Abstract
The histidine biosynthetic pathway is an ancient one found in bacteria, archaebacteria, fungi, and plants that converts 5-phosphoribosyl 1-pyrophosphate to l-histidine in 10 enzymatic reactions. This pathway provided a paradigm for the operon, transcriptional regulation of gene expression, and feedback inhibition of a pathway. l-histidinol dehydrogenase (HisD, EC 1.1.1.23) catalyzes the last two steps in the biosynthesis of l-histidine: sequential NAD-dependent oxidations of l-histidinol to l-histidinaldehyde and then to l-histidine. HisD functions as a homodimer and requires the presence of one Zn2+ cation per monomer. We have determined the three-dimensional structure of Escherichia coli HisD in the apo state as well as complexes with substrate, Zn2+, and NAD+ (best resolution is 1.7 Å). Each monomer is made of four domains, whereas the intertwined dimer possibly results from domain swapping. Two domains display a very similar incomplete Rossmann fold that suggests an ancient event of gene duplication. Residues from both monomers form the active site. Zn2+ plays a crucial role in substrate binding but is not directly involved in catalysis. The active site residue His-327 participates in acid-base catalysis, whereas Glu-326 activates a water molecule. NAD+ binds weakly to one of the Rossmann fold domains in a manner different from that previously observed for other proteins having a Rossmann fold.
Histidine biosynthesis is an ancient pathway found in bacteria, archaebacteria, fungi, and plants and is one of the most extensively studied biochemically and genetically (1–3). Early studies of the genetic structure of this pathway contributed to formulation of several key concepts in molecular biology: the operon hypothesis (4–6), transcriptional regulation of gene expression (reviewed in ref. 7), and feedback inhibition of a pathway (8). The pathway commences with condensation of ATP to form 5′-phosphoribosyl-ATP and encompasses a total of 10 chemical steps to achieve the synthesis of l-histidine. In Salmonella typhimurium and Escherichia coli, these reactions are carried out by products of eight genes, all of which are found on a single his operon (1, 3). Three of these enzymes, HisI, HisB, and HisD, are bifunctional, HisF and HisH form a monofunctional heterodimer, and the remaining enzymes are monofunctional. The organization of the his operon differs somewhat between organisms. For example, in Saccharomyces cerevisiae, some of these genes are fused: hisI and hisD are coded by his4 and hisF and hisH, by his7.
The hisD gene encodes the enzyme l-histidinol dehydrogenase (HisD, EC 1.1.1.23) that catalyzes the last two steps in this pathway. This bifunctional enzyme converts l-histidinol to l-histidine through a l-histidinaldehyde intermediate (Fig. 1). The l-histidinaldehyde molecule is not released from the enzyme during catalysis but undergoes immediate conversion to the final product (9). Like other enzymes of the l-histidine synthesis pathway, the sequence of HisD has been well conserved during evolution from bacteria to fungi to plants. As HisD is absent from mammals, it has become an attractive target for inhibition as part of herbicide development (10, 11).
Figure 1.
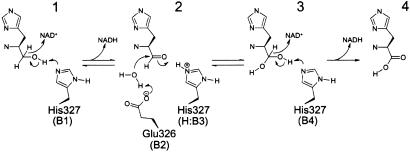
Reactions catalyzed by HisD (after ref. 14). The structure allowed identification of residues Glu-326 as being base B2 and His-327 as B1, B3, and B4. Glu-326 is responsible for the activation of a water molecule that will attack the reactive carbon in step 2 of the reaction mechanism.
HisD is a homodimeric zinc metalloenzyme with one Zn2+-binding site per monomer (12). The Zn2+ ion is essential for enzymatic activity, although other metals such as Mn2+ or Cd2+ can replace Zn2+ in forming an active enzyme (13). Biochemical and mutagenesis studies on HisD from S. typhimurium (12, 14) and Brassica oleracea (15) have identified two histidine residues, corresponding to His-262 and His-419 from E. coli, as part of the active site, although the precise role of His-262 has not been established (14–16). NMR measurements indicated that the substrate participates in Zn2+ coordination through its imidazole ND1 and alcohol oxygen atoms (16), and Zn2+ has been proposed to participate directly in catalysis (14, 16). However, the NAD+-binding site has not been identified. Overall, the enzyme catalyzes a four-electron oxidation reaction. A catalytic mechanism has been proposed for the conversion of l-histidinol to l-histidine, which involves two consecutive oxidation reactions accompanied by a reduction of two NAD+ molecules according to a Bi-Uni-Uni-Bi kinetic mechanism (17–19). On the basis of the available data, a catalytic mechanism has been proposed (14) (Fig. 1).
Although the chemical steps within the histidine pathway are well documented, detailed knowledge of the catalytic machinery at the molecular level is known for only three of them. HisF and its paralog HisA have a triosephosphate isomerase (TIM barrel) fold (20, 21), whereas HisC is structurally similar to aspartate aminotransferases (22, 23). Here we present the crystal structure of E. coli HisD in its apo form and complexed with Zn2+, l-histidinol, and NAD+. The structures provide a detailed view of the active site, allow the assignment of catalytic roles to specific residues, show a mode of NAD+ binding, and implicate domain swapping in dimerization. In addition, an early event of gene duplication is suggested from the structure.
