Abstract
Anti-sigma factors regulate prokaryotic gene expression through interactions with specific sigma factors. The bacteriophage T4 anti-sigma factor AsiA is a molecular switch that both inhibits transcription from bacterial promoters and phage early promoters and promotes transcription at phage middle promoters through its interaction with the primary sigma factor of Escherichia coli, σ70. AsiA is an all-helical, symmetric dimer in solution. The solution structure of the AsiA dimer reveals a novel helical fold for the protomer. Furthermore, the AsiA protomer, surprisingly, contains a helix–turn–helix DNA binding motif, predicting a potential new role for AsiA. The AsiA dimer interface includes a substantial hydrophobic component, and results of hydrogen/deuterium exchange studies suggest that the dimer interface is the most stable region of the AsiA dimer. In addition, the residues that form the dimer interface are those that are involved in binding to σ70. The results promote a model whereby the AsiA dimer maintains the active hydrophobic surfaces and delivers them to σ70, where an AsiA protomer is displaced from the dimer via the interaction of σ70 with the same residues in AsiA that constitute the dimer interface.
Regulation of prokaryotic transcription involves the interaction of sigma (σ) factors, which bind to the core RNA polymerase to impart promoter recognition specificity, with cognate anti-sigma (anti-σ) factors that inhibit σ function. Most of the numerous examples of σ/anti-σ pairs involve alternative σ factors, those activated in response to specific stimuli, as opposed to primary σ factors, which are essential for survival and responsible for the bulk of transcription, including the housekeeping genes (reviewed in refs. 1–4). An important exception is the interaction of σ70 of Escherichia coli, namesake for a family of sequence conserved primary σ factors, with the anti-σ factor AsiA.
The bacteriophage T4-encoded AsiA protein, product of the asiA gene (5), and the first anti-σ factor to be discovered, binds tightly to the σ70 subunit of the E. coli RNA polymerase holoenzyme (6–10), altering the specificity of the complex toward both phage and host promoters. Following infection by bacteriophage T4, the E. coli RNA polymerase is recruited to sequentially transcribe genes from the T4 early, middle, and late promoters. T4 early promoters contain bacterial-like consensus DNA sequences that allow for their immediate recognition by the unmodified RNA polymerase, and the T4 early genes include the asiA gene. Shortly thereafter, transcription at early promoters is inhibited, along with transcription at bacterial promoters, by phage-induced modifications of the RNA polymerase, one of which is the tight association of AsiA with σ70. This interaction inhibits σ70-dependent transcription at early promoters by blocking recognition by σ70 of the conserved sequence element centered at position −35 (11). Furthermore, AsiA not only has the ability to function as an anti-σ factor, it also has the ability to promote transcription. In concert with the T4-encoded MotA protein (product of the T4 early gene motA), AsiA promotes efficient recognition of T4 middle promoters (12–14) by RNA polymerase holoenzyme, again through an interaction with σ70. Thus, direct association of AsiA with σ70 promotes the switch from T4 early to middle transcription (reviewed in refs. 1 and 15). Finally, the highly conserved region of σ70 that interacts with AsiA, denoted region 4, is central to the function and regulation of many aspects of transcription, including specific DNA recognition. Therefore, a detailed structural analysis of AsiA is obligatory for a comprehensive understanding of the initial stages of transcription regulation during T4 infection and prokaryotic transcription in general.
Some structural details of anti-σ factors and their complexes with cognate σ factors have recently been revealed (16–20). However, the only available high resolution structure for any anti-σ factor is that of the heat shock-induced chaperone DnaK, which has been shown to bind to the alternate σ factor σ32 to regulate the heat shock response (21–24). Furthermore, no high resolution structural information has heretofore been available for anti-σ factors that target primary σ factors.
AsiA is a symmetric dimer (25) of small (90 aa, 10.59 kDa) protomers, which are composed of helix and coil regions and are devoid of β-strand/sheet secondary structural elements (16). Although AsiA is a member of a class of cytoplasmic anti-σ factors (3, 4), it has no known sequence homologues, including both the cytoplasmic and membrane-associated anti-σ factors. Herein, we describe the three-dimensional solution structure of the AsiA dimer determined using NMR spectroscopy. In addition, results of studies of the local and global stabilities of AsiA using hydrogen/deuterium exchange are presented. The results provide additional support for an emerging new model of AsiA function and provide intriguing and unexpected evidence for additional biological roles for this novel protein.
Materials and Methods
AsiA Production.
