Abstract
We report that diverse species of bacteria encode a type IB DNA topoisomerase that resembles vaccinia virus topoisomerase. Deinococcus radiodurans topoisomerase IB (DraTopIB), an exemplary member of this family, relaxes supercoiled DNA in the absence of a divalent cation or ATP. DraTopIB has a compact size (346 aa) and is a monomer in solution. Mutational analysis shows that the active site of DraTopIB is composed of the same constellation of catalytic side chains as the vaccinia enzyme. Sequence comparisons and limited proteolysis suggest that their folds are conserved. These findings imply an intimate evolutionary relationship between the poxvirus and bacterial type IB enzymes, and they engender a scheme for the evolution of topoisomerase IB and tyrosine recombinases from a common ancestral strand transferase in the bacterial domain. Remarkably, bacteria that possess topoisomerase IB appear to lack DNA topoisomerase III.
The type IB family of DNA topoisomerases includes eukaryotic nuclear topoisomerase I and the topoisomerases of poxviruses (1, 2). The type IB enzymes relax DNA supercoils via a multistep reaction pathway entailing noncovalent binding of the enzyme to duplex DNA, cleavage of one DNA strand with formation of a covalent DNA-(3′-phosphotyrosyl)-protein intermediate, strand passage, and strand religation. A constellation of conserved amino acid side chains (Arg-130, Lys-167, Lys-220, Arg-223, and His-265 in vaccinia virus topoisomerase) catalyzes the attack of the active site tyrosine nucleophile (Tyr-274) on the scissile phosphodiester. The RKKRH “catalytic pentad” is conserved in all members of the topo IB family. The Arg-130, Lys-167, Arg-223, and His-265 components of the pentad each lend a 102–105 enhancement of the transesterification rate (3–8). The λ Int family of site-specific DNA recombinases (now called tyrosine recombinases) use a similar mechanism to catalyze formation and resolution of Holliday junctions. The catalytic pentad of the tyrosine recombinases (RKHRH) characteristically contains a histidine at the third position occupied by lysine in topo IB (9).
topo IB and tyrosine recombinases consist of two domains that form a C-shaped protein clamp around duplex DNA. The carboxyl-terminal catalytic domains of topo IB and tyrosine recombinases adopt a common fold composed of eight α-helices and a three-strand antiparallel β-sheet (6, 10–15). The constituents of the catalytic pentad occupy similar positions in the tertiary structures of topo IB and tyrosine recombinases. Thus, it is proposed that topo IB and tyrosine recombinases evolved from a common ancestral DNA strand transferase (11), in which case we can consider them distinct branches of an enzyme superfamily defined by a DNA-3′-phosphotyrosyl intermediate.
Remarkably, there is no structural similarity at all between the amino-terminal domains of type IB topoisomerases and those of the tyrosine recombinases. For example, the amino-terminal domain of vaccinia topoisomerase consists almost entirely of β-strands, whereas the amino terminus of Cre is composed entirely of α-helices (10, 16). The analogous domain of human topo IB shares secondary structural elements with vaccinia topoisomerase, but the cellular enzyme is much larger than poxvirus topo IB and is embellished by multiple additional structural modules (17). The key questions are: When and how did the topo IB and tyrosine recombinase branches diverge? What is the evolutionary relationship between the viral and cellular topo IB enzymes?
Clues can be gleaned from the phylogenetic distributions of the two branches. The tyrosine recombinases are represented widely in all three domains of life. They are ubiquitous in bacteria (as Xer proteins involved in resolving multimeric chromosomes); they also are found in bacterial viruses, archaea (e.g., Methanococcus, Thermoplasma, Methanobacterium, and Sulfolobus), and lower eukarya. In contrast, topo IB enzymes have been described only in the eukaryotic domain, with the exception of Topo V, an atypical topo IB-like enzyme in the archaeon Methanopyrus kandleri (18), which has minimal structural similarity to tyrosine recombinases or topo IB and does not fall neatly into either branch of the superfamily. topo IB enzymes are found in every known eukaryal species and in all genera of poxviruses. An outstanding question is whether type IB topoisomerases exist in the other domains of life and, if so, whether inferences can be made concerning their relationship to the eukaryotic topo IB branch.