Materials and Methods
Cloning, Expression, and Purification of E. coli HisD.
The hisD gene was amplified by PCR from E. coli strain MC1061 genomic DNA and cloned into a modified pGEX-4T1 vector (Amersham Pharmacia) as an in-frame N-terminal fusion with Schistosoma japonicum glutathione S-transferase followed by a thrombin cleavage site. The full-length construct after protease cleavage consists of all 434 residues of HisD, with an additional three residues, Gly-Ser-His, at the N terminus resulting from thrombin cleavage. Plasmid DNA was transformed into the strain DL41 and grown in LeMaster medium (24) containing 25 mg/liter of l-selenomethionine and 100 μg/ml of ampicillin at 37°C overnight. The culture was diluted 10-fold into the same medium and grown for an additional 2 h. After transfer to 22°C, protein production was induced by addition of 100 μM isopropyl β-D-thiogalactoside (Sigma).
Cells were harvested by centrifugation (4,000 × g, 4°C, 20 min) and resuspended in 40 ml of lysis buffer [50 mM Tris⋅Cl, pH 7.5/0.4 M NaCl/1 mM EDTA/1% (vol/vol) Triton X-100/5% (vol/vol) glycerol/20 mM DTT] and protease inhibitor mixture (Complete, Roche Diagnostics). Cells were disrupted by sonication (5 cycles, each 30 s on, then 30 s off, 4°C; Heat Systems/Ultrasonics) and the lysate cleared by centrifugation (150,000 × g, 4°C, 45 min). The protein supernatant was passed through a 5-ml DEAE–Sepharose column (Amersham Pharmacia) and the flow-through loaded on a 5-ml glutathione Sepharose 4B column (Amersham Pharmacia). The column was washed with 4 bed volumes of buffer [50 mM Tris⋅Cl, pH 7.5/1 M NaCl/1 mM EDTA/1% (vol/vol) Triton X-100/5% (vol/vol) glycerol] followed by 3 bed volumes of 50 mM Tris⋅Cl, pH 7.5/0.2 M NaCl/5% (vol/vol) glycerol. The glutathione S-transferase tag was removed by incubating the beads with 100 μg of α-thrombin (Hematologic Technologies, Essex Junction, VT) for 75 min at 22°C. HisD protein was eluted from the column and run as a single band of ≈46 kDa by SDS/PAGE. Protein estimation was determined by the method of Bradford (25) by using BSA as a standard.
Crystallization of HisD.
The protein was concentrated by ultrafiltration to 15.4 mg/ml and the buffer changed to 20 mM Tris⋅Cl, pH 7.5/0.2 M NaCl/5 mM DTT/1 mM l-histidine. Dynamic light-scattering measurements (DynaPro 801, Protein Solutions, Charlottesville, VA) indicate that HisD forms dimers in solution, consistent with previous velocity sedimentation experiments (26). Initial crystals were obtained by sparse-matrix screening (Hampton Research, Riverside, CA) by hanging drop vapor diffusion. The best crystals grew at 18°C from droplets containing 2 μl of protein solution and 4 μl of reservoir solution [20% wt/vol polyethylene glycol 3350/7% (vol/vol) glycerol/0.1 M imidazole–malic acid buffer, pH 5.5/0.2 M (NH4)2SO4] and appeared within 2 weeks. Macroseeding was used to obtain larger crystals, typically of dimensions 0.2 × 0.3 × 0.3 mm3. The crystals were orthorhombic, space group P212121 with cell dimensions a = 54.3, b = 107.4, c = 157.0 Å, and two molecules in the asymmetric unit. Because imidazole is an inhibitor of HisD (27), crystals were transferred to 0.1 M sodium acetate buffer, pH 5.5, for preparation of enzyme complexes.
Data Collection, Multiwavelength Anomalous Diffraction Phasing, and Refinement.
Crystals were soaked for ≈30 s in a cryoprotectant solution consisting of reservoir solution supplemented with 28% wt/vol polyethylene glycol 3350 and 20% (vol/vol) glycerol, picked up in a rayon cryo-loop (Hampton Research), and flash-cooled in either liquid nitrogen or in a stream of nitrogen gas at 100 K (Oxford Cryosystems, Oxford). Diffraction data were collected on a Quantum-4 charge-coupled device detector (ADSC, San Diego) at beamline X8C, National Synchrotron Light Source, Brookhaven National Laboratory. In-house diffraction data were collected on an R-axis IIC detector mounted on a RU-300 rotating anode generator (Rigaku, Tokyo). Data indexing, merging, and scaling were performed by using the hkl package (28). Data collection and processing statistics are listed in Table 1.
Table 1.
