Abstract
A general strategy for inactivation of target proteins is presented, which we have termed “oligomerization chain reaction.” This technique is based on the fusion of the self-associating coiled-coil (CC) domain of the nuclear factor promyelocytic leukemia (PML) to target proteins that are able to self-associate naturally. Oligomerization through the CC region of promyelocytic leukemia, and through the natural self-associating domain, triggers the oligomerization chain reaction, leading to formation of large molecular weight complexes and functional inactivation of the target. As a test case, we have chosen the oncosuppressor p53, naturally occurring as a tetramer. Fusion of the CC to p53 leads to formation of stable high molecular weight complexes—as shown by size exclusion chromatography—to which wild-type p53 is recruited with high efficiency. CC-p53 chimeras delocalize wild-type p53 to the cytoplasm and inhibit its transcriptional regulatory properties, resulting in a loss of p53 function. We propose that this strategy may be of general application to self-associating factors and represent a complementary approach to currently used functional inactivation-based strategies.
Functional and genetic approaches have been devised to inactivate cellular proteins with the purpose of studying their function or with therapeutical goals in mind (1, 2). Functional inactivation approaches are widely used in model systems where genetic ablation of target genes is not available and they are especially useful in human cells, where ethical and safety considerations limit the potential of genetic approaches.
Functional inactivation may be reached by (i) interference with protein synthesis (RNA-based antisense and interference technologies), (ii) administration of small molecular weight compounds able to bind and inhibit target proteins, (iii) stimulation of protein degradation, or (iv) by specifically designed “dominant negative” mutants (such as mutant enzymes devoid of catalytic activity or transcription factors deleted for their activating domains; refs. 2–5). These methods have been used successfully in several cases, although none of them can be considered universally applicable. Therefore, the introduction of novel inactivation strategies offers additional targeting possibilities and complements existing technologies. Here, we present evidence to show that oligomerization of target proteins through the coiled-coil (CC) region of the nuclear factor PML may be exploited to reach their functional inactivation.
PML belongs to a family of genes (the TRIM family) containing the so-called tripartite motif, which includes the CC region (6, 7). In acute promyelocytic leukemia (APL), the PML locus is involved in a chromosomal translocation with retinoic acid receptor (RAR), resulting in the generation of the fusion protein PML–RAR (8, 9). PML–RAR behaves as an altered retinoic acid receptor, interfering with normal hematopoietic differentiation. In contrast with RAR, PML–RAR forms oligomeric complexes through the self-associating CC region present in PML (10–13). Oligomerization is the main structural determinant for the alteration in normal function of RAR in the context of the fusion protein. Fusion of the CC region of PML to RAR (in the absence of additional PML sequences) is sufficient for RAR to become leukemogenic, increasing the stability and stoichiometry of its association with the nuclear corepressor/histone deacetylase complex, and leading to aberrant repression of RAR target genes (10–13). Based on these findings, we investigated the possibility that the CC of PML might be used to modify the function of target proteins.
Materials and Methods
Constructs.
A pSG5/CC-p53 plasmid was generated by insertion of the full-length p53 cDNA (amino acids 2–393) in the pSG5-CC plasmid (containing the CC region—amino acids 224–369—of PML; ref. 6). pSG5/p53-CC was obtained by elimination of the stop codon in the p53 coding sequence, and cloning of the CC region of PML (amino acids 224–369) in-frame at the carboxyl terminus, with an artificial stop codon inserted at the end.
Size Exclusion Chromatography.
Whole-cell extracts were prepared from murine embryonic fibroblast (MEF) p53−/− transfected with various expression vectors (5 μg) as indicated. In the CC-p53/p53 cotransfection, a 4:1 ratio of expression vectors was used. The extracts (in buffer 50 mM Tris⋅Cl, pH 8.5/10% glycerol/150 mM NaCl/2 mM DTT/1% Nonidet P-40/10 mg/ml aprotinin/10 mg/ml leupeptin/1mM PMSF) were ultracentrifuged (105,000 × g), and the supernatants were loaded on a Superose 6 HR 10/30 gel filtration column equilibrated in extraction buffer without Nonidet P-40 (11).
Transactivation Assays.
MEF p53−/− cells were transiently transfected by the Lipofectamine Plus reagent as described (11).
