Abstract
A general strategy is described for forcing the engagement of an RNA/protein complex by using small-molecule ligands. A bivalent molecule was created by linking a protein-binding ligand to an RNA-binding ligand. On presentation of the chemical inducer of dimerization to the RNA by the protein, cooperative binding ensued, resulting in higher-affinity complexes. When the chemical inducer of dimerization was used to target the protein to an mRNA template, the resulting RNA/protein complex was sufficiently stable to inhibit mRNA translation. This approach provides a logic to modulate gene expression by using small-molecule ligands to recruit protein surfaces to mRNAs.
RNA plays a key and versatile role in biological processes, carrying information required for biological function and containing complex folded conformations that can participate in sophisticated recognition and catalytic processes. Additionally, RNA can interact with chemically and structurally diverse sets of small molecules, some exerting profound effects on the biological function of the target. Structural studies of antibiotics bound to ribosomal subunits have revealed that rRNA/small-molecule recognition is based on a combination of shape recognition, electrostatic, and hydrogen-bonding interactions (1, 2). Additionally, SELEX (Systematic Evolution of Ligands by Exponential Enrichment) has demonstrated that RNA three-dimensional structures can form a virtually unlimited number of highly specific ligand-binding sites (3). However, the dynamic nature of RNA structures, as well as the presence of associated proteins that can displace all but the most strongly bound ligand, makes RNA an especially difficult species to target with high-affinity ligands.
Small-molecule CIDs, which bind two proteins simultaneously, have been used extensively to modulate the activity of different cellular protein interactions and to demonstrate the feasibility of enlisting nonphysiological interactions to effect physiological changes (for example, see ref. 4). Rapamycin is an example of a well characterized CID that binds to FKBP 12 (FK506-binding protein 12), and this complex then associates with the cell cycle control protein FRAP (FKBP12 rapamycin-associated protein). By itself, rapamycin has no measurable affinity for FRAP; rather, the FKBP/rapamycin complex binds to FRAP with high affinity (5). Rapamycin fits into each protein, allowing alignment and interactions between the two bound proteins to occur. Two important features of CIDs are: (i) their ability to induce proximity—to enhance the activity of a protein by bringing it into close proximity to another protein (6), and (ii) affinity modulation; the trimeric complex is more stable than would be expected on the basis of stabilities of the individual bimolecular components (4).
In the current report, we explore a new logic to target RNA with small molecules. The approach is based on developing CIDs designed to realize a trimolecular complex between an RNA, a protein, and a small-molecule ligand (7). Although the conformationally heterogeneous nature of RNA is thought to make it an intractable drug target, using CIDs to engage an RNA/protein interaction takes advantage of the fact that RNA can undergo induced structural reorganizations when binding to specific protein surfaces (for a review, see ref. 8). We find that appropriate positioning of the RNA/protein complex on an mRNA template abolishes 80S complex formation, resulting in specific inhibition of translation.
Materials and Methods
Plasmid Construction.
Plasmids encoding the desired RNA sequences were generated by using standard cloning techniques (9). All clones generated by ligation of synthetic oligonucleotides or by PCR were sequenced by the chain termination method by using double-stranded DNA templates to ensure the absence of mutations.
Plasmids for in vitro transcription are based on pSP65(A)18, a derivative of pSP65 that contains 18 adenylate residues inserted in the PstI site. Plasmid SP/CAT contains the chloramphenicol acetyltransferase (CAT) ORF inserted into the BamHI site of pSP65(A)18. It is engineered such that the initiation codon is positioned 36 nucleotides from the penultimate 5′ base of the mRNA and in a favorable context (5′CCACCAUG3′). Oligonucleotides encoding the J6f1 or X1 aptamers contained EcoRI ends and were inserted into the EcoR 1 site of pSP65(A)18, to generate pSP/(Tobra) or pSP/X1, respectively. After linearization with BamHI and in vitro transcriptions, SP/(Tobra) and pSP/X1 produced RNA corresponding to J6f1 and X1, respectively, which were used in EMSAs and solution binding. Plasmids SP/T1–3/CAT were derived from pSP/(Tobra)1–3 by digestion the latter with BamHI and transferring the CAT ORF from pKSII/CAT. An oligonucleotide containing sequences derived from the DHFR gene (5′TGAATCACCCAGGCCATCTTAAACTATTTGTGACAAGGATCAAGCAAGACTTTGAAAGTGACACGTTTTTTCCAGAAATTGATTTGGAGAAATATAAACTTCT3′) was inserted either: (i) into the EcoRI site of pSP/T3/CAT to vary the distance of the tobramycin-binding aptamer from the 5′ end of the mRNA [producing pSP/D/T3/CAT and pSP/T3/D/CAT], or (ii) into the 3′ untranslated region (UTR) of pSP/CAT to distance the aptamer from the CAT stop codon (producing pSP/CAT/D). Plasmid SP/CAT(T) was generated by partially digesting pSP/CAT with EcoRI and ligating, in-frame, one copy of the J6f1 aptamer sequence. Plasmids SP/CAT/D/T1–2 were generated by inserting J6f1 aptamer-encoding oligonucleotides (containing XbaI sites) into the XbaI site of pSP/CAT/D.