Statistics for data collection, processing, and refinement
| Data set* | Inflection | Peak | Remote | Apo | l-Histidinol† NAD+, Zn2+ | l-Histidine‡ NAD+, Zn2+ | Histamine§ NAD+, Zn2+ |
|---|---|---|---|---|---|---|---|
| Wavelength, Å | 0.97950 | 0.97934 | 0.97857 | 0.97950 | 0.97945 | 0.97945 | 0.97945 |
| Exposure time, sec | 30 | 30 | 30 | 30 | 20 | 30 | 30 |
| Oscillation, ° | 1.0 | 1.0 | 1.0 | 1.0 | 0.5 | 1.0 | 1.0 |
| Detector distance, mm | 180.0 | 180.0 | 180.0 | 165.0 | 160.0 | 180.0/220.0 | 200.0 |
| No. of frames | 179 | 179 | 180 | 116 | 240 | 240 | 231 |
| Resolution limits, Ŷ | 40.0–2.20 | 40.0–2.20 | 40.0–2.20 | 40.0–1.75 | 40.0–1.70 | 40.0–2.10 | 40.0–2.10 |
| (2.28–2.20) | (2.28–2.20) | (2.28–2.20) | (1.78–1.75) | (1.76–1.70) | (2.18–2.10) | (2.18–2.10) | |
| I/σ(I) after merging¶ | 27.5 (18.6) | 28.6 (19.0) | 26.3 (17.0) | 16.4 (2.2) | 13.0 (1.6) | 15.3 (2.9) | 20.2 (4.0) |
| Completeness, %¶ | 100 (100) | 100 (100) | 100 (100) | 97.3 (79.7) | 92.3 (64.6) | 95.2 (91.3) | 95.2 (91.3) |
| Rsym¶ | 0.067 (0.124) | 0.065 (0.120) | 0.063 (0.129) | 0.060 (0.276) | 0.051 (0.365) | 0.076 (0.342) | 0.076 (0.342) |
| No. of reflections | 342,567 | 337,518 | 344,084 | 402,344 | 425,276 | 372,451 | 366,062 |
| No. of unique reflections | 47,187 | 47,000 | 47,202 | 91,330 | 182,767‖ | 53,978 | 52,600 |
| No. of atoms | — | — | — | 7,391 | 7,249 | 6,468 | 6,462 |
| No. of waters | — | — | — | 927 | 626 | 0 | 0 |
| R | — | — | — | 19.0 (24.4) | 21.3 (36.9) | 25.2 (31.5) | 24.8 (26.6) |
| Rfree | — | — | — | 22.5 (26.2) | 24.1 (37.9) | 28.6 (34.6) | 28.1 (30.7) |
| B factor | — | — | — | 23.4 | 25.9 | 36.2 | 34.2 |
| rms deviation bonds, Å | — | — | — | 0.013 | 0.015 | 0.006 | 0.006 |
| rms deviation bond angles, ° | — | — | — | 1.5 | 1.7 | 1.3 | 1.2 |
Multiwavelength anomalous diffraction and high-resolution apo datasets were collected from the same crystal. All datasets were collected from SeMet-labeled protein crystals.
1 mM ZnCl2, 20 mM l-histidinol, 50 mM NAD+, 2-day soaking.
2 mM ZnCl2, 20 mM l-histidine, 50 mM NAD+, 2-day soaking.
2 mM ZnCl2, 20 mM l-histamine, 50 mM NAD+, 2-day soaking.
Numbers in parentheses refer to the highest-resolution shell.
Processed and refined as anomalous data.
Multiwavelength anomalous diffraction data from a Se-Met-labeled crystal of apo HisD were collected to 2.2-Å resolution at three wavelengths. All 12 expected Se sites in the asymmetric unit were found by using solve (29), and phases were calculated to 2.3 Å, giving an overall figure of merit (FOM) of 0.64. Electron density modification was applied with the program resolve (30), improving the FOM to 0.76. By using the arp/warp program (31), 54% of all residues were built automatically as polyalanines. The remainder of the model was built manually with the program o (32) into 3Fo − 2Fc and 2Fo − Fc electron density maps. The two molecules in the asymmetric unit are related by noncrystallographic 2-fold symmetry along the direction (0.987, 0.162, −0.006), nearly parallel to the crystallographic a-axis.
Refinement of the HisD model was performed with cns version 1.0 (33). Noncrystallographic symmetry (NCS) restraints were applied in the initial cycles of refinement but were removed when diffraction data to 1.7 Å were included. After each cycle of refinement, the model was manually fit to new 3Fo − 2Fc and Fo − Fc electron density maps contoured near 1σ or 3σ, respectively. Water molecules were added with the automatic procedure of arp/warp (31) or by visual inspection. Datasets from crystals soaked with various ligands, alone or in combination, were isomorphous to apo HisD crystals (Table 1). Refinement for each of these models started from the model for the apo HisD molecule. After one cycle of simulated annealing, the model was refitted to the new 3Fo − 2Fc electron density map and ligand and solvent molecules added.
Three short segments in each monomer of the refined structures are poorly defined in the electron density maps: Gly-Ser-His-Met-Ser-Phe at the N terminus and the surface regions containing the residues 26–32 and 57–62. All refined models were validated with the program procheck (34) and have good stereochemistry. The Ramachandran plot shows no outliers, and all main-chain and side-chain parameters were classified as better or inside the expected statistical values. Refinement statistics are shown in Table 1.
Results and Discussion
Overall Structure.