The cloning, overexpression, and purification of AsiA have been described (5, 12, 26), as has preparation of isotopically labeled AsiA using isotopically labeled rich media (16). In addition, the asiA gene was subcloned into the pET-24b expression vector and transformed into E. coli BL21(DE3) for expression. Samples of AsiA produced in this manner were labeled isotopically using minimal media with 13C-glucose and/or 15NH4Cl as the sole carbon or nitrogen source. A sample of ≈10% 13C-labeled AsiA for stereospecific methyl assignments was produced using minimal media with 10% uniformly 13C-labeled and 90% unlabeled glucose as the carbon source. Purification was largely as described previously (12) with minor modifications. A sample of AsiA required to determine the interprotomer contacts consisted of equimolar quantities of uniformly 13C-, 15N-labeled AsiA and unlabeled AsiA. All NMR samples contained 1–2 mM AsiA/50 mM d4-acetate (Na+ or K+)/0.02% sodium azide/10% D2O, pH = 6.2–6.3 (meter reading). In some cases, 50 mM KCl was also present.
NMR Spectroscopy.
All NMR data were acquired at 25°C using Varian INOVA 600 and 750 MHz spectrometers. Main chain (and 13Cβ) resonance assignments for AsiA have been reported (16). Aliphatic side chain resonances were assigned using HCCH-TOCSY (27, 28) and amide-detected heteronuclear resolved TOCSY experiments [H(CCO)NH, C(CO)NH] (29). Aromatic resonances were assigned using a three-dimensional 1H-TOCSY-relayed ct-[13C,1H]HMQC experiment (30), an (Hβ)Cβ(CγCδ)Hδ experiment (31), and nuclear Overhauser effect (spectroscopy) [NOE(SY)] spectra (below). All methyl groups of Val and Leu were assigned stereospecifically using the method of Wüthrich and coworkers (32, 33). Stereospecific assignments for the side-chain −NH2 groups of Gln and Asn residues were also determined (34). Distance restraints for structure calculations were derived from NOE intensities in 13C- and 15N-edited NOESY-HSQC spectra (35, 36). Phi (φ) angle restraints were determined from values of 3JHN,Hα coupling constants from an HNHA experiment (37). Restraints for χ1 angles of aromatic residues were obtained from 3JNCγ and 3JC′Cγ coupling constants (38). A 13C F1-filtered, F3-edited NOESY-HSQC spectrum (39) provided contacts between the protomers (controls with uniformly 13C, 15N-labeled AsiA and with unlabeled AsiA confirmed the integrity of the filtering/editing). The 1H chemical shifts were referenced to external Na+DSS− in D2O (0.00 ppm), whereas 13C and 15N chemical shifts were referenced indirectly (40). Data processing and visualization were accomplished with Felix (Molecular Simulations, Walther, MA).
Structure Calculations.
Structures were calculated by restrained simulated annealing, using NOE-based distance restraints [836 intraresidue, 519 sequential, 384 medium range (2 < |i-j| < 5), 286 long range, and 36 interprotomer, 2,061 total], 3JHN,Hα-based φ angle restraints, 3JNCγ-based χ1 angle restraints, TALOS (41) derived psi (ψ) angle restraints (153 total φ, ψ, and χ1), and hydrogen bond restraints (34 total) based on hydrogen exchange rates (below). The torsion angle dynamics protocols of CNS 1.0 (42) were used to calculate a group of 50 structures that were then refined using Cartesian dynamics. The 25 structures with the lowest total energies were selected for subsequent analyses. None of these violated any distance restraint by more than 0.3 Å or any dihedral angle restraint by more than 5 degrees. Structures were analyzed using PROCHECK-NMR (43) and PROMOTIF (44). All molecular models in the figures were prepared using MOLMOL (45).
Hydrogen/Deuterium Exchange.
Lyophilized 15N-labeled AsiA was dissolved in 70 μl of buffer solution (50 mM d4-acetate/50 mM KCl, pH = 6.3) in H2O. Hydrogen/deuterium exchange was initiated by dilution of the sample with 630 μl of the same buffer in D2O. The rates of exchange of the amide protons were monitored by serial acquisition of two-dimensional 15N-HSQC spectra for ≈1.5 days. Rate constants for exchange (kex) were determined from a fit of the data (peak intensity versus time) to a three-parameter single exponential. Intrinsic rates of exchange (kint) were calculated as described (46) with corrections for temperature, ionization/concentration of H2O and D2O, and the response of the glass pH electrode to D2O (47, 48). The values of kex and kint were used to calculate the slowing/protection factors (kint/kex), the equilibrium constant for the opening event that permits a hydrogen to exchange (Kop = kex/kint) and the free energy change for the opening event (ΔGHX = −RTlnKop). The largest (and equivalent) values of ΔGHX in AsiA (residues 15, 18, 36, 37, and 38) are localized to the dimer interface and show a large variation in kint, indicating exchange in the EX2 regime.