Immunofluorescence.
NIH 3T3 cells were transiently transfected by the Fugene 6 reagent.
Cells were fixed in Pipes pH 6.8/4% paraformaldehyde, permeabilized in 0.1% Triton X-100 for 10 min, and then stained as described (14). The Abs DO-1 (mouse monoclonal anti p53 Ab) and a-CC (mouse polyclonal serum raised against the CC region of PML) were used.
Colony-Forming Assay.
SAOS 2 cells were transiently transfected by calcium phosphate with the indicated expression vectors. Twenty-four hours after transfection, cells were seeded at a concentration of 20,000 cells per 60-mm diameter culture dish, grown in medium containing 500 μg/ml G418 for 2 weeks, and then stained with Crystal Violet dye (15). Colonies were then counted from triplicate plates.
Results
Oligomerization Chain Reaction (OCR).
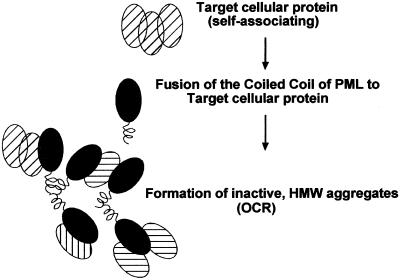
In the case of RAR (a monomeric factor), oligomerization through the CC of PML led to an alteration of its function, enhancing its capacity to recruit transcriptional coregulators (11). We reasoned that in the case of factors oligomeric in nature, the addition of an extra-oligomerization interface would lead to formation of high-order oligomeric complexes, which may result in the formation of nonfunctional aggregates, able to recruit also the natural “target” through its own oligomerization domain. We investigated whether this may constitute a generally applicable approach for functional inactivation and termed this technology “OCR” (Fig. 1).
Figure 1.
A schematic model of the OCR. Fusion of the CC region of PML to an oligomeric protein leads to the formation of nonfunctional aggregates.
The oncosuppressor p53 protein forms tetramers, and oligomerization is required for its function (16–18). The oligomerization domain of PML (CC), fused to the full-length coding sequence of p53, should impose an altered oligomerization state not only of the resulting chimeric protein but also of wild type (wt) , interacting p53. In turn, this association, according to our model, should lead to an improper organization of chimeric protein/wt p53 heterooligomers and to inhibition of p53 function.
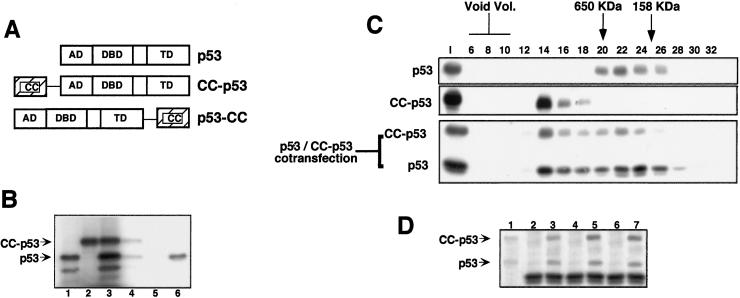
We fused the CC region of PML either amino-terminally or at the carboxyl terminus of the full-length p53 to generate the chimeric CC-p53 and p53-CC proteins (Fig. 2A). A requirement for the chimeric proteins would be the capacity to interact with wt p53. Abs directed against the CC region of CC-p53 were able to immunoprecipitate in vitro-translated p53 only in the presence of the CC-p53 chimera, showing the existence of a CC-p53/wt p53 complex (Fig. 2B). Identical results were obtained for p53-CC (data not shown).
Figure 2.