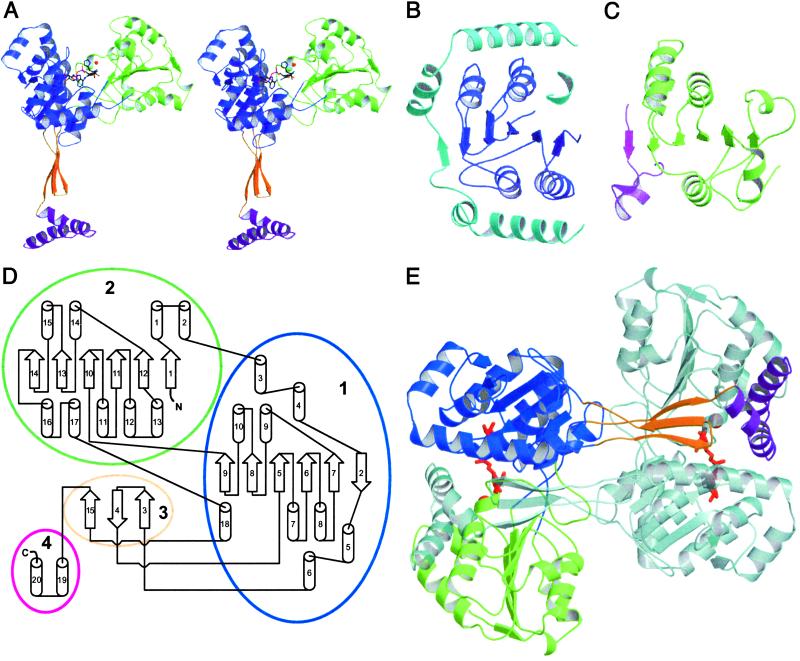
The HisD monomer consists of a globule and a tail (Fig. 2A). The globule is made of two larger domains (1 and 2) and the extending tail of two smaller domains (3 and 4). Domain 1 (residues 25–103, 124–236, and 382–386) and domain 2 (residues 1–24 and 237–381) display an α/β/α topology (Fig. 2 B–D). The cores of both domains (residues 124–236 in domain 1 and 237–381 in domain 2) adopt incomplete Rossmann folds (35) lacking the last strand-helix hairpin (Fig. 2 B and C). They are composed of five-stranded parallel β-sheets, with the first strand at the center of the sheets, and two α-helices on either side oriented nearly antiparallel to the β strands. Segment 25–103, which is noncontiguous with the remainder of domain 1, is folded into four helices arranged into two V-shaped pairs connected by a short extended linker that wrap around three sides of the core Rossmann fold (Fig. 2B). The V-shaped pairs of helices mostly make hydrophobic contacts with the helices of the core segment, whereas the linker forms a sixth strand of the β-sheet running antiparallel to the other five strands on the opposite side of the sheet to the canonical sixth strand of the Rossmann fold. Domain 2 has an extra α-helix (α13) after the second β strand of the Rossmann fold (β11). Residues 1–24 complete this domain forming a strand-helix hairpin, with this β strand providing a sixth strand to the central β-sheet (Fig. 2C). Domain 3 (residues 104–123 and 387–396) forms a three-stranded antiparallel β-sheet (β3, β4, and β15; Fig. 2 A and D) that extends away from domain 1. Domain 4 (residues 397–434) folds into a small V-shaped two-helical hairpin (α19 and α20) that is perpendicular to the domain 3 β-sheet. Together, domains 3 and 4 have an L shape (Fig. 2A).
Figure 2.
Structure and topology of HisD. (A) Stereo view of the monomer. Domains: 1, blue; 2, green; 3, orange; 4, magenta. l-histidinol, NAD+, and the Zn2+ are shown as ball-and-sticks. (B) Domain 1. Rossmann fold shown in blue, V-shaped pairs of helices (residues 25–103) connected by a linker that forms the sixth strand are in cyan. (C) Domain 2. Rossmann fold (green) in similar orientation as B. Strand-helix hairpin completes the β-sheet (residues 1–24, magenta). (D) Topology diagram. Secondary structure elements are numbered consecutively. The chain meanders between domains in the order 2–1-3–1-2–1-3–4. (E) HisD dimer with one molecule colored as in A and the other shown in pale colors. Zn2+ atoms and NAD+ bound to each monomer (red) define the position of the active site. Prepared with MOLSCRIPT (46) and RASTER3D (47).
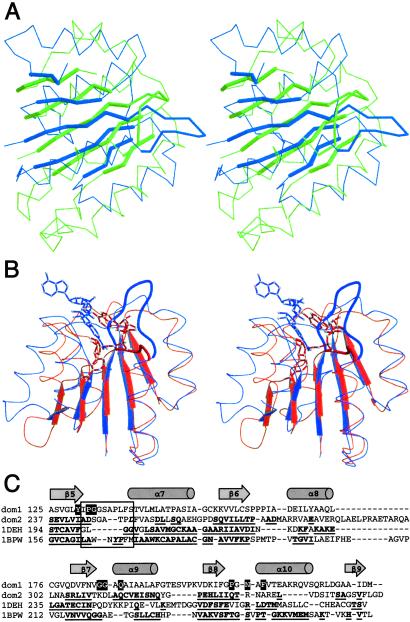
Comparison of the HisD structure to representative proteins by using dali (36) or scop (37) revealed similarity of domains 1 and 2 with other Rossmann fold-containing enzymes. However, these structure-based sequence alignments display less than 20% identity to domain 1 (Fig. 4C), explaining the inability to recognize the Rossmann fold from the sequence alone. Although the core of domain 1 belongs to a highly populated fold family (37), the entire domain 1, including the two V-shaped α helical hairpins, has no counterpart in the structural databases and is a novel tertiary arrangement. Similarly, neither domains 3 and 4 together nor the overall arrangement of domains within the monomer has been previously observed. Two other enzymes that catalyze 4-electron dehydrogenation, hydroxymethyl glutaryl-CoA reductase (38) and UDP-glucose dehydrogenase (39), do not share the same overall fold as HisD, nor do they share similarities in their catalytic mechanisms.
