripeptide anticodon and domain C were cloned in a lac promoter-controlled expression plasmid (pSUIQ) and transformed into wild-type E. coli (W3110). The growth of CmR transformants was monitored at 32°C on YT agar plates (1% bacto-tryptone, 0.1% yeast extract, and 0.25% NaCl) in the presence (i.e., expression of variant RF2 proteins) or absence (i.e., no expression of variant RF2 proteins) of 1 mM IPTG. ++, normal growth; +, slow growth; −, no growth (i.e., growth interference).
E. coli RF2 Specificity for Growth Interference.
Unlike RF2, changing pep-anticodon residues of RF1 to those of RF2 resulted only in the inactivation of RF1; no growth inhibition was exhibited (data not shown). The RF2 protein of the E. coli K-12 strain terminates translation very weakly, and its overexpression interferes with cell growth because of its abnormal Thr residue at position 246 (13, 16, 17), which is close to the universally conserved “GGQ” motif (18), a site for N5-methylation (16). Therefore, we assume that an inaccurate (and slower) decoding of cognate stop codons by the pep-anticodon variants of RF2, in concert with the inefficient catalytic property of E. coli RF2, at least in part via Thr246, may be the basis of growth interference.
Given this assumption is correct, growth interference might be alleviated if the pep-anticodon variant RF2 is altered by an additional mutation that negates detrimental codon recognition. One such known candidate is an RF2 variant, RF2*, that carries a single Glu → Lys change at position 167 (E167K) and terminates translation at all three stop codons (i.e., omnipotent phenotype; ref. 11). This prediction was confirmed when the E167K change was combined with any of the pep-anticodon substitutions; the double-variant RF2 proteins did not interfere with cell growth (data not shown). These findings suggested that the growth interference induced by the variant RF2 protein is caused by an abnormal or inaccurate interaction between the variant pep-anticodon and cognate or noncognate (stop) codons. Our results also establish that RF2* shows an omnipotent phenotype via “loss-of-specificity” of the pep-anticodon by the E167K substitution, not via “gain-of-specificity,” resulting in infidelity of translation termination.
Selection of Compensatory Alterations for Growth Interference.
The above findings of suppression by the E167K change in RF2 prompted us to systematically select and score for potential additional mutations that reverse the growth inhibition by the variant RF2 proteins containing “harmful” tripeptide-anticodon changes. A plasmid pSUIQ derivative encoding the variant RF2 containing the tripeptide-anticodon change F207T was mutagenized in vitro with hydroxylamine and transformed into a temperature-sensitive RF2 strain, RM718 (prfB286). Because hydroxylamine generates both C → T and G → A substitutions, the Thr codon (ACU) in the variant pep-anticodon was unable to revert to either wild-type Phe codon (UUU and UUC). Thus, only additional mutations and not true reversions were selected for. Temperature-resistant and CmR colonies were selected in the presence of IPTG at 42°C and probably harbored additional mutations that rendered the variant pep-anticodon harmless. Plasmid DNAs that gave a reproducible phenotype (i.e., normal growth and RF2 complementation in the presence of IPTG) were thought to harbor secondary suppressor mutations. Seven such plasmids were characterized further by sequence analysis. Interestingly, they sustained the same Glu → Lys changes, similar to E167K, at three distinct positions, 149 (E149K in triplicate), 157 (E157K in single), and 170 (E170K in triplicate), in addition to the F207T change.
These mutations then were monitored for suppression of growth interference caused by the other two variant RF2 proteins harboring the P206A and S205P mutations. Any of these Glu → Lys changes, once combined with P206A and S205P, allowed transformants to grow normally (Table 1), which indicates that the variant RF2 proteins harboring the additional suppressor mutation indeed are innocuous. Therefore, it is likely that negative-to-positive electrostatic alterations, even at a distance of 40–50 amino acids, are able to reverse or negate the abnormal or inaccurate interaction between the variant pep-anticodons of RF2 and the cognate or noncognate (stop) codons in mRNA.
Charge-Flip Changes in Domain C.