Results and Discussion
Solution Structure of the AsiA Dimer.
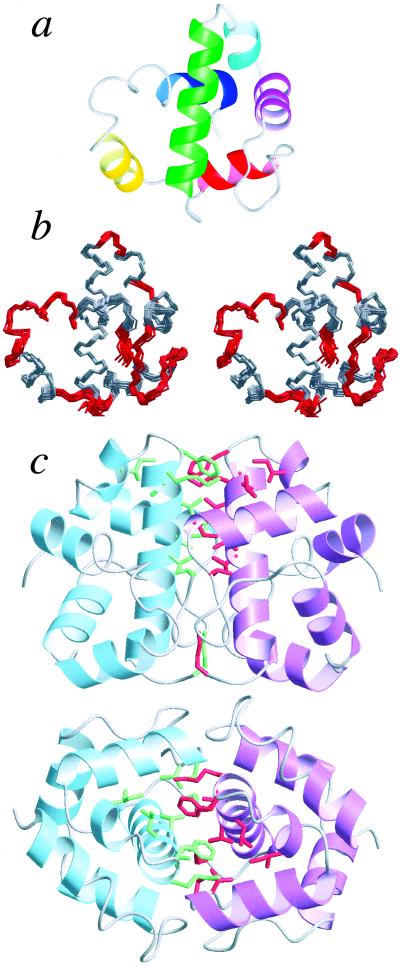
We have shown recently that AsiA is a symmetric dimer in solution (25). High resolution structural models for the AsiA dimer in solution are shown in Fig. 1. These models confirm previous assertions as to the secondary structural elements comprising AsiA based on chemical shifts (16). AsiA is an all-helical protein, composed of six helical segments and intervening loops and turns (Fig. 1b). The helices, as well as the loops and turns in the models, are ordered and well determined (Fig. 1a, Table 1). Deviations from the average structure are very small for the N-terminal half of each protomer and are somewhat larger relatively for the C-terminal half, in part reflecting the relative stabilities of these regions of the protein (below). The large loop between helices 3 and 4 is ordered, as are the series of turns between helices 5 and 6. The two residues at the C terminus are not well ordered in the calculated structures.
Figure 1.
The solution structure of AsiA. (a) Ribbon diagram of an AsiA protomer: helix 1, residues 4–20 (green); helix 2, 24–28 (cyan); helix 3, 30–40 (purple); helix 4, 50–59 (red); helix 5, 63–70 (yellow); helix 6, 82–88 (blue). (b) Stereo view of the superimposed 25 lowest energy AsiA structures (of 50 calculated); residues involved in helices are shown in gray (for clarity, only an AsiA protomer is shown). (c) Ribbon diagrams showing two views of the AsiA dimer. Side chains contributing observed interprotomer contacts are shown in green and red (M1, T13, V14, I17, L18, K20, F21, I26, I40, V42).
Table 1.
Statistics summary for the 25 AsiA structures
| rmsd from experimental restraints | ||
| NOE-based distance restraints, Å | 0.029 ± 0.002* | |
| Dihedral angle restraints, ° | 0.25 ± 0.24* | |
| rmsd from ideal geometry | ||
| Bonds, Å | 0.0048 ± 0.00009 | |
| Angles, ° | 0.69 ± 0.01 | |
| Impropers, ° | 0.51 ± 0.02 | |
| rmsd from the mean structure, Å | Main chain | Heavy atoms |
| Dimer, all residues (1–90) | 0.44 ± 0.10 | 0.96 ± 0.09 |
| Dimer, residues 1–45 | 0.29 ± 0.09 | 0.85 ± 0.09 |
| Dimer, residues 46–90 | 0.52 ± 0.12 | 1.03 ± 0.14 |
| Protomer, all residues (1–90) | 0.39 ± 0.09 | 0.94 ± 0.09 |
| Protomer, residues 1–45 | 0.25 ± 0.09 | 0.84 ± 0.09 |
| Protomer, residues 46–90 | 0.43 ± 0.11 | 0.99 ± 0.13 |
| Ramachandran plot† | ||
| Residues in most favored regions, % | 83.2 | |
| Residues in additional allowed regions, % | 13.1 | |
| Residues in generously allowed regions, % | 3.7 | |
| Residues in disallowed regions, % | 0.0 | |
These values are for all 50 calculated structures.