Electrophoretic Mobility-Shift Assays (EMSAs) and Solution Binding Assays.
EMSA-binding reactions were performed in a total volume of 10 μl in binding buffer (25 mM Hepes⋅KOH, pH 7.5/150 mM KCl/2 mM MgCl2/15% glycerol/0.2 mM EDTA/2 μg of calf liver tRNA) with 32P-labeled RNA (40,000 cpm; 2.4 pmol). Binding reactions were incubated for 10 min at 25°C. Protein/RNA complexes were resolved on 2 × TGE (50 mM Tris⋅HCl, pH 8.3/0.38 M glycine/2 mM EDTA)/5% polyacrylamide gels electrophoresed at 68 V at 4°C. Dried gels were exposed to X-Omat film (Kodak) at room temperature. Quantitations were performed on a Fujix BAS2000 with a Fuji imaging screen.
For solution binding assays, streptavidin (1 mg) was coupled to Affi-Gel 10 (1 ml) (Bio-Rad) by incubating with resin in AC buffer (20 mM Hepes, pH 7.5/10% glycerol/1 mM DTT/1 mM EDTA) at 4°C overnight. After removal of the supernatant, the reaction was stopped by incubating the beads in 80 mM ethanolamine for 1 h at room temperature. The beads were washed in AC-75 (AC + 75 mM NaCl) and stored at 4°C as a 50% slurry. Measurement of protein coupling efficiency indicated that >90% of the input streptavidin was linked to the Affi-Gel beads (data not shown). In 100 μl of EMSA-binding buffer, streptavidin beads (10 μl) were incubated with 10 μM biotinamidocaproate tobramycin amide (BTA) for 10 min, followed by three washes with AC-75 buffer (100 × bed volume). These are referred to as preloaded beads (PLB). J6f1 (2.4 pmol; 40,000 cpm) and increasing concentrations of BTA in the absence or presence of 6.8 μM streptavidin were added to the PLB for 2 h. The binding reactions were centrifuged in a pierced PCR tube to separate the beads from the supernatant, and radioactivity associated with the beads and in the flow-through was quantitated in a Packard 1600CA liquid scintillation analyzer.
In Vitro Translations and Ribosome Bindings.
For translation studies, reporter plasmids were linearized with HindIII. In vitro transcriptions, in the presence of m7GpppG, were performed as described (10). RNA transcripts were quantitated by monitoring incorporation of 3H-CTP (20 Ci/mmol; New England Nuclear) and the quality of each preparation assessed by visualization on formaldehyde/1.2% agarose gels. In vitro translations were performed in wheat germ lysates by using [35S]methionine as directed by the manufacturer (Promega). Unless otherwise indicated, translations were performed at mRNA concentrations of 20 μg/ml and the KOAc adjusted to achieve a final concentration of 150 mM. Trichloroacetic acid precipitation of translation products was performed in duplicate and used to determine the relative translation efficiencies.
Ribosome-binding assays were performed essentially as described (10). Briefly, 32P-labeled mRNA was incubated under conditions of translation in wheat germ extracts, in the presence of either 0.6 mM cyclohexamide or 1 mM 5′-guanylylimidodiphosphate, at 20°C for 10 min. Initiation complexes formed were analyzed by sedimentation through 10–30% glycerol gradients. Centrifugation was for 4 h at 39,000 rpm at 4°C in an SW40 rotor (Beckman Coulter). Fractions of ≈500 μl were collected, and radioactivity was determined by scintillation counting.
Results
CID Design.
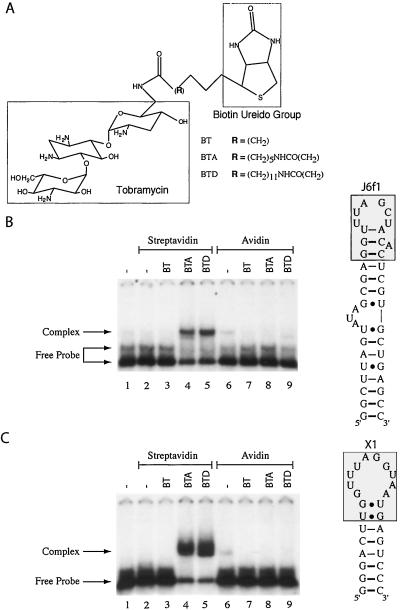
We designed three bifunctional molecules based on biotin and tobramycin. We identified biotin as the protein-binding component of our CID because of the extremely tight binding affinity to avidin and streptavidin (Kd = 10−14–10−15 M). Inspection of the streptavidin- (11, 12) and avidin-biotin (13, 14) three-dimensional structures indicates that the ureido group of biotin is buried in binding pockets with the valeryl carboxyl oxygens partially accessible to solvent. Tobramycin was chosen as the RNA-binding moiety of our CID, because the tobramycin-binding pocket of two RNA aptamers, J6f1 and X1 (Fig. 1), is defined and indicates that the 6′ amino group of tobramycin is solvent accessible and can be used as a coupling point (15–17). Three CIDs were designed in which the length of the spacer between the RNA- and protein-binding groups was varied (Fig. 1A).