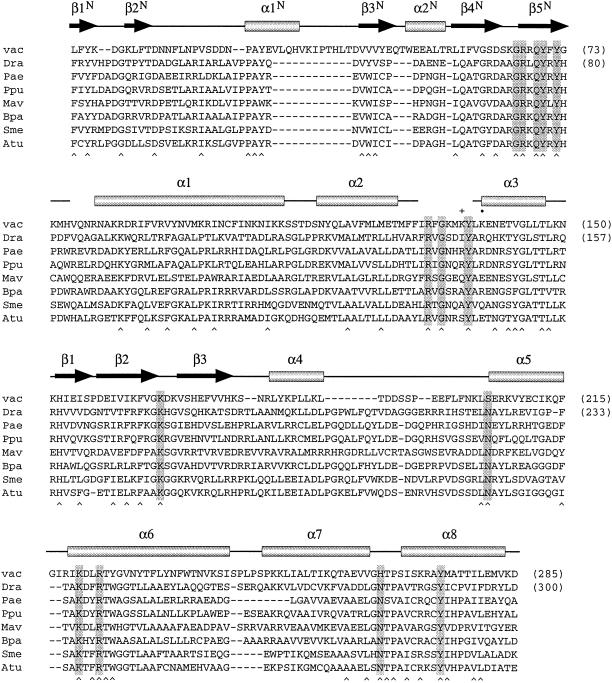
Here we report the identification of a topo IB family in bacteria. The bacterial topo IB proteins were discovered by searching for polypeptides related to vaccinia virus topoisomerase. The bacterial topo IB family embraces polypeptides encoded by diverse genera of bacteria, including Deinococcus, Pseudomonas, Mycobacterium, Bordetella, Agrobacterium, Sinorhizobium, Sphingomonas, and Rhodobacter (Fig. 1 and data not shown). The sizes of the bacterial enzymes (333–351 aa) are remarkably similar to those of the poxvirus topoisomerases (314–333 aa), and the amino acid functional groups implicated in DNA binding and transesterification are well conserved between poxvirus topoisomerases and this bacterial protein family (Fig. 1). The tyrosine nucleophile and four of the constituents of the catalytic pentad of vaccinia topoisomerase (the RKKR residues) are identical in all of the bacterial proteins. Although the fifth catalytic residue of the pentad (His-265 in vaccinia topoisomerase) is replaced by an asparagine in the bacterial proteins, the His–Asn substitution is predicted from mutagenesis studies of vaccinia topoisomerase to be functionally conservative (3). The occurrence of a lysine in the third position of the pentad in all of the bacterial proteins places them tentatively in the same camp as eukaryotic cellular and viral topo IB and distinguishes them from the tyrosine recombinases.
Figure 1.
A family of bacterial type IB topoisomerases related to vaccinia virus topoisomerase. The amino acid sequence of vaccinia virus topoisomerase (vac) from positions 4 to 285 is aligned with the sequences of putative topoisomerases encoded by Deinococcus radiodurans (Dra), Pseudomonas aeruginosa (Pae), Pseudomonas putida (Ppu), Mycobacterium avium (Mav), Bordetella parapertussis (Bpa), Sinorhizobium meliloti (Sme), and Agrobacterium tumefaciens (Atu). The secondary structural elements of vaccinia topoisomerase are shown above its sequence. Conserved residues involved in DNA binding or transesterification by vaccinia topoisomerase are highlighted in shaded boxes. Positions of side-chain identity or similarity in all eight of the proteins are indicated by a ∧ below the sequences.
Even more striking than the conservation of catalytic residues is the similarity of the amino-terminal domains of the vaccinia and bacterial proteins. In particular, the motif GXDXXGRXQYXY is present in all of the proteins aligned in Fig. 1 (in strands β4N and β5N) and includes side chains of vaccinia topoisomerase that contribute to the concave binding interface with the DNA major groove (16, 19, 20). This level of conservation of the amino-terminal domain of the vaccinia protein is unprecedented outside of the immediate subfamily of poxvirus-encoded type IB topoisomerases. Thus, the salient question is whether the bacterial proteins have topoisomerase activity and whether their resemblance to vaccinia topoisomerase is mechanistically informative.
To address these issues, we have expressed, purified, and characterized the putative bacterial type IB topoisomerase encoded by Deinococcus radiodurans. We show that the 346-aa D. radiodurans protein (DraTopIB) relaxes DNA supercoils in the absence of divalent cations or ATP. The relaxation activity requires the conserved active site tyrosine and at least three of the constituents of the catalytic pentad. Structure probing by limited proteolysis suggests tertiary structure conservation between the D. radiodurans enzyme and vaccinia topoisomerase. Taken together, our data suggest that the bacterial, poxviral, and eukaryotic nuclear type IB topoisomerases are derived from a common ancestral lineage of type IB enzymes.
Materials and Methods
Cloning and Site-Directed Mutagenesis of DraTopIB.
D. radiodurans genomic DNA was used as the template for PCR amplification of the DraTopIB ORF. The primers were designed to introduce an NdeI restriction site at the translation start codon and a BamHI site 3′ of the stop codon. The PCR product was digested with NdeI and BamHI and then inserted between the NdeI and BamHI sites of expression plasmid pET21b. Missense mutations Y289F, R137A, R239A, and N280A were introduced into the DraTopIB gene by using the two-stage PCR-based overlap extension method (21). The entire DraTopIB insert was sequenced completely in all clones to ensure that no unwanted mutations had been introduced.
Purification of DraTopIB.