(A) A schematic view of p53, CC-p53, and p53-CC. AD, activation domain; DBD, DNA binding domain; TD, tetramerization domain. (B) Formation of CC-p53/p53 heterooligomers. P53 (lane 1) and CC-p53 (lane 2) were in vitro-translated singly or cotranslated (lane 3). Samples were then immunoprecipitated with Abs against the CC region of PML (lane 4, from cotranslated CC-p53 and p53; and lane 5, from p53 only) or with an anti-p53-specific Ab (lane 6, from p53 only). (C) CC-p53 recruits wt p53 into high molecular weight complexes. Extracts from p53-null MEFs, transiently transfected with the indicated expression vectors, were subjected to SEC. SEC fractions were then analyzed by Western blot, using the appropriate Ab. The elution volume of mass markers is indicated on the top as previously described. I, input. (D) Extract, or pooled SEC fractions from the p53/CC-p53 cotransfection, was immunoprecipitated with Abs against the CC region of PML and then analyzed by Western blot, using an anti-p53 Ab. Lane 1, input; lanes 2 and 3, immunoprecipitates from input extract; lanes 4 and 5, immunoprecipitates from fractions 13–15; lanes 6 and 7, immunoprecipitates from fractions 20–24; lanes 2, 4, and 6, immunoprecipitates with an unrelated Ab; lanes 3, 5, and 7, immunoprecipitates with the anti-CC Ab.
p53 forms stable tetramers and, in agreement with previous reports using purified recombinant p53, is found after size exclusion chromatography (SEC) peaking in fractions corresponding to an apparent molecular mass of 400–500 kDa (Fig. 2C; see also refs. 17 and 18). Consistent with the presence of an additional oligomerization interface, CC-p53 and p53-CC are found in SEC fractions of higher apparent molecular mass, ranging from 600 kDa to the void volume of the column (Fig. 2C and data not shown).
Given the capacity of CC-p53 to associate with wt p53 (Fig. 2B), we measured the apparent molecular mass of the CC-p53/p53 heterooligomeric complex. After interaction with CC-p53, p53 was found to cofractionate with the chimeric protein and was eluted in fractions corresponding to an apparent molecular mass ranging from 250 kDa to the void volume (Fig. 2C). We interpreted the widespread distribution of the two proteins as a result of heterogenous populations of differently sized heterooligomers. Coimmunoprecipitation experiments were performed from SEC fractions corresponding to apparent molecular mass of approximately 250 kDa, 600 kDa, and >1 MDa (Fig. 2D). In all cases, we verified the formation of a heterooligomeric CC-p53/p53 complex, demonstrating that CC-p53 is able to recruit p53 into high-order oligomers (Fig. 2D). Similar results were obtained for the p53-CC/p53 heterooligomeric complex (data not shown).
Inactivation of Target Proteins by OCR.
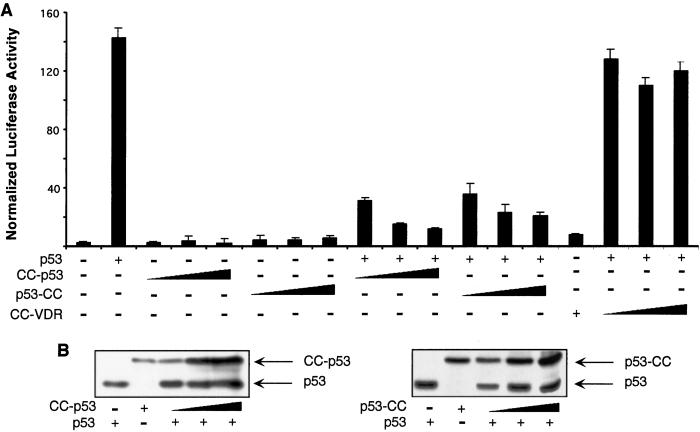
To evaluate the transcriptional properties of CC-p53 and p53-CC, we performed transient transfection assays in MEFs derived from p53−/− mice. In these cells, transfection of a reporter construct containing an array of p53 response elements (pGL13) resulted in minimal levels of transcriptional activity (Fig. 3A). Cotransfection of an expression vector for wt p53 caused a strong increase in transcriptional activity of the reporter construct (approximately 50-fold; Fig. 3A and see ref. 14). Cotransfection of expression vectors for CC-p53 or p53-CC, in contrast, had no significant effect on reporter activity, showing that the chimeric proteins are no longer able to regulate p53 target genes (Fig. 3A). Cotransfection of increasing amounts of CC-p53 or p53-CC with wt p53 (at a fixed amount) resulted in repression of wt p53 transcriptional activity (Fig. 3A). As a control, the chimeric CC-VDR protein, encoding for an unrelated transcription factor (vitamin D receptor), fused to the PML CC, had no effect on p53 wt transcriptional activity (Fig. 3A). CC-p53 and p53-CC, interacting with wt p53, are therefore able to block its transcriptional activity. Western blot analysis of transfected cells show that repression by CC-p53 over wt p53 is already marked at levels of the chimeric protein corresponding to a fraction of the wt target protein, suggesting (as predicted by our model) that CC-p53 may “seed” the OCR efficiently at substoichiometric ratios (Fig. 3B).