Figure 1.
Formation of an RNA/protein complex mediated by a chemical inducer of dimerization. (A) Biotin/tobramycin CIDs used in this study. The nature of the chemical spacer for each CID is denoted (Bottom Right). BT, BTA, and BTD were purchased from Toronto Research Chemicals (Downsview, ON, Canada), and identity was confirmed by 1H-NMR and mass spectrometric analysis. (B) Specificity of RNA/protein complex mediated by CIDs. EMSAs were performed with 240 nM 32P-labeled J6f1 in the presence of 250 ng of streptavidin (1.7 μM) (lanes 2–5) or 250 ng (1.6 μM) of avidin (lanes 6–9). The presence of 10 μM BT, BTA, or BTD in the binding reaction is indicated (Top). Complexes were resolved on a 5% polyacrylamide gel (30/0.8; acrylamide/bisacrylamide) by electrophoresis in 2 × TGE buffer. Gels were dried and exposed to X-Omat x-ray film (Kodak). A schematic diagram of the J6f1 (19) tobramycin-binding RNA aptamer used in this study is presented (Right). The boxed shaded area highlights the tobramycin-binding pocket (16). (C) RNA/protein complex formation with X1 mediated by CIDs. Complexes were generated, resolved, and visualized as described for B. A schematic diagram of the X1 tobramycin-binding RNA aptamer (15) used in this study is presented. The boxed shaded area highlights the tobramycin-binding pocket (17).
Binding Assays.
We tested the ability of these CIDs to induce an interaction between RNA containing the tobramycin-binding aptamers (Fig. 1B, J6f1; Fig. 1C, X1) and streptavidin or avidin. Incubation of J6f1 with only streptavidin (Fig. 1B, lane 2) or avidin (lane 6) did not induce formation of an RNA/protein complex (compare lanes 2 and 6 to lane 1). Additionally, no complex was observed if BT was added to the binding reaction (lanes 3 and 7). However, a single complex was observed when streptavidin, J6f1, and either BTA or N-biotinyl-12-aminododecanoyltobramycin amide (BTD) were present in the same binding reaction (Fig. 1B, lanes 4 and 5), but not if avidin replaced streptavidin in the reaction (Fig. 1B, lanes 8 and 9). The RNA aptamer X1 demonstrated a similar pattern of behavior, forming an RNA/protein complex only with streptavidin in the presence of BTA or BTD (Fig. 1C, lanes 4 and 5). No complex was observed if J6f1 or X1 was incubated with only BT, BTA, or BTD (data not shown). These results indicate that a CID can induce the formation of an RNA/protein complex between partners that do not normally interact in a physiological context, and that complex formation depends on specific structural properties of the CID.
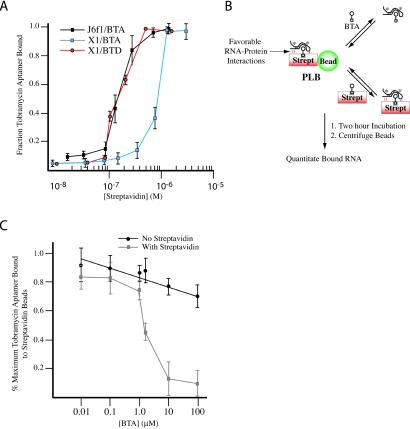
We measured the dissociation constant of J6f1 and X1 RNA/protein complexes formed in the presence of BTA or BTD (streptavidin/BTA or streptavidin/BTD) are treated as a single complex given the high affinity of streptavidin for biotin). The streptavidin/BTA/J6f1 and streptavidin/BTD/X1 complexes show apparent equilibrium dissociation constants (Kd) of 1.5 ± 0.5 × 10−7 M, whereas the streptavidin/BTA/X1 complex shows an apparent equilibrium dissociation constant (Kd) of 9 ± 0.5 × 10−7 M (Fig. 2A). If the engagement of J6f1 and streptavidin by BTA results in new interactions forming between the two macromolecules, then the complex should be more stable than would be expected solely on the basis of the interaction between J6f1 and the tobramycin portion of BTA. One manner in which to assess this hypothesis is to use a competition assay, where the ability of BTA to compete with streptavidin/BTA for J6f1 is compared (Fig. 2B). In the event of cooperative binding between J6f1 and streptavidin/BTA, streptavidin/BTA should be a more effective competitor than BTA. For this purpose, streptavidin was covalently coupled to Affi-Gel beads and preloaded with BTA (PLB). The beads were then presented with 32P-J6f1 and increasing concentrations of either BTA or streptavidin/BTA for 2 h (Fig. 2B). After sedimentation of the beads, the amount of J6f1 associated with the beads was quantified. BTA is not an effective competitor because at 100 μM, 71% of radiolabeled J6f1 was still associated with the PLBs (Fig. 2C). Streptavidin alone did not effectively compete, because 75% of the J6f1 RNA was still associated with the PLBs when incubated with 7 μM streptavidin (data not shown). However, streptavidin/BTA efficiently competed J6f1 from the PLBs (at concentrations at least 100-fold lower than BTA) (Fig. 2C). These results indicate that J6f1 cooperatively interacts with streptavidin/BTA, consistent with BTA mediating an affinity-enhancing effect between streptavidin and J6f1.