The compensatory mutations were localized in domain C adjacent to the pep-anticodon segment, domain D. Domain C is rich in positive and negative charges regularly spaced by aromatic residues, a feature that is well conserved in prokaryotic RF1 and RF2 proteins (see Fig. 1). This unique and conservative structure led us to speculate that domain C might interact with negative or positive charges in the terminating ribosomal complex such as at the site of rRNA or ribosomal proteins and might play a vital role in allowing the accurate interaction between the pep-anticodon and stop codons within the A site of the ribosome. To understand the significance of negative charge residues in domain C and their charge-flip changes better, five other Glu residues at positions 140, 164, 168, 172, and 195 were changed to Lys. First, these substitutions were examined for their capacity to reverse the growth inhibition by the harmful tripeptide-anticodon changes. Three of them, E164K, E168K, and E172K, were able to reverse the growth interference by the F207T or P206A mutant, whereas the other two, E140K and E195K, failed to do so (Table 2). Second, these single Glu → Lys variants of E. coli RF2 were examined for their capacity to complement the RF2 knockout strain RM789B (prfB∷CmR; ref. 11) by plasmid transformation. Again, plasmids carrying the same three variants, E164K, E168K, and E172K, allowed RF2 knockout cells to grow, whereas the other two did not (Table 2). These results suggest that Glu-140 and Glu-195, the charge-flipping of which simply nullified the RF2 activity, are separated functionally from the other seven Glu residues between positions 149 and 172, which probably constitute a common functional unit to influence the pep-anticodon function.
Table 2.
Complementation of RF1/RF2 knockout strains by domain C variant RF2 proteins
| Domain C variants of RF2 | Suppression of pep-anticodon mutants† | Complementation
|
|
|---|---|---|---|
| prfA∷KmR | prfB∷CmR | ||
| Wild type | No | − | ++ |
| E140K | No | − | − |
| E149K | Yes | − | ++ |
| E157K | Yes | + | ++ |
| E164K | Yes | − | ++ |
| E167K | Yes | + | ++ |
| E167R | Yes | + | ++ |
| E167Q | Yes | − | ++ |
| E168K | Yes | − | ++ |
| E170K | Yes | − | ++ |
| E172K | Yes | ++ | ++ |
| E195K | No | − | − |
| E149K E167K | Yes | ++ | ++ |
| E149K E170K | Yes | + | ++ |
The RF1 knockout (RM789A prfA∷KmR) and RF2 knockout (RM789B prfB∷CmR) test strains contained pSUIQT-RF2* (11) as a maintainer plasmid. Hence, they grow only in the presence of IPTG. Complementation tests were carried out by transformation of these RF1 and RF2 mutants with pBR322 plasmids carrying the indicated charge-flip variant RF2 genes (without the pep-anticodon changes). Transformants (containing the two compatible plasmids) were selected at 37°C on LB agar plates supplemented with 1 mM IPTG and examined for their growth in the absence of IPTG. ++, normal growth (i.e., complementation); +, slow growth (i.e., weak complementation); −, no growth (i.e., no complementation).
Suppression of growth interference was monitored using pSUIQ-RF2 variants containing the indicated changes in domain C and either pep-anticodon change, P206A, or F207T, as described in Table 1. Yes, normal growth of transformants in the presence of IPTG; No, no growth of transformants in the presence of IPTG.
If the reduction in negative charges in domain C is required to reverse the growth interference, we might be able to substitute Arg or Gln for Lys. Because hydroxylamine mutagenesis only allows the Glu → Lys change, Arg and Gln substitutions for Glu-167 (E167R, E167Q) were manipulated by site-directed mutagenesis and examined. The data indicated that both changes retained the RF2-complementing activity and negated the F207T-generated growth interference (Table 2). E167Q showed a slightly reduced ability to alleviate the RF1 defect compared with E167K and E167R (see below).
Omnipotence Acquired by Glu → Lys Changes in Domain C.