Figure 3.
CC-p53 and p53-CC inhibit p53 transcriptional activity. (A) p53-null MEFs were transiently transfected with the indicated expression vectors (50 ng for p53; 50, 100, and 200 ng for CC-p53, p53-CC, and CC-VDR) and pGL13, a luciferase-based reporter vector for p53 transcriptional activity. Twenty-four hours after transfection, cells were collected and analyzed for reporter activity. A β-galactosidase expression vector was used to normalize for transfection efficiency. (B) Western blot analysis with an anti-p53 Ab of p53-null MEFs cotransfected with the indicated expression vectors.
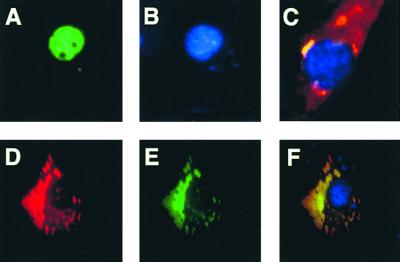
Next, we investigated in greater details the mechanism(s) underlying the dominant negative effect of CC-p53 over the wt p53 protein. The wt p53 protein is posttranslationally regulated at several levels: stability, phosphorylation, and acetylation (19–21). We first asked whether the heterooligomeric CC-p53/wt p53 complexes are less stable than the wt p53 protein: Western blot analysis of cells transiently transfected with the expression vector for wt p53 or cotransfected with the expression vectors for wt p53 and CC-p53 or p53-CC showed no significant difference in wt p53 levels, suggesting that CC-p53 and p53-CC are not targeting wt p53 for degradation (in conditions where p53 transcriptional activity is strongly repressed; Fig. 3B). Next, we checked for proper localization of the heterooligomeric complexes. NIH 3T3 cells were transiently transfected with expression vectors for wt p53, CC-p53, p53-CC, and, in some experiments, a GFP-p53 fusion protein, to allow visualization of p53 prior fixation of the cells and to distinguish unambiguously p53 from CC-p53/p53-CC (Fig. 4). GFP-p53 behaves identically to wt p53 in all functional assays tested (data not shown). GFP-p53 and p53 displayed a nuclear localization pattern, as described (Fig. 4 A and B and data not shown). In contrast, CC-p53 was almost entirely localized in the cytoplasm (Fig. 4D). CC-p53 caused almost complete delocalization of either wt p53 or GFP-p53 (Fig. 4 D–F and data not shown). Similar results were obtained for p53-CC (Fig. 4C). These results suggest that the dominant negative effect of CC-p53 (and of p53-CC) over wt p53 is mainly achieved through formation of delocalized heterooligomeric complexes.
Figure 4.
CC-p53 and p53-CC delocalize p53. NIH 3T3 cells were transiently transfected with the following expression vectors: (A and B) GFP-p53 [(A) GFP-p53; (B) 4′,6-diamidino-2-phenylindole (DAPI) staining]; (C) p53-CC and GFP-p53 at a 1:1 ratio of expression vectors (merge of the immunofluorescence signal for p53-CC by using an anti-CC Ab, of the GFP-p53 signal, and of DAPI staining); (D–F) CC-p53 and GFP-p53 at a 1:1 ratio of expression vectors [(D) CC-p53 after immunofluorescence with an anti-CC Ab; (E) GFP-p53; (F) merge]. In parallel experiments, p53 was used in place of the GFP-p53 chimeric protein with identical results (data not shown).