Figure 2.
Enhanced affinity of J6f1 for streptavidin mediated by BTA. (A) Binding isotherm of J6f1 and X1 to streptavidin in the presence of 2 μM BTA or 2 μM BTD. Reported values are the average of three independent experiments with SE. (B) Model of the competition-binding assay. Favorable RNA/protein-enhancing interactions are schematically denoted and predicted to result in complexes that can be more efficiently competed by streptavidin/BTA than by BTA. Asterisks denote radiolabeled RNA. (C) Competition-binding curves. Reported values represent the average of four experiments with SD shown. The maximum radioactivity associated with the beads was 11,961 cpm (100%) (PLB incubated with RNA). Less than 10% of the input radioactivity was retained if streptavidin beads were incubated with 32P-labeled J6f1 in the absence of BTA. Incubation of PLB, 32P-J6f1 RNA, and 7 μM streptavidin (no BTA) for 2 h resulted in only ≈25% of the RNA being lost from the PLB.
Forced RNA/Protein Engagement by a CID Inhibits Translation.
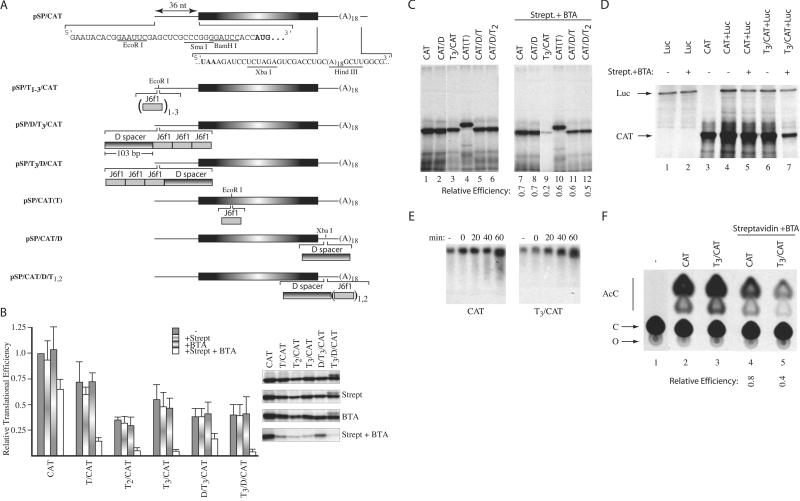
We wished to assess whether CID-mediated RNA/protein complex formation can be used as a molecular switch to modulate gene expression. We engineered one, two, or three copies of the J6f1 aptamer into the 5′ UTR (pSP/T1/CAT, pSP/T2/CAT, pSP/T3/CAT,pSP/D/T3/CAT, pSP/T3/D/CAT), coding region [pSP/CAT(T)], and 3′ UTR of the CAT reporter gene (pSP/CAT/D, pSP/CAT/D/T1–2) (Fig. 3A). Of the possible points for interdicting protein expression by interfering with mRNA metabolism, we chose to analyze the effect on the process of translation, because physical barriers (secondary structure or bound proteins) within mRNA 5′ UTRs are known to be inhibitory to mRNA expression (10, 18).
Figure 3.
Inhibition of eukaryotic translation by forced protein/RNA interaction. (A) The nucleotide sequence within the 5′ and 3′ UTRs of the CAT reporter used in this study is presented, and the CAT initiation and termination codons are shown in bold. The J6f1 aptamer is denoted by a gray shaded box, and the sequence used as a spacer (labeled D spacer) is denoted as a box with horizontal shading. Subscripts denote the number of copies inserted within a reporter. (B) Relative translation efficiencies of reporters containing tobramycin-binding sites within their 5′ UTRs. Translations were performed with RNA alone (20 μg/ml) or in the presence of 16.6 μg/ml of streptavidin, 10 μM BTA, or 16.6 μg/ml of streptavidin + 10 μM BTA, and the efficiencies standardized to the efficiency obtained with CAT mRNA, which was set at one. In these experiments, streptavidin and BTA were first mixed with the translation extracts, and mRNA templates were added last. All values represent the average of at least four independent experiments, with trichloroacetic acid precipitation counts performed in duplicate for each experiment. Standard deviations are shown. To the right of the bar graph is a representative autoradiograph from one experiment, illustrating the translation products obtained with each reporter. (C) Relative translational efficiencies of reporters containing tobramycin-binding sites within the CAT coding or 3′ UTR. The identity of the input RNA and the presence of 16.6 μg/ml of streptavidin + 10 μM BTA are indicated (Top). Shown below are the relative translational efficiencies compared with the input mRNA translated in the absence of streptavidin + BTA (average of two experiments). The largest SE for this data set was ± 0.1. (D) Cotranslation of luciferase and CAT reporters in the presence or absence of 16.6 μg/ml of streptavidin/10 μM BTA (indicated above lanes). The positions of migration of luciferase and CAT products are indicated. After electrophoresis in a 10% SDS/polyacrylamide gel, the gel was treated with EN3Hance, dried, and exposed to X-Omat (Kodak) film. (E) Stability of CAT and T3/CAT mRNA in translation extracts. 3H-labeled CAT and T3/CAT mRNA were translated in the presence of 16.6 μg/ml of streptavidin/10 μM BTA. At the indicated times, an aliquot of the translation was removed (10 μl) and incubated with 50 μg of Proteinase K at 37°C for 15 min, after which the sample was phenol/chloroform extracted. After ethanol precipitation, RNA samples were fractionated on a 1.2% agarose/formaldehyde gel. The gel was treated with En3Hance, dried, and exposed to X-Omat x-ray film (Kodak) at −70°C with an intensifying screen. (F) Effect of BTA on translation of CAT and T3/CAT in vivo. Translations in Xenopus oocytes consisted of microinjecting 10 oocytes with 50 nl of [3H]CTP-labeled mRNA (5 ng) alone or in combination with streptavidin (17 ng) and BTA. Given an average oocyte diameter of 1.2 mm (≈0.7 μl) (31), this achieved a final concentration of ≈7 μg/ml of mRNA, 24 μg/ml of streptavidin, and 10 μM