Figure 4.
Superposition of (A) domain 1 (blue) and domain 2 (green) of HisD performed with SPDBV (48). β strands are in thick and P loop in medium lines. (B) Domain 1 (blue) and Rossmann fold domain of human β1 alcohol dehydrogenase (orange, Protein Data Bank ID code 1DEH). The NAD+ molecules are shown in darker shades. The phosphate-binding loops are in thicker lines. (C) Structure-based sequence alignment of Rossmann folds of domains 1 and 2, β1 alcohol dehydrogenase (1DEH), and betaine aldehyde dehydrogenase (1BPW). The amino acids are in one-letter codes; bold underline letters mark residues structurally aligned to domain 1. The P loop is enclosed in a box. White letters on black background in the sequence of domain 1 indicate residues within 3.5 Å of NAD+.
Dimer Formation and Domain Swapping.
The crystal structure shows a functional homodimer in the asymmetric unit (Fig. 2E) with a large surface area, 10,240 Å2, buried on dimerization. The L-shaped arm extending from each monomer (domains 3 and 4) forms a latch-like structure that closes over domains 1* and 2* (asterisk indicates the second monomer) forming part of the substrate-binding site. The arm makes intimate contacts primarily with domain 2*, following along the depression between the domains, and acts to tie the two molecules together. There are a total of 24 polar contacts (<3.2 Å) between the arm and domain 2*. The three-stranded β-sheet of domain three packs against the six-stranded β-sheet of domain 2*, forming a single nine-stranded β-sheet (Fig. 2E). This extended arm conformation (Fig. 2A) is unlikely to exist in a single monomer during the initial folding process. The twisting around pivotal points located at the end of the β-sheet of domain 3 near domain 2 (Leu-104, Thr-121, Arg-389) would bring the arm toward domains 1 and 2 along the surface that is involved in the dimerization. Dimer formation most likely involves swapping of domains 3 and 4 between the two monomers, leading to an increase in the buried surface area. Approximately 90% of the surface area buried on dimer formation is contributed by contacts between domains 1 and 2 from one molecule and the arm from the other molecule (and their counterparts). Domain swapping on formation of oligomeric structures has been observed for several proteins (40).
Substrate and Zn2+-Binding Sites.
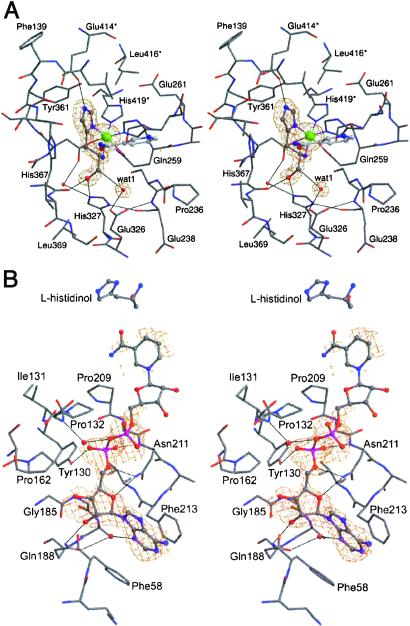
The structure of HisD complexed with Zn2+, l-histidinol, and NAD+ unequivocally identified the location of the active site (Fig. 3A). Although electron density for l-histidinol and Zn2+ was clearly defined in monomer A, the density at the same site in monomer B was interpreted as an imidazole moiety and a nearby water molecule, the latter coordinating to Zn2+. The substrate binds in a deep pocket formed at the dimer interface between domains 1, 2, and 4*, with most interactions with residues at the N-terminal end of the β-sheet within domain 2. The hydroxyl group of l-histidinol forms hydrogen bonds to the backbone carbonyl of His-367 and to the His-327NE2 atom (Fig. 3A). The amino group and ND1 atom of the imidazole ring provide coordination to Zn2+, whereas the NE2 atom forms a hydrogen bond to Glu-414OE1*.
Figure 3.
(A) Octahedral coordination of Zn2+ in the active site with the simulated annealing omit map calculated without l-histidinol and Zn2+, contoured at the 4.5σ level. Asterisk (*) marks residues from the neighboring monomer. (B) NAD+-binding site with the simulated annealing omit map calculated without NAD+ contoured at the 3σ level.