A 100-ml culture of Escherichia coli BL21(DE3)/pET21-DraTop1 was grown at 37°C in LB medium containing 0.4 mg/ml ampicillin and 0.4% glucose until the OD600 reached 0.6. The culture was chilled on ice for 30 min and then adjusted to 0.4 mM isopropyl β-d-thiogalactoside. Incubation was continued for 17 h at 17°C. Cells were harvested by centrifugation, and the pellet was stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 100 ml of lysis buffer (50 mM Tris⋅HCl, pH 7.5/1 mM DTT/10 mM EDTA/10% sucrose) containing 0.15 M NaCl and 0.1 mg/ml lysozyme. After incubation on ice for 40 min, Triton X-100 was added to 0.1%, and then the lysate was sonicated to reduce viscosity. Insoluble material was removed by centrifugation. The supernatant was applied to a 1-ml column of phosphocellulose that had been equilibrated in buffer A (50 mM Tris⋅HCl, pH 7.5/2.5 mM DTT/1 mM EDTA/10% glycerol/0.1% Triton X-100) containing 0.1 M NaCl. The column was washed serially with 10 ml of buffer A containing 0.1 and 0.35 M NaCl. The bound DraTopIB was then step-eluted with 5 ml of 1 M NaCl in buffer A; 0.5-ml fractions were collected during this step. Mutant proteins Y289F, R137A, R239A, and N280A were purified by using the same protocol and recovered with similar yields (≈3 mg of protein in the peak 1 M NaCl phosphocellulose eluate fraction).
Glycerol Gradient Sedimentation.
Aliquots (≈1 mg) of the phosphocellulose preparations of wild-type and mutant DraTopIB were applied to 4.8-ml 15–30% glycerol gradients containing 50 mM Tris⋅HCl (pH 7.5), 2.5 mM DTT, 1 mM EDTA, 0.4 M NaCl, and 0.1% Triton X-100. The gradients were centrifuged at 50,000 rpm for 40 h at 4°C in a Beckman SW50 rotor. Fractions were collected from the bottoms of the tubes.
DNA Relaxation.
Reaction mixtures containing (per 20 μl) 0.3 μg of pUC19 DNA, DraTopIB, and other components as specified were incubated at 37°C. The reactions were initiated by adding enzyme and quenched with SDS. The products were electrophoresed through a horizontal 1% agarose gel in 90 mM Tris borate/2.5 mM EDTA, pH 8.3. The gels were stained in 0.5 μg/ml ethidium bromide and photographed under short-wave UV illumination.
Results
Recombinant DraTopIB Relaxes Supercoiled DNA.
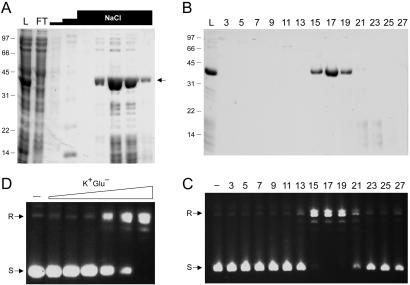
Recombinant DraTopIB was produced in E. coli and purified from the soluble lysate by phosphocellulose column chromatography (Fig. 2A) and glycerol gradient sedimentation (Fig. 2B). The 40-kDa DraTopIB polypeptide sedimented as a discrete peak in fractions 15–19 and was resolved from multiple lower molecular weight proteins sedimenting in fractions 21–23. Aliquots of the gradient fractions were assayed for relaxation of supercoiled pUC19 plasmid DNA. A single peak of EDTA-resistant topoisomerase activity was detected in fractions 15–21 (Fig. 2C), coincident with the sedimentation profile of the DraTopIB protein. We surmise that DraTopIB is a bona fide DNA topoisomerase. To accurately gauge the native size of DraTopIB, we mixed an aliquot of the phosphocellulose DraTopIB preparation with marker proteins BSA (66 kDa), soybean trypsin inhibitor (22 kDa), and cytochrome c (12 kDa) and then sedimented the mixture in a glycerol gradient. The 40-kDa DraTopIB polypeptide sedimented between BSA and soybean trypsin inhibitor (not shown). This result suggests that DraTopIB is a monomeric enzyme.
Figure 2.
Purification and supercoil relaxation activity of DraTopIB. (A) Aliquots (15 μl) of the soluble bacterial lysate (L), the phosphocellulose flow-through (FT), the 0.1 and 0.35 M NaCl washes (first two NaCl “steps”), and sequential fractions from the 1 M NaCl elution (top NaCl step) were analyzed by SDS/PAGE. The gel was stained with Coomassie blue dye. The positions and sizes (in kDa) of marker polypeptides are indicated on the left. The DraTopIB polypeptide is denoted by the arrow at right. (B) Glycerol gradient sedimentation. Aliquots (20 μl) of the odd-numbered gradient fractions were analyzed by SDS/PAGE. An aliquot of the phosphocellulose load fraction is included in lane L. (C) DNA relaxation reaction mixtures containing 50 mM Tris⋅HCl (pH 7.5), 2.5 mM EDTA, 100 mM NaCl, 0.3 μg of supercoiled pUC19 DNA, and 0.5 μl of the indicated gradient fractions were incubated for 2 h at 37°C. The DNA products were analyzed by agarose gel electrophoresis. A photograph of the ethidium bromide-stained gel is shown; the positions of supercoiled (S) and relaxed (R) circular DNAs are indicated on the left. (D) Stimulation of relaxation by potassium glutamate. Reaction mixtures containing 50 mM potassium phosphate buffer (pH 8.0), 0.3 μg of pUC19 DNA, 350 ng of DraTopIB, and 5, 10, 20, 50, 100, or 200 mM potassium glutamate were incubated for 15 min at 37°C. DraTopIB was omitted from the control reaction in lane −.