BTA. Oocytes were incubated at 20°C for 2 h, homogenized in lysis buffer (20 mM Tris⋅HCl, pH 7.6/0.1 M NaCl/1% Triton X-100/1 mM phenylmethylsulfonyl fluoride), centrifuged in a microfuge for 5 min at 14,000 ×g, and the supernatant used to measure CAT activity (32). An autoradiograph of a representative TLC for the CAT assays performed is shown. The relative expression was calculated by comparing the relative conversion for each sample in the presence of streptavidin/BTA to the conversion obtained with the same mRNA in the absence of streptavidin/BTA, which was set as one. The nature of the injected mRNA and the presence of streptavidin and BTA are indicated (Top). Shown below is the relative translational efficiencies compared with the mRNA injected in the absence of streptavidin + BTA (average of two experiments). AcC, acetylated forms of chloramphenicol; C, chloramphenicol; O, origin.
The effect of complex formation on translation was assessed as a function of copy number and position of the J6f1 aptamer within the 5′ UTR (Fig. 3B). In these experiments, BTA and streptavidin were added to the translation mix before addition of mRNA template. The relative translation efficiencies of T2/CAT, T3/CAT, D/T3/CAT, and T3/D/CAT, in the absence of exogenously supplied streptavidin or BTA, are ≈2- to 3-fold lower than CAT mRNA expression (Fig. 3B, gray bars) and most likely reflect the increased secondary structure within the 5′ UTRs because of multiple copies of J6f1 aptamers (predicted ΔG = −18 kcal/mol/aptamer inserted). When translated in the presence of either streptavidin or BTA, the relative efficiency of each reporter transcript, compared with the same reporter translated in the absence of streptavidin or BTA, does not change significantly (Fig. 3B). However, in the presence of streptavidin and BTA, the translation of T1–3/CAT and T3/D/CAT transcripts was significantly impaired (5- to 10-fold) (Fig. 3B, open bars). The expression of CAT and D/T3/CAT mRNAs was only slightly decreased (≈1.6- and 2-fold, respectively). Reporter transcripts containing J6f1 aptamers within the CAT coding or 3′ UTR showed a <2-fold effect on translation efficiency when translated in the presence of streptavidin/BTA (Fig. 3C, compare lanes 10–12 to lanes 4–6). These results indicate that the forced engagement of a protein/RNA complex by a small molecule can be used to inhibit eukaryotic protein synthesis when targeted to the 5′ proximal cap sequences of the mRNA.
A formal possibility for the CID-mediated inhibitory effect on translation is that a transdominant inhibitor, formed in the context of the T1–3/CAT reporter mRNAs, leads to nonspecific inhibition. To exclude this possibility, luciferase mRNA was translated alone (Fig. 3D, lanes 1 and 2), cotranslated with CAT (lanes 4 and 5), or cotranslated with T3/CAT mRNA (lanes 6 and 7). The translation of luciferase mRNA was not affected by the presence of streptavidin/BTA (compare lane 2 to lane 1). There was no significant difference in the translation of CAT and luciferase mRNAs, when cotranslated in the absence or presence of streptavidin/BTA (compare lanes 5, 4, and 3). However, when luciferase and T3/CAT mRNAs were cotranslated, T3/CAT mRNA expression was reduced 4-fold, whereas luciferase expression was not affected (compare lane 7 to lane 6).
Kinetic analysis of CAT and T3/CAT mRNAs, translated in the presence of streptavidin/BTA, revealed that both transcripts are equally stable over a 1-h translation time course (Fig. 3E). The results rule out differences in stability between CAT and T3/CAT mRNAs, in the presence of streptavidin/BTA, as a possible reason for the observed differences in translation (Fig. 3B). To assess the effects of CID-mediated complex formation on translation in vivo, we used the Xenopus oocyte system, because direct microinjection of BTA and streptavidin overcomes delivery and permeability issues inherent to mammalian cell culture experiments. Translation of CAT and T3/CAT mRNAs, in the absence or presence of streptavidin/BTA, was determined by analysis of CAT activity after injection of mRNA. In these experiments, mRNA, BTA, and streptavidin were premixed and injected simultaneously. Extracts from oocytes not injected with mRNA showed no CAT activity (Fig. 3F, lane 1), whereas those injected with CAT or T3/CAT mRNA exhibited similar activity (lane 2, 20%; lane 3, 24% conversion). Coinjection of CAT with streptavidin/BTA resulted in a slight decrease in translation (compare lane 4 to lane 2), whereas injection of T3/CAT mRNA with streptavidin/BTA resulted in significant inhibition of translation (2.5-fold less than injection of T3/CAT alone; compare lane 5 to lane 3). These results demonstrate that CIDs can be used to inhibit translation of a target mRNA in vivo.