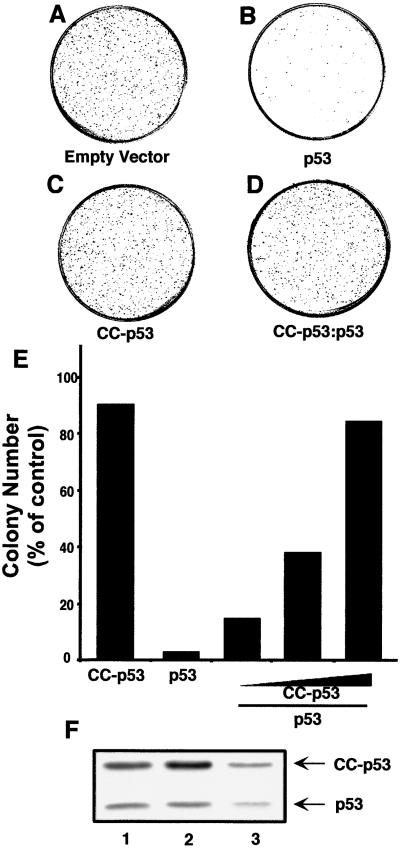
Finally, we measured the capacity of CC-p53 and p53-CC to inhibit the biological function of p53. Expression of wt p53 in p53 null SAOS cells results in cell growth arrest and apoptosis and loss of colony-forming capacity (compare Fig. 5 A with B; see also ref. 15). CC-p53 and p53-CC had no effect on cell viability (Fig. 5C and data not shown). CC-p53 and p53-CC almost completely abrogated the growth suppression effect by wt p53 (Fig. 5 D and E and data not shown). The colonies obtained after cotransfection of CC-p53 and wt p53 continue to express wt p53 (Fig. 5F). These results demonstrate that the dominant negative effect on wt p53 activity is sufficient to inhibit its biological function.
Figure 5.
p53-null SAOS cells were transfected with a control vector (empty vector) or with vectors encoding for p53 (6 μg), CC-p53, or both (CC-p53 and p53 at 1:1, 2:1, and 4:1 ratio). Twenty-four hours after transfection, cells were split and plated in medium containing G418 to select for transfected cells. Two weeks after plating, G418-resistant colonies were counted. (A–D) Representative plates after Crystal Violet dye staining of the colonies [(D) CC-p53/p53 at a 4:1 ratio]. (E) Colony number (expressed as % of colonies obtained with the control vector) from a representative experiment performed in triplicate. (F) Three random colonies from a CC-p53/p53 cotransfection experiment were picked and analyzed for expression of p53 and CC-p53 by Western blot analysis with an anti-p53 Ab.
Discussion
Our results show that in the case of an oligomeric protein (p53), addition of an extra oligomerization domain (the CC of PML) results in an oligomerization chain reaction that alters its localization and function. The chimeric protein retains the ability to associate with the wt target, behaving as a powerful dominant negative mutant. The OCR technology may therefore be applied to inactivate natural oligomeric proteins.
The CC behaves as a general functional modifier of target proteins: it may act as a dominant negative for oligomeric targets in the OCR (this work) but we have shown that it may also function as an “enhancing” tool in the case of a nonoligomeric factor (RAR), where oligomerization led to increased association with the nuclear corepressor/histone deacetylase complex (10, 11). Because we have observed that the CC domain may mediate oligomerization also in the extracellular environment (unpublished data), this technique may be applied to both intra- and extracellular target proteins.
In our study, we have presented as a test case the inactivation of a self-associating target (p53), but heterooligomeric complexes may also be considered as valid targets. The full-length p53 protein was used in the chimeric CC-p53 and p53-CC constructs to show unambiguously that the dominant negative effect was a result of the addition of the CC domain, but the isolated self-association domain of the target protein may also suffice. Isolated self-association domains of target proteins have been used for their own dominant negative activity (being able to bind endogenous full-length targets and titrating them out of physiological interactions with DNA/other proteins). The OCR triggered by the additional oligomerization domain, however, offers some advantages over this inactivation strategy. (i) Being applicable to full-length proteins, it does not require the characterization of the domains required for self-association. (ii) Being an exponential reaction, the ratio chimeric construct:wt protein may be low (even <1, see Fig. 3B). (iii) The OCR may further potentiate dominant negative effects caused by other mechanisms (i.e., titration of partners bound by a protein–protein interaction domain).
Alternative CC regions, or oligomerization domains of a different structure, may be used in the design of inactivating constructs. CC modules are extremely stable and therefore result in the formation during the OCR of macromolecular aggregates resistant to dissociation under a large variety of experimental conditions. Up to five-stranded CC modules have been identified (22). The isolated CC domain of PML mediates the formation of trimers, and it is therefore possible that other CC domains (with a >3 self-associating number) may be even more effective in triggering the OCR.