For the 25 structures, using Procheck-NMR (43).
The dimer interface is composed of stable contacts, deduced from observed intermolecular NOEs, involving primarily residues in the N-terminal helix, with additional contacts involving residues in helices 2 and 3 (Fig. 1c). The highest density of observed contacts is localized to the C-terminal half of helix 1 and helix 3 of each protomer. The contacts observed involve predominantly hydrophobic side chains, and the dimer interface seems to constitute the hydrophobic core of the protein (below); however, the relative contributions of the hydrophobic and electrostatic components to the affinity of the dimer are unknown. The C-terminal halves of helix 1 from both protomers constitute a short, parallel coiled-coil. It should be noted that whereas the structures reported were solved without inclusion of ambiguous restraints for the interprotomer contacts, inclusion of these restraints as ambiguous (49) resulted in no substantial changes in the structures.
Of particular significance is the fact that many of the residues comprising the dimer interface show large chemical shift changes when AsiA binds to the AsiA binding domain(s) of σ70. We have shown recently (25) that many residues in the N-terminal half of AsiA, including many in helices 1, 2, and 3, show large chemical shift changes upon binding to the AsiA binding domain(s) of σ70. These residues almost certainly are involved in σ70 binding, and from the structures in Fig. 1 it is now clear that they also comprise the dimer interface. These and other observations suggest AsiA interacts with σ70 as a monomer via the same residues used for dimerization (below).
A Novel Helical Fold and a DNA Binding Motif in AsiA.
Using the DALI (50) (http://www.ebi.ac.uk/dali) and 3dSearch/LOCK (ref. 51; see http://cmgm.Stanford.edu/∼brutlag/Papers/singh00.pdf and http://gene.stanford.edu/3dsearch) tertiary structure similarity search and superposition protocols, no proteins with an arrangement of six helices as in the AsiA protomer were found. No more than three of the helices of AsiA were simultaneously superimposable with those of known proteins. By these criteria, the AsiA six-helix bundle (five excluding the short single turn helix 2) represents a novel helical fold.
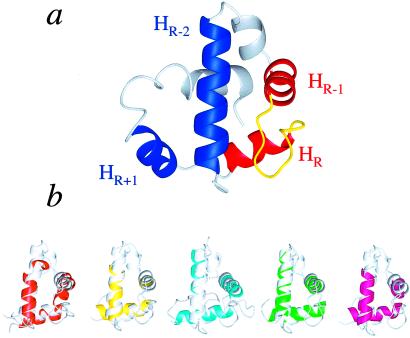
One of the most intriguing facets of the AsiA structure is the presence of a helix–turn–helix (HTH) DNA binding motif (Fig. 2), although neither free AsiA nor AsiA bound to σ70 has been shown to interact directly with DNA, and there is no significant sequence homology between AsiA and other DNA binding proteins containing this motif. However, predictive methods based on profile–profile matching to sequence profiles of proteins in the Protein Data Bank (Fold and Function Assignment System, ref. 52) predict similarities between AsiA and small DNA binding proteins, whereas the DALI and 3dSearch protocols both find significant numbers of DNA binding proteins with tertiary structural similarity to AsiA (superposition of three helices of the HTH motif).
Figure 2.
The DNA binding motif in AsiA. (a) Helices 3 and 4 of AsiA (red) and the intervening turn (loop, yellow) form the helix (HR-1), turn (loop), and DNA recognition helix (HR) of the HTHloop motif in AsiA. Helices 1 and 5 (blue, HR-2 and HR+1, respectively) are the potential third helices of this motif. (b) Helices 1, 3, and 4 of AsiA (light gray) superimpose well on HTH motifs of some DNA binding proteins (brown, yeast heat shock factor HSF, PDB identifier 2HTS; yellow, right origin binding protein Rob, 1D5Y; cyan, Ets domain of the Friend leukemia integration protein FLI-1, 1FLI; green, diptheria toxin repressor DtxR, 2DTR; magenta, multiple antibiotic resistance protein MarA, 1BL0).