The Zn2+ cation is located at the bottom of the cavity occupied by the substrate and is octahedrally coordinated by Gln-259OE1 (2.6 Å), His-262NE2 (2.1 Å), Asp-360OD2 (2.2 Å), His-419NE2* (2.0 Å), and two ligands from l-histidinol, ND1 (2.0 Å) and N (2.2 Å) (Fig. 3A). The Zn2+ ligand atom distances are within the expected limits (41). The presence of His-262 and His-419 in the Zn2+ coordination environment is consistent with previous mutagenesis experiments (15). NMR experiments (16) suggested that the amino group of the ligand (including l-histidinol, histamine, and l-histidine) is not involved in Zn2+ coordination. However, our results clearly show that l-histidinol binds with its amino group directed toward the cation. The electron density maps for HisD complexed with l-histidine or histamine, although less well defined at this end of the molecule, indicate the same arrangement of the NH2 group, coordinating Zn2+. In monomer A, l-histidinol binding is accompanied by a small rearrangement of residues 233–239 and a reorientation of the Gln-259 side chain toward Zn2+ compared with apo HisD. In monomer B, the segment 233–239 and the Gln-259 side chain adopt two conformations, one as in the apo structure, the other as in monomer A with bound l-histidinol. Although the Zn2+ cation binds independently of l-histidinol, the binding of l-histidinol is Zn2+-dependent.
NAD+-Binding Site.
Extended electron density was observed near the C-terminal ends of β strands of the β-sheet within domain 1 for all NAD+-soaked crystals and was absent in the crystals not soaked in NAD+. The best-defined electron density corresponds to the phosphate groups and to the adenosine moiety, whereas the density for the nicotinamide is poorly defined (Fig. 3B). This weak binding is not too surprising in view of the high Km ≈ 1 mM for NAD+ association with S. typhimurium HisD (17). There are no domain–domain movements within HisD comparing the apo form to the complex with NAD+, l-histidinol, and Zn2+. Each NAD+ molecule lies in a groove in the protein surface making contact with only one monomer (Fig. 2E). The phosphate groups are partially exposed on the surface making hydrogen bonds to the protein residues Asn-211, Tyr-130, and via water molecule to Gly-133 (Figs. 3B and 4). Additionally, the Gln-188 side chain hydrogen bonds to the adenosine sugar O2′, whereas O4′ interacts with Asn-211. The adenine N3 atom makes a hydrogen bond to Gln-188 via a water molecule. The adenine ring stacks between the aromatic side chains Phe-213 and Phe-58, the latter being poorly ordered in the structure. As modeled in a fully extended conformation, which is common to active complexes of NAD+ found in the Protein Data Bank, the NAD+ nicotinamide C4 atom is ≈4.5 Å from the l-histidinol reactive carbon. This distance is too long for a productive complex, possibly reflecting the low pH used in crystallization (6.5) compared with the pH optimum [8.5–9.0; ref. 9], explaining why the enzyme–substrate complex was trapped in the active site. We reason that the correct positioning of the NAD+ molecule may require a small structural adjustment that is prevented by the crystal lattice and that did not occur during soaking experiments.
A recent survey of Rossmann fold–NAD+ complexes showed that NAD+-binding sites share a common mode of binding to the phosphate and adenosine groups, whereas the nicotinamide adopts a variety of conformations to fulfill different functions (42). To compare the mode of NAD+ binding in HisD with the canonical binding mode, we superimposed domain 1 of HisD with the Rossmann fold of human β1 alcohol dehydrogenase (Fig. 4B) (43). Although the secondary structural elements superimpose well, there are differences in two regions important for NAD+ binding. HisD has a six-residue insertion within the P loop and lacks the pattern of glycine residues common to NAD+-binding sites. This insertion creates a longer loop between the first β strand and the following helix of the Rossmann fold, which extends through the region usually occupied by NAD+. A second difference is the substitution of Ser-161 in HisD for the usually observed acidic residue that makes a bidentate interaction with the ribose hydroxyl groups of adenosine. Overall, the position occupied by NAD+ in HisD is different from that observed in 1DEH by a translation of ≈10 Å sideways in the direction perpendicular to the β strands (Fig. 4B). The mode of binding of NAD+ to HisD resembles to some extent the mode of binding observed in the aldehyde dehydrogenases [1BPW; ref. 44], which also have an insertion in the P loop, although shorter than in HisD.
Reaction Mechanism.
Our structural data are in agreement with the overall mechanism of action for HisD proposed by Teng and Grubmeyer (14), where four bases (B1–B4) would be involved in the acid-base catalysis (Fig. 1). The three-dimensional structures of apo and complexed HisD allowed us to identify residues Glu-326 as being base B2 and His-327 as B1, B3, and B4.