We have shown previously that the E167K-variant RF2 protein terminates translation at all three stop codons (11). Hence, we examined whether the newly isolated variant RF2 proteins respond to all three stop codons in the in vivo complementation test by using an RF1 mutant. First, the chromosomal copy of RF1 was nullified by the insertion of the KmR gene cassette (prfA∷KmR; refs. 6 and 11) in a strain harboring a second plasmid-based functional copy of omnipotent RF2* under the control of the lac promoter. Hence, the resulting strain RM789A is only viable in the presence of IPTG. This test strain was transformed with pBR322 derivatives encoding the Glu → Lys variant RF2 proteins and monitored for the growth of transformants. Variant RF2 proteins containing E157K, E172K, and E167K restored the growth of RM789A in the absence of IPTG, showing that these three variants acquired the omnipotent ability to terminate translation at three stop codons. On the other hand, the other four Glu → Lys single variants at positions 149, 164, 168, and 170 did not (Table 2). Under these conditions, the cellular level of these variant RF2 proteins did not alter appreciably by Western blotting (data not shown). However, these seemingly “silent” changes seem to possess potential to alleviate the RF defect because the E149K change enhanced the growth of the RF1 knockout strain when combined with another silent E170K change or with E167K (Table 2). It is noteworthy also that E167R conferred the same complementation activity as E167K on RF2, but E167Q failed to complement the RF1 knockout strain even though it reversed growth inhibition (Table 2), showing that the phenotype depends on the degree of electrostatic charge. These results are interpreted as indicating that Glu → Lys charge-flip changes in domain C reduced the selectivity or accuracy of RF2 in deciphering the stop codons, thereby creating a “phenotypically” omnipotent RF variant.
Codon Bypassing by Variant RF2.
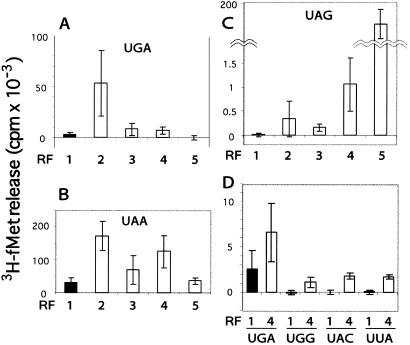
Next, wild-type and three variant RF2 proteins containing E149K, E170K, and the double E149K-E170K substitutions were purified, and their activities in vitro were monitored by f[3H]Met release from ribosomes complexed with f[3H]Met-tRNAfMet and a 9-mer minimessenger RNA (5′-UUC AUG-3′, followed by stop or test triplets). Compared with wild type, the three variant RF2 proteins responded more efficiently to the cognate stop codons UGA and UAA, with the E149K single mutant showing the greatest enhancement of termination (Fig. 2 A and B). The same set of variant RF2 proteins showed a drastic increase in peptide termination even at the noncognate UAG stop codon (Fig. 2C). It is noteworthy that the E149K-E170K double mutant RF2 responded differently to UGA and UAG, probably reflecting that native RF2 is designed to be responsive to UGA but not to UAG. The enhanced termination was seen not only at the noncognate stop codon, UAG, but also at sense codons. The E149K-E170K variant RF2 significantly enhanced fMet release at UGG (Trp), UAC (Tyr), and UUA (Leu) codons in vitro (Fig. 2D) as well as at UCA (Ser) and CAA (Gln) codons (data not shown).
Figure 2.
Polypeptide release activity of the charge-flip variant RF2 proteins at cognate and noncognate codons. After addition of the same molar amount of each variant RF protein, f[3H]Met release from the (f[3H]Met-tRNAfMet-minimessenger-ribosome) complex was determined. Minimessenger RNAs used in these reactions were 9-mer sequences consisting of 5′-UUC AUG-3′ followed by stop (or test) triplets. RF samples: 1, wild-type RF2; 2, E149K variant RF2; 3, E170K variant RF2; 4, E149K E170K variant RF2; 5, wild-type RF1. Experiments were performed independently at least three times, and the mean values are expressed. (A) fMet release at UGA. (B) fMet release at UAA. (C) fMet release at UAG (to magnify release at UAG, the specific activity of f[3H]Met-tRNAfMet used in this assay was 30 times higher than that used for UGA and UAA). (D) fMet release at UGG, UAC, and UUA sense codons.