We must mention the possibility that the CC-based chimeric constructs may interact with wt PML and interfere with its function. The CC region of PML, however, is not sufficient to target appropriately itself—or chimeric proteins—to the subnuclear compartments containing the full-length PML (nuclear bodies), and CC-p53—or p53-CC—is not able to disrupt PML nuclear bodies (data not shown).
As for other inactivating strategies, several applications may be envisioned for the OCR. The possibility to generate recombinant chimeric proteins for extracellular use make it an immediate, attractive strategy for targeted inactivation of extracellular proteins involved in pathogenesis of a wide variety of diseases. Thus, the discovery of a molecular property of a fusion protein (PML–RAR oligomerization) responsible for leukemia may help to generate artificial factors with therapeutic potential.
Acknowledgments
We acknowledge Mario Faretta for technical help, and Sabrina Giavara for discussions. This work was supported by grants from the Fondazione Italiana per la Ricerca sul Cancro (to S.M.), the European Economic Community (to P.G.P.), and the Associazione Italiana per la Ricerca sul Cancro (to P.G.P.).
Abbreviations
- OCR
oligomerization chain reaction
- CC
coiled coil
- SEC
size exclusion chromatography
- wt
wild type
- MEF
murine embryonic fibroblast
- RAR
retinoic acid receptor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Capecchi M R. Nat Genet. 2000;26:159–161. doi: 10.1038/82825. [DOI] [PubMed] [Google Scholar]
- 2.Herskowitz I. Nature (London) 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 3.Fire A. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- 4.Hung D T, Jamison T F, Schreiber S L. Chem Biol. 1996;3:623–639. doi: 10.1016/s1074-5521(96)90129-5. [DOI] [PubMed] [Google Scholar]
- 5.Zhou P, Bogacki R, McReynolds L, Howley P M. Mol Cell. 2000;6:751–756. doi: 10.1016/s1097-2765(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 6.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong S, Salomoni P, Pandolfi P P. Nat Cell Biol. 2000;2:E85–E90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
- 8.Melnick A, Licht J D. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- 9.Minucci S, Pelicci P G. Semin Cell Dev Biol. 1999;10:215–225. doi: 10.1006/scdb.1999.0303. [DOI] [PubMed] [Google Scholar]
- 10.Lin R J, Evans R M. Mol Cell. 2000;5:821–830. doi: 10.1016/s1097-2765(00)80322-6. [DOI] [PubMed] [Google Scholar]
- 11.Minucci S, Maccarana M, Cioce M, De Luca P, Gelmetti V, Segalla S, Di Croce L, Giavara S, Matteucci C, Gobbi A, et al. Mol Cell. 2000;5:811–820. doi: 10.1016/s1097-2765(00)80321-4. [DOI] [PubMed] [Google Scholar]
- 12.Minucci S, Nervi C, Coco F L, Pelicci P G. Oncogene. 2001;20:3110–3115. doi: 10.1038/sj.onc.1204336. [DOI] [PubMed] [Google Scholar]
- 13.Salomoni P, Pandolfi P P. Nat Med. 2000;6:742–744. doi: 10.1038/77459. [DOI] [PubMed] [Google Scholar]
- 14.Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi P P, Pelicci P G. Nature (London) 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 15.Luo J, Su F, Chen D, Shiloh A, Gu W. Nature (London) 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 16.Chene P. Oncogene. 2001;20:2611–2617. doi: 10.1038/sj.onc.1204373. [DOI] [PubMed] [Google Scholar]
- 17.Friedman P N, Chen X, Bargonetti J, Prives C. Proc Natl Acad Sci USA. 1993;90:3319–3323. doi: 10.1073/pnas.90.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P, Reed M, Wang Y, Mayr G, Stenger J E, Anderson M E, Schwedes J F, Tegtmeyer P. Mol Cell Biol. 1994;14:5182–5191. doi: 10.1128/mcb.14.8.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Appella E, Anderson C W. Eur J Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 20.Gottifredi V, Prives C. Trends Cell Biol. 2001;11:184–187. doi: 10.1016/s0962-8924(01)01983-3. [DOI] [PubMed] [Google Scholar]
- 21.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 22.Lupas A. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]