The topoisomerase activity was characterized by using the peak glycerol gradient enzyme fraction. Initially we had observed that addition of NaCl or magnesium stimulated relaxation by DraTopIB (not shown). We found subsequently that potassium glutamate was superior to NaCl and all other solutes tested in its stimulatory effect. Whereas activity was virtually nil at 0–20 mM glutamate, the addition of 50 and 100 mM glutamate stimulated supercoil relaxation incrementally, and 200 mM glutamate resulted in quantitative relaxation of the input plasmid DNA (Fig. 2D). The relaxation activity of DraTopIB at pH 8.0 in 200 mM glutamate was optimal between 23°C and 45°C. No activity was observed at 2°C or at ≥50°C (not shown). The pH optimum in 50 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane (BTP) buffer was between pH 6.5 and 8.0. No activity was detected in BTP buffer between pH 8.5 and 10 or in Mes buffer below pH 6.5 (not shown). DNA relaxation by DraTopIB was unaffected by 100 μM camptothecin (not shown).
Active-Site Tyrosine of DraTopIB.
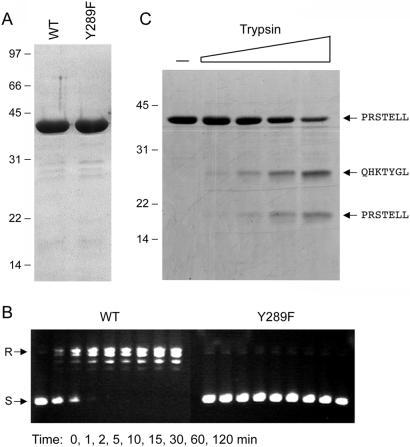
The active-site tyrosine of eukaryotic topo IB is located within a conserved peptide motif (SKXXY) in helix α8 of the catalytic domain (Fig. 1). A similar motif (TRXXY289) is found in DraTopIB. To test the role of Tyr-289 in catalysis by DraTopIB, we mutated this residue to phenylalanine. The recombinant Y289F protein was purified by phosphocellulose chromatography and glycerol gradient sedimentation (Fig. 3A). A kinetic analysis of DNA relaxation by wild-type DraTopIB and the Y289F mutant is shown in Fig. 3B. Whereas wild-type DraTopIB relaxed the pUC19 plasmid to completion in 5 min, an equivalent amount of Y289F had no effect on DNA topology during a 2-h reaction. Indeed, the Y289F mutant elicited no changes in substrate topology when the incubation was extended for up to 2 d (not shown). We conclude from these results that: (i) The observed topoisomerase activity of the wild-type DraTopIB preparation is intrinsic to the recombinant DraTopIB protein; (ii) the hydroxyl moiety of Tyr-289 is required absolutely for topoisomerase activity and likely serves as the nucleophile in the DNA transesterification reaction; and (iii) DraTopIB proteins resemble the poxvirus and nuclear type IB topoisomerases with respect to the location and sequence context of the active site tyrosine.
Figure 3.
Probing the mechanism and structure of DraTopIB. (A) Aliquots (5 μg) of wild-type DraTopIB and the Y289F mutant were analyzed by SDS/PAGE. A Coomassie blue-stained gel is shown. (B) Relaxation reaction mixtures containing (per 20 μl) 50 mM potassium phosphate (pH 8.0), 200 mM potassium glutamate, 0.3 μg of pUC19, and 350 ng of enzyme were incubated at 37°C. Aliquots (20 μl) were withdrawn at the times specified and quenched immediately with SDS. The DNA products were analyzed by agarose gel electrophoresis. (C) Mixtures (20 μl) containing 50 mM Tris⋅HCl (pH 7.5), 4 μg of wild-type DraTopIB, and 0, 10, 20, 40, or 80 ng of trypsin were incubated on ice for 15 min. The digestion products were resolved by SDS/PAGE. The gel contents were electroblotted to a poly(vinylidene difluoride) membrane. A picture of the Coomassie blue-stained membrane is shown. The amino-terminal amino acid sequences of intact DraTopIB and the two major tryptic fragments were determined by automated Edman chemistry and are indicated on the right.
Structure Probing by Limited Proteolysis.
Recombinant DraTopIB was subjected to proteolysis with increasing amounts of trypsin (Fig. 3C). Sequencing of the undigested DraTopIB polypeptide confirmed that its amino terminus (PRSTELL) corresponded to that of the recombinant gene product starting from the second residue of the protein. Although there are 53 potential cleavage sites for trypsin in DraTopIB, the protease appeared to cleave preferentially at a single site, yielding two polypeptide fragments of ≈25 kDa and ≈18 kDa (Fig. 1). Sequencing of the tryptic cleavage products revealed that the smaller species originated from the amino terminus of DraTopIB and the larger fragment arose from cleavage between amino acids Arg-145 and Gln-146 (Fig. 3C).