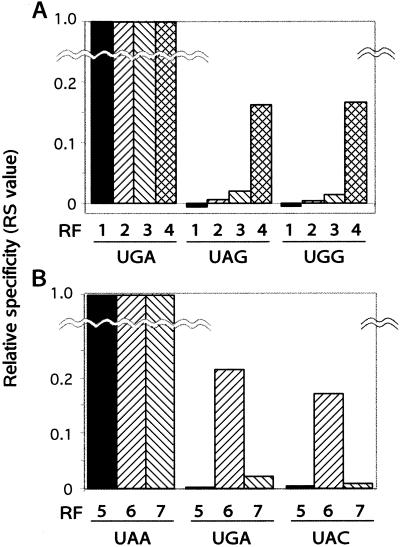
Because the increase in the termination at cognate stop codons was unexpected, we estimated the “altered” frequency of termination at noncognate codons relative to the cognate codon as relative specificity values. Fig. 3A demonstrates such a normalized estimation of the degree to which variant RF2 proteins terminated translation at noncognate UAG (stop) and UGG (sense) codons. Each Glu → Lys change was remarkably error-prone in decoding both noncognate codons, and the E149K-E170K double mutant erred by decoding these noncognate codons at one-sixth the level of the cognate UGA codon. These results led us to conclude that Glu → Lys changes in domain C confer on RF2 the capacity of triggering peptidyl-tRNA hydrolysis by bypassing the decoding step or, less likely, by recognizing more strongly not only stop codons but also sense codons in the A site of the ribosome.
Figure 3.
Codon bypassing by the charge-flip variant RF proteins. In vitro peptide release activity of charge-flip variants of RF2 (A), as shown in Fig. 2, and RF1 (B) were used to estimate the relative specificity (RS) value representing the relative termination frequency at each indicated codon compared with the relevant cognate codon (i.e., UGA for RF2 variants and UAA for RF1 variants). RF samples: 1, wild-type RF2; 2, E149K variant RF2; 3, E170K variant RF2; 4, E149K E170K variant RF2; 5, wild-type RF1; 6, E140K variant RF1; 7, E153K variant RF1.
Codon Bypassing in an in Vitro S-30 System.
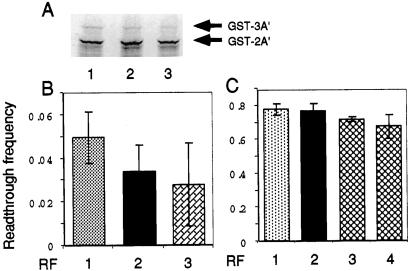
Evidence for the increased peptide termination by charge-flip variant RF2 was demonstrated also in an in vitro S-30 translation system by using a gene that codes for three identical engineered antibody-binding B domains of protein A from Staphylococcus aureus (15, 19). The UGA and UCA sequences were inserted into a linker between the segments coding for the second and third identical IgG binding domains (i.e., A domains) of plasmid pAB94 (15). (The 3A′ and glutathione S-transferase coding sequences contain only a single UCA codon.) The predicted influence of Glu → Lys variant RF2 on RT of UGA was monitored by the appearance of the three-domain (3A′) protein (i.e., an RT product) vs. the two-domain (2A′) protein (i.e., a terminated product). As shown in Fig. 4 A and B, the RT of UGA was reduced after the addition of 1 pmol of wild-type RF2 and reduced further after the addition of 0.5 pmol of E149K-E170K variant RF2. Similarly, the E149K-E170K variant RF2 gave rise to a small but significant (i.e., reproducible) decrease in RT translation of the UAG codon (data not shown) and the stretch of UCA codons (Fig. 4C), probably reflecting the weak activity of the variant RF2 to compete with RF1 and tRNAs at the cognate codons.
Figure 4.