The basic DNA binding unit of the HTH DNA binding motif comprises three helices (53). Ignoring the small, single turn helix 2, the HTH DNA binding motif in AsiA can be classified as both the HR-2, HR-1, HR type and the HR-1, HR, HR+1 type, where HR-2 is helix 1, HR-1 is helix 3, HR (the DNA recognition helix) is helix 4, and HR+1 is helix 5 (Fig. 2a). The turn between HR-1 and HR in AsiA is a loop (HTHloop) (53). We were unable to find examples of proteins with helices superimposable with helices 5, 4, and 3 of AsiA. There are, however, many examples of HTH DNA binding motifs in small DNA binding proteins with HR-2, HR-1, and HR helices, which superimpose well on helices 1, 3, and 4, respectively, of AsiA (Fig. 2b). In AsiA, two helices of the HTH motif, helix 1 (HR-2) and helix 3 (HR-1), function as both an interface for dimerization and as a surface for interaction with σ70 (25). Helix 4 (HR) is physically located such that it is apparently not involved directly in these capacities.
The presence of the HTH DNA binding motif in AsiA, along with previous results, suggests a reexamination of the role of AsiA during middle mode, and potentially early mode, transcription initiation. Productive recognition of T4 phage middle promoters by the prokaryotic RNA polymerase requires both AsiA and MotA (26). Interestingly, binding of middle promoter DNA by the RNA polymerase holoenzyme in the presence of MotA is enhanced significantly in the presence of AsiA, and the RNA polymerase/MotA DNase I footprint on middle promoter DNA is altered substantially again in the presence of AsiA (11, 14). Whereas AsiA most certainly interacts tightly with σ70 in these complexes, the available data indicate that this interaction occurs through residues in helices 1 and 3 of monomeric AsiA, without affecting helix 4 (HR), which is positioned on the opposite side of AsiA, leaving helix 4 free to contact DNA. These observations suggest the hypothesis that AsiA can interact directly with DNA during middle mode transcription to increase the affinity of the polymerase/MotA/AsiA complex for the promoter DNA and possibly to influence specificity.
Stability of AsiA.
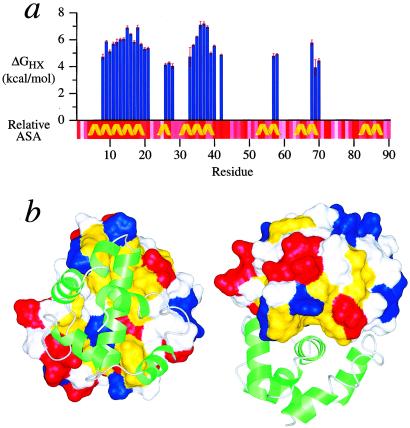
Results of hydrogen/deuterium exchange studies reveal the local and global stabilities of AsiA (Fig. 3a). Overall, the N-terminal half of AsiA is somewhat more stable than the C-terminal half. Many of the observed hydrogen exchange stabilities for residues in helices 1 and 3 are significantly larger than the lower limit of ≈4 kcal/mol that can be detected by the method used, whereas only a relatively small number of residues in the C-terminal half exhibited observable hydrogen exchange stabilities. According to the structural models, the helices in the C-terminal half of AsiA are short, and the C-terminal half is relatively more solvent exposed than the N-terminal half, leading in part to the reduced protection from hydrogen exchange. Residues in the C-terminal helix (helix 6) show no observable protection from hydrogen exchange with solvent.
Figure 3.
Stability of AsiA. (a) The residue-specific hydrogen exchange stabilities, ΔGHX, for the slowly exchanging amide protons in AsiA are plotted. The relative residue-specific accessible surface area (relative ASA) is also shown (calculated with naccess), darker red indicating less accessibility. The helices of AsiA are indicated. (b) The hydrophobic cleft formed by helix 3 and the C-terminal region of helix 1 of one AsiA protomer (van der Waals surface) and its interaction with the other protomer (ribbon diagram, green) in the AsiA dimer is shown. Hydrophobic (Leu, Ile, Val, Phe, and Ala) side chain atoms are mapped to the surface and colored yellow, revealing the hydrophobic cleft. Side chain atoms of charged residues are mapped to the surface and colored red (Glu and Asp) and blue (Lys and Arg).