The trypsin-accessible site in DraTopIB (denoted by • in Fig. 1) aligns to a protease-sensitive segment of the catalytic domain of vaccinia topoisomerase located between helices α2 and α3, which is disordered in the crystal structure (11, 22). The trypsin-sensitive site of vaccinia topoisomerase (Lys-135, indicated by + in Fig. 1) is located 3 aa upstream of the trypsin-sensitive site of DraTopIB, suggesting that the fold of the bacterial topoisomerase is similar to that of the poxvirus enzyme. The disordered segment of poxvirus topoisomerase that punctuates the catalytic domain includes residues Arg-130 and Tyr-136, which are important for DNA cleavage by vaccinia topoisomerase and are conserved in all of the bacterial topo IB proteins (Fig. 1). Whereas the Arg-130 residue of vaccinia topoisomerase is found in all eukaryotic topo IB and tyrosine recombinase superfamily members, the Tyr-136 position is uniquely conserved in the poxvirus and bacterial proteins. Such local sequence identities can provide clues to the evolutionary links among subgroups of the DNA strand transferase superfamily (discussed below).
Sedimentation analysis of the tryptic digestion products under native conditions showed that the two proteolytic fragments cosedimented at a position similar to that of the intact DraTopIB; however, the fractions containing the cosedimenting fragments evinced no DNA relaxation activity (not shown). We surmise that scission of the polypeptide backbone between Arg-145 and Gln-146 did not disrupt the global tertiary structure but did affect the disposition of nearby catalytic residues. These findings for DraTopIB recapitulate faithfully the effects of breaking the backbone of vaccinia topoisomerase with chymotrypsin between Tyr-136 and Leu-137 (22). DraTopIB was not cleaved by trypsin at a position equivalent to the other major tryptic site in vaccinia topoisomerase (Arg-80) that demarcates the boundary between the amino-terminal and the carboxyl-terminal structural domains. We attribute this difference to the fact that DraTopIB has a glycine (Gly-87) at the position corresponding to Arg-80 of the vaccinia protein.
Conservation of the Active Site in Bacterial and Poxvirus topo IB.
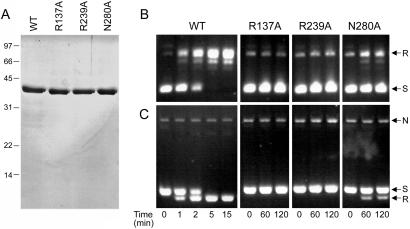
Four of five of the constituents of the catalytic pentad of vaccinia topoisomerase (Arg-130, Lys-167, Lys-220, and Arg-223) are identical in DraTopIB (Arg-137, Lys-174, Lys-236, and Arg-239). The fifth catalytic residue of the vaccinia pentad (His-265) is replaced by an asparagine in DraTopIB (Asn-280). To address whether the active site and catalytic mechanism of DraTopIB is similar to that of vaccinia topoisomerase, we introduced alanine substitutions for DraTopIB residues Arg-137, Arg-239, and Asn-280 and gauged their effects on topoisomerase activity. The recombinant R137A, R239A, and N280A proteins were produced in E. coli and purified from soluble lysates by phosphocellulose chromatography and glycerol gradient sedimentation (Fig. 4A). A kinetic analysis of relaxation by equivalent amounts of the enzymes is shown in Fig. 4B. Whereas wild-type DraTopIB relaxed the plasmid DNA to completion in 5 min, the R137A and R239A mutants were inactive over the course of a 2-h incubation. The N280A mutant displayed a low level of relaxation (Fig. 4B). A parallel analysis of the reaction products by electrophoresis in the presence of ethidium bromide resolved the supercoiled, nicked, and relaxed forms of circular pUC19 DNA (Fig. 4C). This experiment showed that: (i) the wild-type DraTopIB converted the negatively supercoiled plasmid DNA exclusively into relaxed covalently closed circles (i.e., there was no detectable DNA endonuclease activity associated with DraTopIB); (ii) the R137A and R239A mutants were effectively inert; and (iii) the N280A mutations diminished, but did not abolish, the topoisomerase activity. Limited digestion of the purified Arg-137, Arg-239, and Asn-280 mutants with trypsin revealed the identical pattern of two proteolytic fragments observed for wild-type DraTopIB (not shown), from which we surmise that the alanine substitutions did not cause a global disruption of the protein fold. The mutational effects at DraTopIB positions Arg-137, Arg-239, and Asn-280 agree well with the mutational data for vaccinia topoisomerase, whereby alanine substitutions for the conserved arginines reduced the transesterification rate by a factor of 10−5 (effectively abolishing DNA relaxation) and the H265A change reduced activity by a factor of 10−2. Activity of vaccinia topoisomerase was restored to near wild-type levels when His-265 was replaced by asparagine (3), which effectively installs the catalytic pentad of DraTopIB. We infer from these results that the active site and catalytic mechanism of DraTopIB are similar to those of vaccinia topoisomerase.