S-30 translation assay of codon bypassing by the charge-flip variant RF2 proteins. (A) 3A′ and 2A′ products synthesized in an in vitro S-30 translation system. The 3A′ mRNA containing an insert of the UGA codon at the junction between the second and third A domains was synthesized in vitro and added into the in vitro translation reaction in the presence of wild-type and variant RF2 proteins. 35S-labeled glutathione S-transferase (GST)-3A′ and -2A′ products were analyzed by SDS/PAGE. RF samples: 1, none; 2, wild-type RF2 (1 pmol); 3, E140K-E170K variant RF2 (0.5 pmol). (B) Influence on UGA RT of the variant RF2. RT values, i.e., molar amounts of 3A′ domain protein (translation RT) relative to 2A′ domain protein (translation termination), were calculated as described (13) by using the quantified intensities shown in A. RF samples are the same as those in A. Experiments were performed independently at least five times, and the values are expressed with standard deviations. (C) Influence on UCA-pentatriplet RT of the variant RF2. Experimental conditions and procedures were the same as described for A and B except that the UCA-pentatriplet sequence was inserted at the second- and third-domain junction of the 3A′ mRNA. RF samples were the same as for A except for sample 4, E140K-E170K variant RF2 (1 pmol).
Codon Bypassing by Charge-Flip Variant RF1.
Charged amino acids in domain C are highly conserved not only in RF2 but also in RF1 (see Fig. 1). Two variant RF1 proteins containing E140K and E153K changes, equivalent to E157K and E170K in RF2, were generated by site-directed mutagenesis (see Fig. 1). These variant RF1 proteins acquired the ability to terminate translation at noncognate stop (UGA) codons as well as at sense (UAC) codons in the in vitro release assay (Fig. 3B). Notably, the E140K-variant RF1 showed 20% peptide release activity at noncognate codons compared with the cognate UAA codon. The E153K variant showed the similar but less profound effect. These results pointed out that Glu → Lys charge-flip changes in domain C generally reduce the selectivity of cognate stop codons, thereby triggering peptidyl-tRNA hydrolysis via bypassing the decoding step for the cognate or noncognate codons in the A site of the ribosome.
Discussion
Similar to the sense codon decoding within the ribosome, the stop codon recognition is designed to be extremely accurate. But unlike the sense codon decoding, stop codon recognition does not require any energy-driven proofreading, at least not in the bacterial system (9). This means that the architecture of the ribosomal decoding pocket is so precise that it dictates highly accurate interaction between the pep-anticodon of RF and the stop codon of mRNA. In this study, we found that the accuracy of stop codon recognition is impaired severely by several Glu → Lys charge-flip changes in domain C of RF. To our surprise, these changes do not impair the peptidyl-tRNA hydrolysis but rather enhance or (newly) induce the hydrolysis at cognate stop codons and also at noncognate stop codons and sense codons. These results are well explained by assuming that the variant RFs can bypass the decoding step for any codons at the A site of the ribosome and trigger peptidyl-tRNA hydrolysis in the peptidyltransferase center of the 50S subunit. The drastic increase in peptide release at cognate stop codons can be accounted for by this bypassing, because the stop codon decoding step is rate-limiting.
All the relevant Glu → Lys changes are localized in domain C, >40 amino acids apart from the tripeptide-anticodon, and charge-flip variant RF1 and RF2 proteins appear to reduce the accuracy in quite a general fashion. By these changes, the ribosome-RF complex is altered to terminate translation even at sense codons in vitro 15–20% as efficiently as the termination at cognate stop codons (see Fig. 3), showing that the Glu → Lys variant is a loss-of-specificity variant. Therefore, we assume that the growth interference by the pep-anticodon change of E. coli RF2 could be attributed to an abnormal harmful interaction, not a simple loss of interaction, between the pep-anticodon and stop codons and that Glu → Lys secondary changes in domain C rendered this interaction harmless, at a distance of 40–50 amino acids, through bypassing stop codon recognition. The growth interference by the pep-anticodon changes is specific to E. coli K-12 RF2 and thus at least in part is caused by Thr-246. Wilson et al. (8) have pointed out that position 229 of RF1, equivalent to position 246 of RF2, is located at the peptidyltransferase center of 23S rRNA within the terminating ribosomal complex. Therefore, although the biological meaning of the exceptional Thr is not immediately obvious, we speculate that the inactive or interfering amino acid(s) at position 246, in conjunction with unusual methylation of Gln-252 (16), induces disordered topology and interferes with the normal peptidyl-tRNA hydrolysis.