The hydrogen/deuterium exchange results suggest that the most stable region of AsiA is the dimer interface. The C-terminal end of helix 1 from each protomer is aligned with the hydrophobic cleft between helices 1 and 3 of the opposing protomer to form the interface (Fig. 3b). The interface is predominantly hydrophobic and relatively solvent inaccessible (Fig. 3b). The hydrogen exchange stabilities for residues in these regions of the AsiA dimer are the largest overall and include those for residues 15, 18, 36, 37, and 38, which are statistically identical. It has been established that, in most cases, the overall global or conformational stability of a protein can be estimated from the largest hydrogen exchange stabilities (54, 55) because the most stable amide hydrogens, in general, require global unfolding of the protein to exchange with solvent. For the AsiA dimer, the five largest amide hydrogen exchange stabilities correspond to residues at the dimer interface, and the mean of these gives a global/conformational stability (in D2O) of 7.0 ± 0.4 kcal/mol. Thus, the dimer interface of AsiA is clearly very stable, and complete dissociation of the free dimer may require global unfolding of the protein.
As shown previously (25), AsiA binds tightly to peptides corresponding to AsiA binding determinants of σ70 based on the slow exchange behavior of the complexes on the NMR time scale. However, significant excess peptide is necessary to titrate completely all of the AsiA, indicating a reduced apparent or overall affinity. The modest apparent affinity stems from the necessity to reorganize the dimer interface to accommodate binding of the peptides because the same residues that form the dimer interface are also involved in peptide binding. The high affinity interaction of the peptide with these residues drives the energetically unfavorable rearrangement of the dimer interface, resulting in the modest overall affinity. The hydrogen exchange results support these suggestions because the dimer interface is demonstrably very stable.
Mechanism of AsiA Interaction with σ70.
Until recently, AsiA was proposed to interact exclusively with a single highly conserved region of σ70, denoted region 4.2 (approximately residues 570–599, an HTH DNA binding motif). Both the ability of AsiA to inhibit transcription at phage early promoters and its ability to facilitate transcription at phage middle promoters were considered to result from this single interaction (56, 57). We have shown recently (25) that AsiA also exhibits a high affinity for conserved region 4.1 of σ70 (approximately residues 540–565). Moreover, regions 4.1 and 4.2 can interact individually or simultaneously with AsiA (25). These results indicate the potential for discrete functional states resulting from the interaction of AsiA with these regions of σ70. Importantly, as described above, these regions of σ70 bind to AsiA via high affinity interactions with residues that comprise the dimer interface, and these interactions drive the reorganization of the dimer interface. These and other observations (25) indicate that the dimer interface reorganization is most likely a simple dimer dissociation. This model is consistent with the reported stoichiometry for the AsiA/σ70 complex (11) and the lack of observed structural asymmetry deduced from the NMR spectra of AsiA bound to σ70-derived peptides as detailed previously (25), both of which indicate that a single AsiA monomer binds to σ70 (or σ70-derived peptides). In addition, preliminary attempts (not shown) to observe interprotomer contacts in complexes of AsiA and σ70-derived peptides using NMR indicate these contacts are not present, signifying that the dimer is not intact in these complexes.
This model for the interaction of AsiA with σ70 is compelling in terms of the structure and stability studies detailed above. The dimer interface of AsiA is very stable and hydrophobic, whereas interaction of AsiA with σ70 apparently dissociates the dimer. Most likely, σ70 interacts with AsiA by direct displacement of one of the AsiA protomers, which would minimize the exposure of the hydrophobic dimer interface to solvent and minimize the activation energy for the process. In this scenario, the biological role of the AsiA dimer is to deliver the active hydrophobic surface(s), harbored in the dimer interface, to σ70.
The previous accepted model for AsiA function depicted AsiA as monofunctional in its interactions with σ70. The emerging new model for AsiA function, rooted in recent results (25) and those herein, portrays regulation of σ70 by AsiA as rich and complex, involving multiple sites of interaction on σ70, and, most likely, multiple functional states, and a potential direct AsiA–DNA interaction.
Acknowledgments
We thank A. Joshua Wand for his advice and support. Konstantin Severinov is acknowledged for discussions of results before publication. Lewis E. Kay and the University of Toronto are gratefully acknowledged for providing Varian NMR pulse programs. Leland Mayne and S. Walter Englander are acknowledged for spreadsheets to calculate intrinsic hydrogen exchange rates. This work was supported in part by National Institutes of Health Grants GM54998 (to J.L.U.), GM50700 (to E.N.B.), and DK39806 (to A.J.W.), National Science Foundation Grant DBI-0088931 (to J.L.U.), and a National Science Foundation Graduate Fellowship (to K.A.). In addition, this research is based upon work supported by National Science Foundation Experimental Program to Stimulate Competitive Research (EPS-9874732) and matching support from the State of Kansas.
Abbreviations
- NOE(SY)
nuclear Overhauser effect (spectroscopy)
- HTH
helix–turn–helix
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1JR5). The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 4040).