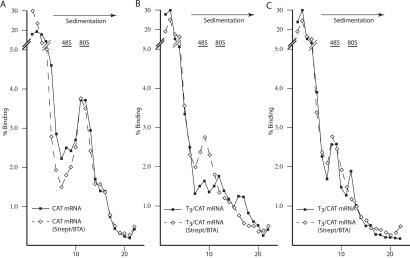
Inhibition of 80S Complex Formation.
Cis-acting mRNA structures can inhibit translation initiation by (i) perturbing the interaction of initiation factors with the mRNA template, (ii) preventing binding of the 40S ribosome at or near the cap structure, and/or (iii) preventing migration of the 40S ribosome along the mRNA 5′ UTR. To determine at which of these steps streptavidin/BTA exerted their effects on T3/CAT translation, we performed a series of ribosome-binding studies. The formation of 80S complexes on the CAT mRNA template is not affected by streptavidin/BTA (Fig. 4A), consistent with previous translation results (Fig. 3). The ribosome-binding pattern of T3/CAT (in the absence of streptavidin/BTA) was more complex than that of CAT and suggested the possible presence of: (i) a “trapped” upstream 40S ribosome (Fig. 4B, peak in the 48S region), (ii) 80S ribosomes, and (iii) a heavier complex that may represent an 80S ribosome with a loaded upstream 40S ribosome. Simply interpreted, these results suggest that secondary structure (because of the J6f1 aptamers) reduces the ability of 40S ribosomes to scan the 5′ UTR and traps some of these upstream of the J6f1 aptamer. In the presence of streptavidin/BTA, T3/CAT is unable to form 80S complexes and accumulates only 40S ribosomes (Fig. 4B).
Figure 4.
Effects of forced RNA/protein engagement on 80S ribosome assembly. 32P-labeled CAT (A) or T3/CAT (B, C) was assayed in the absence (solid line) or presence (dashed line) of 160 ng/μl of streptavidin + 8 μM BTA. Ribosome bindings were performed in the presence of 0.6 mM cyclohexamide (A, B) or 0.6 mM 5′-guanylylimidodiphosphate (C). Fractions were collected by using a Brandel Tube Piercer (Gaithersburg, MD) connected to an ISCO fraction collector (Lincoln, NE) and radioactivity determined by Cherenkov counting. The total counts recovered from each gradient were: (A) CAT mRNA (59,489 cpm); CAT mRNA + strept/BTA (46,482 cpm); (B) T3/CAT mRNA (69,451cpm); T3/CAT mRNA + strept/BTA (50,124 cpm); (C) T3/CAT mRNA (68,119 cpm); and T3/CAT mRNA + strept/BTA (50,553 cpm).
To evaluate directly 40S ribosome binding to T3/CAT, we performed ribosome bindings in the presence of 0.6 mM 5′-guanylylimidodiphosphate (which inhibits the joining of the 60S ribosome subunit as well as release of eIF-2) (Fig. 4C). In the absence of streptavidin/BTA, two complexes are observed, possibly reflecting the presence of one and two 40S ribosomes on T3/CAT (Fig. 4C). In the presence of streptavidin/BTA, a single 48S peak is observed. Thus, although streptavidin/BTA appears to exert a minor effect on loading of the 40S ribosome onto the mRNA template (Fig. 4C), the major effect appears to prevent conversion of 40S to 80S complexes (Fig. 4B). A possible mechanism by which this could be achieved would be to sterically inhibit migration of the 40S ribosome.
Discussion
RNA offers several tactical advantages as a drug target: it lacks a known cellular repair mechanism, contains a large repertoire of structural diversity, and its manipulation has the potential to prevent or enhance gene expression, as well as to achieve regulation at levels that would be difficult to obtain by other approaches (i.e., allele- or isoform-specific modulation of gene expression). Our results indicate that CIDs can be engineered to force the engagement of an RNA/protein interaction, and that this complex can be sufficiently stable to exert a biological effect on translation. Although we have not formally demonstrated that BTA remains part of the complex formed between streptavidin and the tobramycin-binding aptamers, it is unlikely to be discarded during complex formation, given the high affinity between biotin and streptavidin.