Figure 4.
Defining the active site of DraTopIB. (A) The indicated DraTopIB proteins (2 μg) were analyzed by SDS/PAGE. A Coomassie blue-stained gel is shown. (B and C) Relaxation reaction mixtures containing (per 20 μl) 50 mM potassium phosphate (pH 8.0), 200 mM potassium glutamate, 0.3 μg of pUC19 DNA, and 350 ng of enzyme were incubated at 37°C. Aliquots (20 μl) were withdrawn at the times specified and quenched immediately with SDS. One-half of each sample was analyzed by agarose gel electrophoresis, after which the gels were stained with ethidium bromide (B). The other one-half of each sample was electrophoresed through an agarose gel containing 0.5 μg/ml ethidium bromide (C). The positions of supercoiled (S), relaxed (R), and nicked (N) circular DNAs are indicated on the right.
Discussion
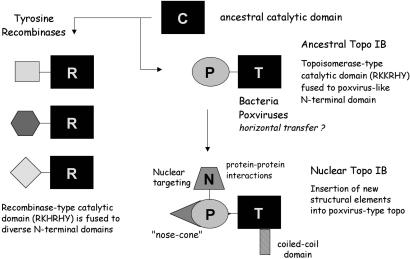
Here we have shown that diverse species of bacteria encode a type IB topoisomerase that resembles poxvirus DNA topoisomerase. Biochemical analysis of DraTopIB showed it to be capable of relaxing negatively supercoiled DNA in the absence of divalent cations or a high-energy cofactor. DraTopIB also relaxed positive supercoils introduced by the addition of ethidium bromide to relaxed covalently closed circular DNA (unpublished data). These properties are characteristic of the type IB topoisomerase family. Like vaccinia topoisomerase (23), DraTopIB has a compact size, is a monomer in solution, and is resistant to camptothecin, a cytotoxic plant alkaloid that poisons the relaxation activity of nuclear topo IB. A distinctive feature of vaccinia topoisomerase is its specificity for cleavage at the pentapyrimidine sequence (C/T)CCTT↓ (28). DraTopIB does not share this sequence specificity, insofar as the recombinant DraTopIB protein did not form a covalent adduct on synthetic CCCTT-containing duplexes that are cleaved with high efficiency by vaccinia topoisomerase (unpublished data). Nonetheless, the active site of DraTopIB is composed of a constellation of catalytic side chains similar to that of the vaccinia enzyme, with the catalytic residues being located at equivalent positions in the primary structures. These findings suggest a close evolutionary relationship between the poxvirus and bacterial enzymes, and they engender a plausible scheme for the evolution of topoisomerase IB and tyrosine recombinases from a common ancestor (Fig. 5).
Figure 5.
Evolution of the topo IB family. See text for details.
The similar folds and active sites of the catalytic domains of topo IB and tyrosine recombinases indicate that they derive from a progenitor strand transferase that acted via a tyrosyl-3′-phosphodiester intermediate. We envision that the ancestral enzyme was a “stand-alone” catalytic domain (“C” in Fig. 5) analogous to the carboxyl-terminal domains of present-day tyrosine recombinases or poxvirus topoisomerases. The ancestor C domain would have contained the tyrosine nucleophile and the four essential side chains (equivalent to Arg-130, Lys-167, Arg-223, and His-265 of vaccinia topo IB) that function in transition state stabilization and general acid catalysis. The fact that the catalytic repertoire of present day topo IB and tyrosine recombinases embraces transesterification at a ribonucleotide phosphodiester, resulting in either formation of a 2′,3′-cyclic phosphodiester that can serve as a substrate for topoisomerase-mediated ligation to another polynucleotide (24, 25) or formation of a 3′ phosphate (26), raises the prospect that an ancestral C domain predated the emergence of DNA-based genomes. Indeed, it is attractive to think that a world based on RNA and protein would devise a means to recombine RNA strands by a topoisomerase-like mechanism that eschews a requirement for a high-energy cofactor.
The discovery of type IB topoisomerases in bacteria overcomes earlier difficulties in tracing a connection between tyrosine recombinases and topo IB, i.e., because their distributions appeared not to overlap much until now. We posit that the two families evolved by: (i) differentiation of the C domain into recombinase-type (R) and topoisomerase-type (T) catalytic domains and (ii) the acquisition by gene fusion of distinct amino-terminal domain modules that impart biological specificity. The R and T catalytic domains are distinguished by active sites composed of RKHRH vs. RKKRH pentad arrays. (The His vs. Lys status of the third position of the pentad in the ancestral enzyme cannot be surmised.) The structural diversity of the amino-terminal domains among the various tyrosine recombinases argues that they evolved by independent gene fusion events that captured different amino-terminal modules.