References
- 1.Brody E N, Kassavetis G A, Ouhammouch M, Sanders G M, Tinker R L, Geiduschek G P. FEMS Microbiol Lett. 1995;128:1–8. doi: 10.1111/j.1574-6968.1995.tb07491.x. [DOI] [PubMed] [Google Scholar]
- 2.Brown K L, Hughes K T. Mol Microbiol. 1995;16:397–404. doi: 10.1111/j.1365-2958.1995.tb02405.x. [DOI] [PubMed] [Google Scholar]
- 3.Hughes K T, Mathee K. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- 4.Helmann J D. Curr Opin Microbiol. 1999;2:135–141. doi: 10.1016/S1369-5274(99)80024-1. [DOI] [PubMed] [Google Scholar]
- 5.Orsini G, Ouhammouch M, LeCaer J P, Brody E N. J Bacteriol. 1993;175:85–93. doi: 10.1128/jb.175.1.85-93.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevens A. Proc Natl Acad Sci USA. 1972;69:603–607. doi: 10.1073/pnas.69.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevens A. Biochemistry. 1974;13:493–503. doi: 10.1021/bi00700a015. [DOI] [PubMed] [Google Scholar]
- 8.Stevens A. In: RNA Polymerase. Losick R, Chamberlin M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1976. pp. 617–627. [Google Scholar]
- 9.Stevens A. Biochim Biophys Acta. 1977;475:193–196. doi: 10.1016/0005-2787(77)90355-0. [DOI] [PubMed] [Google Scholar]
- 10.Stevens A, Rhoton J. Biochemistry. 1975;14:5074–5079. doi: 10.1021/bi00694a007. [DOI] [PubMed] [Google Scholar]
- 11.Adelman K, Orsini G, Kolb A, Graziani L, Brody E N. J Biol Chem. 1997;272:27435–27443. doi: 10.1074/jbc.272.43.27435. [DOI] [PubMed] [Google Scholar]
- 12.Ouhammouch M, Adelman K, Harvey S R, Orsini G, Brody E N. Proc Natl Acad Sci USA. 1995;92:1451–1455. doi: 10.1073/pnas.92.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adelman K, Brody E N, Buckle M. Proc Natl Acad Sci USA. 1998;95:15247–15252. doi: 10.1073/pnas.95.26.15247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinton D M, March-Amegadzie R, Gerber J S, Sharma M. J Mol Biol. 1996;256:235–248. doi: 10.1006/jmbi.1996.0082. [DOI] [PubMed] [Google Scholar]
- 15.Hinton D M, March-Amegadzie R, Gerber J S, Sharma M. Methods Enzymol. 1996;274:43–57. doi: 10.1016/s0076-6879(96)74007-7. [DOI] [PubMed] [Google Scholar]
- 16.Urbauer J L, Adelman K, Brody E N. J Biomol NMR. 1997;10:205–206. doi: 10.1023/a:1018343223888. [DOI] [PubMed] [Google Scholar]
- 17.Daughdrill G W, Chadsey M S, Karlinsey J E, Hughes K T, Dahlquist F W. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- 18.Daughdrill G W, Hanely L J, Dahlquist F W. Biochemistry. 1998;37:1076–1082. doi: 10.1021/bi971952t. [DOI] [PubMed] [Google Scholar]
- 19.Campbell E A, Darst S A. J Mol Biol. 2000;300:17–28. doi: 10.1006/jmbi.2000.3838. [DOI] [PubMed] [Google Scholar]
- 20.Minakhin L, Camarero J A, Holford M, Parker C, Muir T W, Severinov K. J Mol Biol. 2001;306:631–642. doi: 10.1006/jmbi.2001.4445. [DOI] [PubMed] [Google Scholar]
- 21.Pellecchia M, Montgomery D L, Stevens S Y, Vander Kooi C W, Feng H P, Gierasch L M, Zuiderweg E R. Nat Struct Biol. 2000;7:298–303. doi: 10.1038/74062. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Kurochkin A V, Hu W, Flynn G C, Zuiderweg E R. Biochemistry. 1998;37:7929–7940. doi: 10.1021/bi9800855. [DOI] [PubMed] [Google Scholar]
- 23.Harrison C J, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 24.Zhu X, Zhao X, Burkholder W F, Gragerov A, Ogata C M, Gottesman M E, Hendrickson W A. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbauer J L, Adelman K, Bieber Urbauer R J, Simeonov M F, Gilmore J M, Zolkiewski M, Brody E N. J Biol Chem. 2001;276:41128–41132. doi: 10.1074/jbc.M106400200. [DOI] [PubMed] [Google Scholar]