The complex formed between streptavidin, BTA, and J6f1 appears more stable than expected based on the sum of the stabilities of the individual components, because a ≈100-fold difference in concentration of BTA is required to compete with preloaded streptavidin/BTA beads for J6f1, compared with streptavidin/BTA (Fig. 2C). Although, the Kd for tobramycin and J6f1 has been reported to be 0.8–5 nM (19, 20), much lower than what we measured for the streptavidin/BTA/J6F1 complex, it is known that chemical addition at the 6′ amino group of tobramycin can significantly lower the Kd value [e.g., Kd = 0.2 μM after addition of carboxytetramethyl rhodamine at the 6′ position of tobramycin (20)]. Measurements of the Kd between J6f1 and BTA by polyacrylamide affinity coelectrophoresis (21) have indicated a value >1 μM (data not shown). Thus, in the case of streptavidin, BTA, and J6f1, the complex is more stable than would be expected based on the stabilities of the individual components.
One likely explanation for the increase in affinity observed with the trimeric complex is that streptavidin/J6f1 interactions make a significant direct energetic contribution to the stability of the complex. Although our data do not directly demonstrate novel protein/RNA interactions resulting from the covalent linkage of biotin to tobramycin, the observed change in affinity is related to both the structure of the RNA (Fig. 2A, compare J6f1/BTA and X1/BTA) and the nature of the protein surface (Fig. 1 B and C; compare streptavidin and avidin). These results identify the surface between the RNA and protein domains as the origin of the altered affinity. There are several ways in which RNA could interact with a proximal protein surface. An induced fit mechanism would involve the reorganization of local elements of RNA secondary structure and/or the formation of structure from disordered single-stranded regions, with the result being stabilization of a defined three-dimensional conformation. In this case, the protein moiety would provide a rigid scaffold around which the RNA could mold. Additionally, local disordered protein regions could gain secondary structure through intermolecular interactions with an RNA scaffold (22), making protein folding dependent on specific sequences of the RNA target. The contribution of the CID to the RNA/protein complex is critical because, in its absence, no interaction occurs (Fig. 1), and it likely plays a role in the early steps of ligand recognition. One interesting mechanism here is a combination of induced fit and rigid docking, with the target molecule being recognized by rigid binding (provided by the BTA/J6f1 interaction) and then maximizing the interaction potential through induced fit, as suggested for some protein/RNA complexes (23, 24). Thus, although the flexible nature of RNA is disadvantageous for small-molecule targeting, it is advantageous in allowing adaptive binding when presented a protein surface by a small-molecule CID. These models provide an understanding of how nonphysiological partners can be made to interact in a specific fashion and form high-affinity complexes. Ultimately, structural and molecular dynamic studies will be helpful for elucidating the mechanism of complex formation and the nature of the contacts in the complex.
There are a number of possible reasons why avidin does not appear to interact with J6f1 or X1 in a CID-dependent fashion. An appropriate local surface area for binding to RNA may be absent on this protein (either because of a lack of amino acids that can interact with RNA or the presence of amino acids that are energetically unfavorable to binding). Alternatively, the CIDs we designed may not favor avidin/RNA interaction. Indeed, there are several features of our strategy that could be optimized to produce trimeric complexes of improved affinities: (i) Varying the nature of the recruited protein by altering one portion of the bivalent CID. In the case provided herein, we used a protein (streptavidin) unlikely to participate in RNA metabolism and thus whose surface has not evolved to accommodate RNA binding. The appropriate recruitment of an RNA-binding protein may provide a surface that can better accommodate RNA. (ii) Optimizing the nature of the spacer of the bivalent CID to provide the appropriate spacing, rigidity, or tilt for correct alignment of the protein with the target RNA. (iii) Altering the region of the target RNA such that the appropriate degree of single and double strandedness is available to allow for protein binding. (iv) For this targeting approach to be applicable in vivo, one would also want the protein and RNA partners to be in the same subcellular compartment and in relatively close proximity.
Although there are many steps at which mRNA expression can be regulated, the only ones where stable higher-order complexes are known to reproducibly and predictably inhibit mRNA function are at the level of splicing (25) and translation (10, 18, 26). The observed positional effects of CID-mediated inhibition of translation are consistent with what is known concerning binding of iron regulatory protein 1 (IRP-1) to the 5′ UTR of the ferritin and eLAS (5-aminolevulinate synthase) mRNAs (27, 28), which prevents the recruitment of the 43S preinitiation complex by eIF-4F. Increasing the distance between the binding site for IRP-1 and the cap structure has been shown to decrease the inhibitory effects of IRP-1 (29, 30). In our case, T3/CAT is able to load 40S ribosomes (Fig. 4C) but cannot form functional 80S complexes (Fig. 4B) in the presence of streptavidin/BTA. Therefore, we postulate that ribosome scanning is blocked by streptavidin/BTA/J6f1 interaction. Our results provide a new logic to modulate gene expression by demonstrating that CID-mediated engagement of a protein/RNA complex can be used to inhibit translation, and that the effect is most profound when the protein/RNA interaction targets cap-proximal sequences.
Acknowledgments
We thank Dr. Nahum Sonenberg for critical reading of the manuscript and for support, enthusiasm, and insights during the course of this work. We are grateful to Dr. Edward Meighen for helpful discussions regarding cooperative binding and to Drs. David Housman (Massachusetts Institute of Technology) and Vincent Stanton (Variagenics) for their enthusiasm during this study. J.P. is a Canadian Institutes of Health Research (CIHR) Senior Investigator. This work was supported by grants from the Canadian Institutes of Health Research and from the National Cancer Institute of Canada to J.P.