In the first step of this mechanism, one proton and one hydride are abstracted from l-histidinol. The role of a base that abstracts the proton from the hydroxyl group (B1 in ref. 14) is played by His-327, which becomes transiently protonated at the NE2 atom. The pH optimum of HisD of 8.5–9.0 (9) correlates well with the observation of an active site residue having a pKa of between 7.7 and 8.4 (45) and with our proposed role for His-327 in catalysis (Fig. 1). The NAD+ molecule accepts the hydride from the reactive carbon (the carbon bound to the hydroxyl group), which now adopts the sp2 configuration concluding the transformation of l-histidinol to l-histidinaldehyde. Next, the reduced NADH cofactor leaves and is replaced by a second NAD+ molecule. A neighboring water molecule is activated by Glu-326 (B2) and makes a nucleophilic attack on the reactive carbon, leading to the formation of a new bond. Concomitantly, protonated His-327 (H:B3) donates its proton to the aldehyde oxygen. The configuration of the reactive carbon changes back to sp3 forming l-histidindiol (gem-diol). This reactive water molecule is likely WAT1 found in the l-histidinol complex and located 2.6 Å from the Glu-326OE2 atom (Fig. 3A). The proximity to the negatively charged side chain polarizes the water molecule for a nucleophilic attack on the reactive carbon. The water–carbon distance is 3.6 Å, suggesting that this molecule moves closer to the reactive carbon. The third step is a repetition of the first step. His-327 (B4) abstracts a proton from the hydroxyl group, and the second NAD+ molecule is reduced by a hydride bound to the reactive carbon. This event leads to the formation of l-histidine, with the reactive carbon now forming a carboxylate group with an sp2 configuration. Protonated His-327+ is restored to its neutral state by donating the proton to a nearby water molecule. The weak binding of NAD+ (26) seems necessary for a fast exchange at the active site.
Previous 113Cd NMR studies on B. oleracea HisD (16) and biochemical studies on S. typhimurium HisD (14) indicated that zinc plays a catalytic role. According to the mechanism we propose, Zn2+ plays a crucial role only in proper positioning of l-histidinol and other reaction intermediates such that the reactive carbon and its hydroxyl group are presented to the water molecule activated by Glu-326 and to His-327, respectively. Another difference from the previous proposal is the assignment of Glu-326, rather than His-327, as B2. The complex with l-histidinol shows that no space is available for a water molecule between the reactive carbon and the His-327 imidazole ring.
The stereochemistry of the NAD+ reductions has been previously assigned as R for both steps of the reaction. The hydride transfers would be to the pro-R position of NAD+ by sequentially removing the pro-S and then pro-R hydrogens of the l-histidinol reactive carbon (25). Our structural results differ somewhat from the previous proposal (25) and indicate, on the basis of the positions of the nicotinamide moiety and l-histidinol, that NAD+ sequentially accepts first the pro-R then pro-S hydrides at its pro-R position.
Gene Duplication.
The superposition of domains 1 and 2 shows that their cores are highly similar, with 50 Cα atoms having an rms deviation of only 1.55 Å (Fig. 4A). Although the structures of the two domains are very similar, their sequences show a very small number of identical residues. The structure-based alignment of the two domains indicates that there is only 11% amino acid identity in their sequences. However, when the amino acid similarity is taken into account, the fraction of similar residues between the two domains increases to 41%. Clearly noticeable is the preservation of the hydrophobic character of the residues forming the core of these domains (Fig. 4C). This sequence similarity was not previously noticed. The likeness of the three-dimensional structure of the two domains, low but discernable sequence homology, and their tandem occurrence with only two residues between the end of the first and the start of the second provide a convincing argument for gene duplication, which must have occurred early in the evolution of this enzyme. Only one of the two Rossmann-fold domains retained the ability to bind NAD+, whereas the other evolved to bind the Zn2+ ion and the substrate.
Acknowledgments
We thank Leon Flaks for assistance with data collection. J.A.R.G.B. thanks the Government of Canada for a Natural Sciences and Engineering Research Council (Canada) Visiting Fellowship.
Abbreviation
- HisD
l-histidinol dehydrogenase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes 1K75 (native), 1KAE (l-histidinol-NAD), 1KAR (l-histamine), and 1KAH (l-histidine)].
References
- 1.Brenner M, Ames B N. In: Metabolic Regulation. Vogel H J, editor. New York: Academic; 1971. pp. 349–387. [Google Scholar]
- 2.Winkler M E. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 485–505. [Google Scholar]
- 3.Alifano P, Fani R, Lio P, Lazcano A, Bazzicalupo M, Carlomagno M S, Bruni C B. Microbiol Rev. 1996;60:44–69. doi: 10.1128/mr.60.1.44-69.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ames B N. In: Amino Acid Metabolism. McElroy W D, Glass B, editors. Baltimore: Johns Hopkins Univ. Press; 1955. pp. 357–372. [Google Scholar]