- 26.Ouhammouch M, Orsini G, Brody E N. J Bacteriol. 1994;176:3956–3965. doi: 10.1128/jb.176.13.3956-3965.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bax A, Clore M, Gronenborn A M. J Magn Reson. 1990;88:425–431. [Google Scholar]
- 28.Kay L E, Xu G-Y, Singer A U, Muhandiram D R, Forman-Kay J D. J Magn Reson B. 1993;101:333–337. [Google Scholar]
- 29.Grzesiek S, Anglister J, Bax A. J Magn Reson B. 1993;101:114–119. [Google Scholar]
- 30.Zerbe O, Szyperski T, Ottiger M, Wüthrich K. J Biomol NMR. 1996;7:99–106. doi: 10.1007/BF00203820. [DOI] [PubMed] [Google Scholar]
- 31.Yamazaki T, Kay J D, Kay L E. J Am Chem Soc. 1993;115:11054–11055. [Google Scholar]
- 32.Neri D, Otting G, Wüthrich K. Tetrahedron. 1990;46:3287–3296. [Google Scholar]
- 33.Neri D, Szyperski T, Otting G, Senn H, Wüthrich K. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 34.McIntosh L P, Brun E, Kay L E. J Biomol NMR. 1997;9:306–312. doi: 10.1023/A:1018635110491. [DOI] [PubMed] [Google Scholar]
- 35.Fesik S W, Zuiderweg E R P. J Magn Reson. 1988;78:588–593. [Google Scholar]
- 36.Marion D, Kay L E, Sparks S W, Torchia D A, Bax A. J Am Chem Soc. 1989;111:1515–1517. [Google Scholar]
- 37.Vuister G, Bax A. J Am Chem Soc. 1993;115:7772–7777. [Google Scholar]
- 38.Hu J-S, Grzesiek S, Bax A. J Am Chem Soc. 1997;119:1803–1804. [Google Scholar]
- 39.Zwahlen C, Legault P, Vincent S J F, Greenblatt J, Konrat R, Kay L E. J Am Chem Soc. 1997;119:6711–6721. [Google Scholar]
- 40.Wishart D S, Bigam C G, Yao J, Abildgaard F, Dyson H J, Oldfield E, Markley J L, Sykes B D. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 41.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 42.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Laskowski R A, Rullman J A, MacArthur M W, Kaptein R, Thronton J M. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 44.Hutchinson E G, Thornton J M. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konradi R, Billeter M, Wuthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 46.Bai Y, Milne J S, Mayne L, Englander S W. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Covington A K, Robinson R A, Bates R G. J Phys Chem. 1966;70:3820–3824. [Google Scholar]
- 48.Schowen K J B. In: Transition States of Biochemical Processes. Gandour R D, Schowen R L, editors. New York: Plenum; 1978. p. 283. [Google Scholar]
- 49.O'Donoghue S L, King G F, Nilges M. J Biomol NMR. 1996;8:193–206. doi: 10.1007/BF00211165. [DOI] [PubMed] [Google Scholar]
- 50.Holm L, Sander C. J Mol Biol. 1993;233:123–128. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 51.Singh A P, Brutlag D L. Proceedings of the Fifth International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1997. pp. 284–293. [PubMed] [Google Scholar]
- 52.Rychlewski L, Jaroszewski L, Li W, Godzik A. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wintjens R, Rooman M. J Mol Biol. 1996;262:294–313. doi: 10.1006/jmbi.1996.0514. [DOI] [PubMed] [Google Scholar]
- 54.Huyghues-Despointes B M P, Pace C N, Englander S W, Scholtz J M. Methods Mol Biol. 2001;168:69–92. doi: 10.1385/1-59259-193-0:069. [DOI] [PubMed] [Google Scholar]
- 55.Huyghues-Despointes B M P, Scholtz J M, Pace C N. Nat Struct Biol. 1999;6:910–912. doi: 10.1038/13273. [DOI] [PubMed] [Google Scholar]
- 56.Severinova E, Severinov K, Darst S A. J Mol Biol. 1998;279:9–18. doi: 10.1006/jmbi.1998.1742. [DOI] [PubMed] [Google Scholar]
- 57.Colland F, Orsini G, Brody E N, Buc H, Kolb A. Mol Microbiol. 1998;27:819–829. doi: 10.1046/j.1365-2958.1998.00729.x. [DOI] [PubMed] [Google Scholar]