Abbreviations
- CID
chemical inducer of dimerization
- UTR
untranslated region
- EMSA
electrophoretic mobility-shift assay
- PLB
preloaded beads
- CAT
chloramphenicol acetyltransferase
- BTA
biotinamidocaproate tobramycin amide
- BTD
N-biotinyl-12-aminododecanoyltobramycin amide
References
- 1.Carter A P, Clemons W M, Brodersen D E, Morgan-Warren R J, Wimberly B T, Ramakrishnan V. Nature (London) 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 2.Brodersen D E, Clemons W M, Jr, Carter A P, Morgan-Warren R J, Wimberly B T, Ramakrishnan V. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 3.Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 4.Briesewitz R, Ray G T, Wandless T J, Crabtree G R. Proc Natl Acad Sci USA. 1999;96:1953–1958. doi: 10.1073/pnas.96.5.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi J, Chen J, Schreiber S L, Clardy J. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 6.Crabtree G R, Schreiber S L. Trends Biochem Sci. 1996;21:418–422. doi: 10.1016/s0968-0004(96)20027-1. [DOI] [PubMed] [Google Scholar]
- 7.Ling, S., Verdine, G., Basilion, J. P. & Stanton, V. P., Jr. (2000) Eur. Patent Org. WO 0052210.
- 8.Leulliot N, Varani G. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J, Russell D W. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 10.Pelletier J, Sonenberg N. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 11.Weber P C, Ohlendorf D H, Wendoloski J J, Salemme F R. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 12.Hendrickson W A, Pahler A, Smith J L, Satow Y, Merritt E A, Phizackerley R P. Proc Natl Acad Sci USA. 1989;86:2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pugliese L, Coda A, Malcovati M, Bolognesi M. J Mol Biol. 1993;231:698–710. doi: 10.1006/jmbi.1993.1321. [DOI] [PubMed] [Google Scholar]
- 14.Livnah O, Bayer E A, Wilchek M, Sussman J L. Proc Natl Acad Sci USA. 1993;90:5076–5080. doi: 10.1073/pnas.90.11.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Rando R R. Chem Biol. 1995;2:281–290. doi: 10.1016/1074-5521(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 16.Jiang L, Suri A K, Fiala R, Patel D J. Chem Biol. 1997;4:35–50. doi: 10.1016/s1074-5521(97)90235-0. [DOI] [PubMed] [Google Scholar]
- 17.Jiang L, Patel D J. Nat Struct Biol. 1998;5:769–774. doi: 10.1038/1804. [DOI] [PubMed] [Google Scholar]
- 18.Stripecke R, Hentze M W. Nucleic Acids Res. 1992;20:5555–5564. doi: 10.1093/nar/20.21.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Killian J, Hamasaki K, Rando R R. Biochemistry. 1996;35:12338–12346. doi: 10.1021/bi960878w. [DOI] [PubMed] [Google Scholar]
- 20.Cho J, Hamasaki K, Rando R R. Biochemistry. 1998;37:4985–4992. doi: 10.1021/bi972757h. [DOI] [PubMed] [Google Scholar]
- 21.Cilley C D, Williamson J R. Methods Mol Biol. 1999;118:129–141. doi: 10.1385/1-59259-676-2:129. [DOI] [PubMed] [Google Scholar]
- 22.Legault P, Li J, Mogridge J, Kay L E, Greenblatt J. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 23.Allain F H, Howe P W, Neuhaus D, Varani G. EMBO J. 1997;16:5764–5772. doi: 10.1093/emboj/16.18.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoldt M, Wohnert J, Ohlenschlager O, Gorlach M, Brown L R. EMBO J. 1999;18:6508–6521. doi: 10.1093/emboj/18.22.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clouet d'Orval B, d'Aubenton Carafa Y, Sirand-Pugnet P, Gallego M, Brody E, Marie J. Science. 1991;252:1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- 26.Stripecke R, Hentze M W. Mol Cell Biol. 1994;14:5898–5909. doi: 10.1128/mcb.14.9.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray N K, Hentze M W. EMBO J. 1994;13:3882–3891. doi: 10.1002/j.1460-2075.1994.tb06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muckenthaler M, Gray N K, Hentze M W. Mol Cell. 1998;2:383–388. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- 29.Goossen B, Caughman S W, Harford J B, Klausner R D, Hentze M W. EMBO J. 1990;9:4127–4133. doi: 10.1002/j.1460-2075.1990.tb07635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goossen B, Hentze M W. Mol Cell Biol. 1992;12:1959–1966. doi: 10.1128/mcb.12.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colman A. In: Translation of Eukaryotic Messenger RNA in Xenopus Oocytes. Hames B D, Higgins S J, editors. Oxford: IRL; 1984. pp. 271–302. [Google Scholar]
- 32.Gorman C. In: High Efficiency Gene Transfer into Mammalian Cells. Glover D M, editor. Oxford: IRL; 1984. pp. 143–190. [DOI] [PubMed] [Google Scholar]