- 5.Ames B N, Garry B, Herzenberg L A. J Gen Microbiol. 1960;22:369–378. doi: 10.1099/00221287-22-2-369. [DOI] [PubMed] [Google Scholar]
- 6.Ames B N, Goldberger R F, Hartman P E, Martin R G, Roth J R. In: Regulation of Nucleic Acid and Protein Biosynthesis. Konigsberger V V, Bosch L, editors. Amsterdam: Elsevier; 1967. pp. 272–287. [Google Scholar]
- 7.Blasi F, Bruni C B. Curr Top Cell Regul. 1981;19:1–45. doi: 10.1016/b978-0-12-152819-5.50018-x. [DOI] [PubMed] [Google Scholar]
- 8.Martin R G. J Biol Chem. 1963;238:257–268. [Google Scholar]
- 9.Adams E. J Biol Chem. 1955;217:325–344. [PubMed] [Google Scholar]
- 10.Dancer J E, Ford M J, Hamilton K, Kilkelly M, Lindell S D, O'Mahony M J, Saville-Stones E A. Bioorg Med Chem Lett. 1996;6:2131–2136. [Google Scholar]
- 11.Gohda K, Ohta D, Iwasaki G, Ertl P, Jacob O. J Chem Inf Comput Sci. 2001;41:196–201. doi: 10.1021/ci000332n. [DOI] [PubMed] [Google Scholar]
- 12.Grubmeyer C, Skiadopoulos M, Senior A E. Arch Biochem Biophys. 1989;272:311–317. doi: 10.1016/0003-9861(89)90224-5. [DOI] [PubMed] [Google Scholar]
- 13.Kanaori K, Ohta D, Nosaka A Y. FEBS Lett. 1997;412:301–304. doi: 10.1016/s0014-5793(97)00765-5. [DOI] [PubMed] [Google Scholar]
- 14.Teng H, Grubmeyer C. Biochemistry. 1999;38:7363–7371. doi: 10.1021/bi982758p. [DOI] [PubMed] [Google Scholar]
- 15.Nagai A, Ohta D. J Biochem (Tokyo) 1994;115:22–25. doi: 10.1093/oxfordjournals.jbchem.a124298. [DOI] [PubMed] [Google Scholar]
- 16.Kanaori K, Uodome N, Nagai A, Ohta D, Ogawa A, Iwasaki G, Nosaka A Y. Biochemistry. 1996;35:5949–5954. doi: 10.1021/bi951659y. [DOI] [PubMed] [Google Scholar]
- 17.Bürger E, Görisch H. Eur J Biochem. 1981;116:137–142. doi: 10.1111/j.1432-1033.1981.tb05311.x. [DOI] [PubMed] [Google Scholar]
- 18.Grubmeyer C T, Chu K W, Insinga S. Biochemistry. 1987;26:3369–3373. doi: 10.1021/bi00386a018. [DOI] [PubMed] [Google Scholar]
- 19.Kheirolomoom A, Mano J, Nagai A, Ogawa A, Iwasaki G, Ohta D. Arch Biochem Biophys. 1994;312:493–500. doi: 10.1006/abbi.1994.1337. [DOI] [PubMed] [Google Scholar]
- 20.Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M. Science. 2000;289:1546–1550. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]
- 21.Fani R, Lio P, Chiarelli I, Bazzicalupo M. J Mol Evol. 1994;38:489–495. doi: 10.1007/BF00178849. [DOI] [PubMed] [Google Scholar]
- 22.Sivaraman J, Li Y, Larocque R, Schrag J D, Cygler M, Matte A. J Mol Biol. 2001;311:761–776. doi: 10.1006/jmbi.2001.4882. [DOI] [PubMed] [Google Scholar]
- 23.Haruyama K, Nakai T, Miyahara I, Hirotsu K, Mizuguchi H, Hayashi H, Kagamiyama H. Biochemistry. 2001;40:4633–4644. doi: 10.1021/bi002769u. [DOI] [PubMed] [Google Scholar]
- 24.Hendrickson W A, Horton J R, LeMaster D M. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Bürger E, Görisch H, Lingens F. Biochem J. 1979;181:771–774. doi: 10.1042/bj1810771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grubmeyer C T, Insinga S, Bhatia M, Moazami N. Biochemistry. 1989;28:8174–8180. doi: 10.1021/bi00446a032. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger T C. Acta Crystallogr D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrakis A, Morris R, Lamzin V S. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 32.Jones T A, Zhou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;4:7110–7119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 33.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 35.Rossmann M G, Liljas A, Brandën C-I, Banaszak L J. In: The Enzymes: Oxidation-Reduction. Boyer P D, editor. New York: Academic; 1975. pp. 61–102. [Google Scholar]
- 36.Holm L, Sander C. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 37.Murzin A G, Brenner S E, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 38.Istvan E S, Palnitkar M, Buchanan S K, Deisenhofer J. EMBO J. 2000;19:819–830. doi: 10.1093/emboj/19.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell R E, Mosimann S C, van De Rijn I, Tanner M E, Strynadka N C. Biochemistry. 2000;39:7012–7023. [PubMed] [Google Scholar]
- 40.Bennett M J, Schlunegger M P, Eisenberg D. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harding M M. Acta Crystallogr D. 2001;57:401–411. doi: 10.1107/s0907444900019168. [DOI] [PubMed] [Google Scholar]
- 42.Carugo O, Argos P. Proteins. 1997;28:10–28. doi: 10.1002/(sici)1097-0134(199705)28:1<10::aid-prot2>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 43.Hurley T D, Bosron W F, Stone C L, Amzel L M. J Mol Biol. 1994;239:415–429. doi: 10.1006/jmbi.1994.1382. [DOI] [PubMed] [Google Scholar]
- 44.Johansson K, El Ahmad M, Ramaswamy S, Hjelmqvist L, Jornvall H, Eklund H. Protein Sci. 1998;7:2106–2117. doi: 10.1002/pro.5560071007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grubmeyer C, Teng H. Biochemistry. 1999;38:7355–7362. doi: 10.1021/bi982757x. [DOI] [PubMed] [Google Scholar]
- 46.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 47.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 48.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]