E. coli grows normally after expression from plasmids of any charge-flip variant RF2 proteins. This seems to be inconsistent with the view that sense codon termination should be harmful to cells. This apparent conflict can be accounted for by assuming either that variant RF proteins cannot efficiently compete at any sense codons with the cognate tRNAs because of much lower affinity to the ribosome or that the synthesis of RF2 is regulated by autogenous +1 frameshift control at the internal UGA signal (20). Of these two possibilities, the former is more likely, because the sense codon termination associated with the domain C variants was evident in an in vitro fMet release assay but less evident in a S-30 system (compare Figs. 3 and 4).
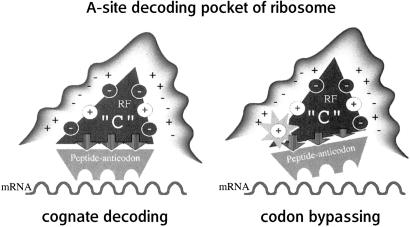
The conservative multiple-charged residues in domain C may be involved in electrostatic interactions not only with other parts of RF but also with negative or positive charges in rRNA or ribosomal proteins. Recent crystallographic solutions of ribosome structures have pointed out that the decoding pocket of the 30S subunit is rich in negative charges contributed by 16S rRNA (21–24). Hence, we speculate that the increased positive charge(s) in domain C should strengthen the binding of RF to the ribosome codon independently and/or influence the docking position or orientation of RF to the ribosome as to reduce the stringency of the pep-anticodon codon interaction (Fig. 5). These two possibilities may not necessarily be mutually exclusive.
Figure 5.
Predicted electrostatic interactions necessary for the accurate recognition of stop codons by the RF in the A site of the ribosome. This model assumes that multiple electrostatic interactions between charged amino acids in domain C of RF and negative or positive charges in rRNA or ribosomal proteins play a vital role for the correct docking of RF into the decoding pocket. The accurate positioning of the tripeptide-anticodon in the A site of the 30S subunit allows the selective recognition of cognate stop codons (Left), whereas Glu → Lys change(s) affect this electrostatic balance, leading to relaxed recognition (i.e., bypassing) of any codons including sense codons (Right).
How codon bypassing triggers peptidyl-tRNA hydrolysis is puzzling. It is believed generally that the catalytic mode of the peptidyltransferase center in the 50S subunit is converted to peptidyl-tRNA hydrolysis after binding of the RFs and the recognition of cognate stop codons in the A site of the 30S subunit. The mechanism behind this action at a distance is unknown. Here we propose two scenarios to account for this puzzle. In one, the codon bypassing mimics the commitment of the correct recognition, and this “pseudo” commitment is signaled to the peptidyltransferase center through RF or ribosomal structure (the pseudo commitment model). In the other, more positive charge(s) in domain C, in concert with codon bypassing, increases the duration of the occupation of the A site of the ribosome by RF in a codon-independent fashion, and this prolonged state of peptidyl-tRNA in the P site is more susceptible to natural hydrolysis (the “prolonged occupation” model). These two possibilities remain to be investigated.
In summary, the present study reveals that electrostatic interactions between domain C of RF and the decoding pocket of the ribosome or other parts of RF controls the accuracy of the stop codon recognition. By changing this electrostatic balance, the variant RF proteins acquired a phenotype of triggering peptide release at the noncognate stop codon and also at sense codons by virtue of codon bypassing. This, in turn, provides us with the potential to create an RF variant that terminates protein synthesis at a certain class of sense codons. Here we are only aware that the charge-flip variant RF proteins can bypass the decoding step with at least five sense codons: UGG, UAC, UUC, UCA, and CAA. Obviously, further analysis will be needed to resolve the specificity if any of each Glu → Lys change affecting the codon spectrum. Finally, given that a single-charge flip change is able to cause codon bypassing, we should not rule out the possibility that a novel, yet undiscovered, “recoding” (25) mechanism of translational termination at sense codon(s) is programmed already in some gene(s) in nature.