In contrast, we envision that the bacterial, poxviral, and nuclear branches of the topo IB family evolved serially, rather than in parallel as invoked for the recombinases. We propose that the ancestral topo IB arose by fusion of the catalytic T domain to a small poxvirus-like amino-terminal domain (P) (Fig. 5). The P domain of vaccinia topoisomerase is structurally similar to the amino-terminal domains of the bacterial topo IB group (Fig. 1). It seems probable that the initial split into recombinase and topoisomerase families occurred in the bacterial domain of life. Whether a once-ubiquitous bacterial topo IB was lost from some bacterial species or whether the earliest topo IB was dispersed among present bacterial species by horizontal transfer (perhaps by a virus) is not clear. We suspect that horizontal gene transfer did play a key role in propelling the topo IB proteins into the eukaryal domain.
Indeed, a novel feature of the scheme we propose is that nuclear topoisomerase I enzymes evolved from a bacterial/poxvirus-like precursor, rather than the other way around, as suggested previously (17). Human topoisomerase I, an exemplary nuclear topo IB, is a 765-aa polypeptide; it is significantly larger than vaccinia topoisomerase (314 aa) or DraTopIB (346 aa). Nuclear topo IB contains multiple discrete structural modules that appear to have been added to a protein scaffold that is otherwise quite similar to that of the vaccinia protein (6, 11, 17). In human topoisomerase I, a coiled-coil domain has been inserted into the T domain at a position between helices α7 and α8 of vaccinia topoisomerase (Fig. 5). A large amino-terminal addition to the P-like segment of human topoisomerase has occurred upstream of what would be the amino terminus of the vaccinia or DraTopIB proteins. The amino-terminal embellishments of nuclear topo IB include a large hydrophilic segment (N in Fig. 5) responsible for nuclear targeting and for protein–protein interactions with other nuclear components (2). The N segment would obviously not be relevant to bacterial topo IB or the topoisomerases of cytoplasmic poxviruses. The amino-terminal portion of human topoisomerase I also contains a pair of helices that extend like a nose cone over the DNA duplex; this nose cone structure is not present in the poxvirus topoisomerases. It now seems simpler to posit that the core components of bacterial/poxvirus enzymes were supplemented by fusions and insertions during eukaryotic evolution than to invoke a process of elimination from the nuclear topoisomerase I of all but the catalytic nuts and bolts to yield a stripped-down topoisomerase common to poxviruses and the diverse bacteria described here.
The discovery of a family of bacterial topoisomerases raises interesting questions about their physiological role, their connections to known pathways of DNA replication, repair, and recombination, and their functional relationship to other topoisomerases resident in bacteria. Makarova et al. (27) in their recent review of the D. radiodurans genome cite their unpublished findings that a strain deleted for the DraTopIB gene is sensitized to UV, but not to ionizing radiation. It will be of interest to evaluate whether and how the catalytic activity of DraTopIB is connected to its apparent function in UV damage—e.g., by eventually testing the in vivo repair activity of the catalytically defective proteins characterized here. Given that topo IB proteins are present in many bacteria, it is conceivable that the requirement (or lack thereof) for topo IB for bacterial growth may vary from one species to another, just as it does for eukaryotic nuclear topo IB (2).
It is instructive to compare the repertoire of topoisomerases in bacteria that encode a type IB topoisomerase to that of an exemplary bacterium like E. coli that lacks a poxvirus-like topo IB polypeptide. E. coli has four topoisomerases that fall into two mechanistically distinct families (2). Topo I and topo III are members of the type IA family of monomeric topoisomerases that incise single strands and form a covalent tyrosyl-5′-phosphodiester intermediate. Topo II (DNA gyrase, a tetramer of GyrA and GyrB subunits) and topo IV (a tetramer of ParE and ParC subunits) are members of the type IIa family of ATP-dependent topoisomerases, which have two active sites that make staggered double-strand DNA breaks and form covalent tyrosyl-5′-phosphodiester intermediates. E. coli also has Xer recombinases, which we classify mechanistically as type IB strand transferases. We searched the completed proteomes of four bacteria that encode members of the type IB topoisomerase group described here (D. radiodurans, Pseudomonas aeruginosa, Sinorhizobium meliloti, and Agrobacterium tumefaciens) for polypeptides related to the standard E. coli enzymes. The salient findings were as follows: (i) All four topo IB+ species encode a homolog of Xer recombinase, from which we infer that topo IB is not simply replacing Xer function in resolving multimers of the bacterial chromosome; (ii) the topo IB+ bacteria all have a single type IA enzyme homologous to E. coli topoisomerase I; and (iii) none of the four topo IB+ bacteria encodes a type IA enzyme homologous to E. coli topoisomerase III. The correlation between the presence of topo IB and the lack of topoisomerase III is provocative and raises the prospect that these two enzymes might be either functionally redundant or functionally incompatible in the bacterial cell. Analysis of additional topo IB+ bacterial proteomes as they are completed will clarify how well this correlation holds.