Acknowledgments
We thank Charles Yanofsky, Feng Gong, and Colin Crist for critical reading of the manuscript and valuable comments. This work was supported in part by grants from The Ministry of Education, Sports, Culture, Science and Technology of Japan, the Human Frontier Science Program, the Basic Research for Innovation Biosciences Program of the Bio-oriented Technology Research Advancement Institution (BRAIN), and the Mitsubishi Foundation.
Abbreviations
- RF
release factor
- pep-anticodon
peptide-anticodon
- IPTG
isopropyl-1-thio-β-d-galactoside
- fMet
N-formylmethionine
- RT
read-through
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nakamura Y, Ito K, Isaksson L A. Cell. 1996;87:147–150. doi: 10.1016/s0092-8674(00)81331-8. [DOI] [PubMed] [Google Scholar]
- 2.Tate W P, Poole E S, Mannering S A. Prog Nucleic Acids Res. 1996;52:293–335. doi: 10.1016/s0079-6603(08)60970-8. [DOI] [PubMed] [Google Scholar]
- 3.Kisselev L L, Buckingham R H. Trends Biochem Sci. 2000;25:561–567. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- 4.Ito K, Ebihara K, Uno M, Nakamura Y. Proc Natl Acad Sci USA. 1996;93:5443–5448. doi: 10.1073/pnas.93.11.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark B F C, Nyborg J. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 6.Ito K, Uno M, Nakamura Y. Nature (London) 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura Y, Ito K, Ehrenberg M. Cell. 2000;101:349–352. doi: 10.1016/s0092-8674(00)80845-4. [DOI] [PubMed] [Google Scholar]
- 8.Wilson K, Ito K, Noller H, Nakamura Y. Nat Struct Biol. 2000;7:866–870. doi: 10.1038/82818. [DOI] [PubMed] [Google Scholar]
- 9.Freistroffer D V, Pavlov M Y, MacDougall J, Buckingham R H, Ehrenberg M. EMBO J. 1997;16:4126–4133. doi: 10.1093/emboj/16.13.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshizawa S, Fourmy D, Puglisi J D. Science. 1999;285:1722–1725. doi: 10.1126/science.285.5434.1722. [DOI] [PubMed] [Google Scholar]
- 11.Ito K, Uno M, Nakamura Y. Proc Natl Acad Sci USA. 1998;95:8165–8169. doi: 10.1073/pnas.95.14.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawakami K, Inada T, Nakamura Y. J Bacteriol. 1988;170:5378–5381. doi: 10.1128/jb.170.11.5378-5381.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uno M, Ito K, Nakamura Y. Biochimie. 1996;78:935–943. doi: 10.1016/s0300-9084(97)86715-6. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Mottagui-Tabar S, Björnsson A, Isaksson L A. EMBO J. 1994;13:249–257. doi: 10.1002/j.1460-2075.1994.tb06255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinçbas-Renqvist V, Engström Å, Mora L, Heurgué-Hamard V, Buckingham R, Ehrenberg M. EMBO J. 2000;24:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura Y, Kawazu Y, Uno M, Yoshimura K, Ito K. In: The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. Garrett R A, Douthwaite S R, Liljas A, Matheson A T, Moore P B, Noller H F, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 519–526. [Google Scholar]
- 18.Frolova L Y, Tsivkovskii R Y, Sivolobova G F, Oparina N Y, Serpinsky O I, Blinov V M, Tatkov S I, Kisselev L L. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Björnsson A, Isaksson L A. J Mol Biol. 1993;232:1017–1029. doi: 10.1006/jmbi.1993.1457. [DOI] [PubMed] [Google Scholar]
- 20.Craigen W J, Cook R G, Tate W P, Caskey C T. Proc Natl Acad Sci USA. 1985;82:3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wimberly B T, Brodersen D E, Clemons W M, Jr, Morgan-Warren R J, Carter A P, Vonrhein C, Hartsch T, Ramakrishnan V. Nature (London) 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 22.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 23.Yusupov M M, Yusupova G Z, Baucom A, Lieberman K, Earnest T N, Cate J H D, Noller H F. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 24.Ogle J M, Brodersen D E, Clemons W M, Jr, Tarry M J, Carter A P, Ramakrishnan V. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 25.Gesteland R F, Atkins J F. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]