The pharmacological properties of the bacterial and poxvirus topoisomerases are distinct from those of the eukaryotic nuclear enzyme with respect to camptothecin sensitivity. Thus, it is worth considering the bacterial/poxvirus type IB topoisomerases as targets for antibiotic and antiviral drug discovery, especially the discovery of new topoisomerase poisons specific for this branch of the topo IB family. Among the bacteria that encode poxvirus-like type IB topoisomerases are Mycobacterium avium and Pseudomonas aeruginosa, which are human pathogens that cause significant morbidity and are difficult to treat. Novel antipoxvirus drug targets are also a pressing issue, given the very real concern that smallpox might be used as a bioterror weapon against an unvaccinated population.
Acknowledgments
We thank Mike Cox for a starter D. radiodurans culture and instructions for the preparation of genomic DNA. This research was supported by National Institutes of Health Grant GM46330.
Abbreviations
- topo IB
DNA topoisomerase type IB
- DraTopIB
Deinococcus radiodurans topo IB
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Shuman S. Biochim Biophys Acta. 1998;1400:321–337. doi: 10.1016/s0167-4781(98)00144-4. [DOI] [PubMed] [Google Scholar]
- 2.Champoux J J. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Petersen B Ø, Shuman S. J Biol Chem. 1997;272:3891–3896. doi: 10.1074/jbc.272.7.3891. [DOI] [PubMed] [Google Scholar]
- 4.Wittschieben J, Shuman S. Nucleic Acids Res. 1997;25:3001–3008. doi: 10.1093/nar/25.15.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng C, Wang L K, Sekiguchi J, Shuman S. J Biol Chem. 1997;272:8263–8269. doi: 10.1074/jbc.272.13.8263. [DOI] [PubMed] [Google Scholar]
- 6.Redinbo M R, Stewart L, Kuhn P, Champoux J J, Hol W G J. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 7.Stivers J T, Jagadeesh G J, Nawrot B, Stec W J, Shuman S. Biochemistry. 2000;39:5561–5572. doi: 10.1021/bi992429c. [DOI] [PubMed] [Google Scholar]
- 8.Krogh B O, Shuman S. Mol Cell. 2000;5:1035–1041. doi: 10.1016/s1097-2765(00)80268-3. [DOI] [PubMed] [Google Scholar]
- 9.Nunes-Düby S E, Kwon H J, Tirumalai R S, Ellenberger T, Landy A. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo F, Gopaul D N, Van Duyne G D. Nature (London) 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 11.Cheng C, Kussie P, Pavletich N, Shuman S. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Narendra U, Iype L E, Cox M M, Rice P A. Mol Cell. 2000;6:885–897. [PubMed] [Google Scholar]
- 13.Kwon H J, Tirumalai R, Landy A, Ellenberger T. Science. 1997;276:126–131. doi: 10.1126/science.276.5309.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hickman A B, Waninger S, Scocca J J, Dyda F. Cell. 1997;89:227–237. doi: 10.1016/s0092-8674(00)80202-0. [DOI] [PubMed] [Google Scholar]
- 15.Subramanya H S, Arciszewska L K, Baker R A, Bird L E, Sherratt D J, Wigley D B. EMBO J. 1997;16:5178–5187. doi: 10.1093/emboj/16.17.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma A, Hanai R, Mondragon A. Structure (London) 1994;2:767–777. doi: 10.1016/s0969-2126(94)00077-8. [DOI] [PubMed] [Google Scholar]
- 17.Redinbo M R, Champoux J J, Hol W G J. Curr Opin Struct Biol. 1999;9:29–36. doi: 10.1016/s0959-440x(99)80005-0. [DOI] [PubMed] [Google Scholar]
- 18.Belova G I, Prasad R, Kozyavkin S A, Lake J A, Wilson S H, Slesarev A I. Proc Natl Acad Sci USA. 2001;98:6015–6020. doi: 10.1073/pnas.111040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekiguchi J, Shuman S. EMBO J. 1996;15:3448–3457. [PMC free article] [PubMed] [Google Scholar]
- 20.Sekiguchi J, Shuman S. Nucleic Acids Res. 1997;25:3649–3656. doi: 10.1093/nar/25.18.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 22.Sekiguchi J, Shuman S. J Biol Chem. 1995;270:11636–11645. doi: 10.1074/jbc.270.19.11636. [DOI] [PubMed] [Google Scholar]
- 23.Shuman S, Golder M, Moss B. J Biol Chem. 1988;263:16401–16407. [PubMed] [Google Scholar]
- 24.Sekiguchi J, Shuman S. Mol Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 25.Shuman S. Mol Cell. 1998;1:741–748. doi: 10.1016/s1097-2765(00)80073-8. [DOI] [PubMed] [Google Scholar]
- 26.Xu C, Grainge I, Lee J, Harshey R M, Jayaram M. Mol Cell. 1998;1:729–739. doi: 10.1016/s1097-2765(00)80072-6. [DOI] [PubMed] [Google Scholar]
- 27.Makarova K S, Aravind L, Wolf Y I, Tatusov R L, Minton K W, Koonin E V, Daly M J. Microbiol Mol Biol Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuman S, Prescott J. J Biol Chem. 1990;265:17826–17836. [PubMed] [Google Scholar]