Abstract
The Tap protein mediates the sequence nonspecific nuclear export of cellular mRNAs as well as the sequence-specific export of retroviral mRNAs bearing the constitutive transport element (CTE). Previously, the structures of individual Tap subdomains, including ribonucleoprotein and leucine-rich repeat domains, have been described. Here, we report the crystal structure of a functional CTE RNA-binding domain of human Tap, including the N-terminal arm of the ribonucleoprotein domain and interdomain linking polypeptide. To identify residues that interact with the CTE, we have introduced 38 alanine substitutions for surface residues in the Tap CTE-binding domain and tested these mutants for their ability to support CTE-dependent nuclear RNA export and CTE binding. Four residues that cluster on a concave surface in the leucine-rich repeat domain were found to be critical for CTE binding and define a CTE-interacting surface on this domain. The second critical CTE-interacting surface on Tap is defined by three previously identified residues on the surface of the ribonucleoprotein domain. The structural and mutational data define a novel RNA-binding site on the Tap protein.
Retroviral replication requires the nuclear export and cytoplasmic translation of both fully and incompletely spliced viral mRNAs. The ability to express mRNAs that retain one or more introns contrasts sharply with cellular mRNAs, which are exported in a fully processed form. In fact, unspliced viral mRNAs must overcome cellular retention mechanisms that normally prevent the inappropriate export of incompletely spliced cellular pre-mRNAs. To accomplish this task, retroviruses have developed at least two mechanisms to export unspliced viral transcripts (reviewed in ref. 1). Complex retroviruses, e.g., HIV type 1, encode an adapter protein, termed Rev, which recruits the nuclear export factor Crm1 to viral mRNAs (2–5). In contrast, several simple retroviruses, such as Mason–Pfizer monkey virus (MPMV), encode an RNA element, termed the constitutive transport element (CTE), which accesses a cellular RNA export pathway directly (6). The human Tap/NXF1 protein can mediate the sequence-specific nuclear export of mRNAs bearing the MPMV CTE and is also thought to play a critical role in the sequence nonspecific export of cellular mRNAs (7–11).
Tap/NXF1 bears at least three distinct functional domains, i.e., a CTE RNA-binding domain (96–372), a central binding domain (≈370–550) for an essential cellular cofactor termed p15 or NXT-1 (10, 12, 13), and finally a carboxyl-terminal domain that directly interacts with several nucleoporins and also functions as a nuclear export signal (≈550–619) (10, 14, 15). Two crystal structures of a Tap molecule comprising residues 102–372 have been determined (16). One structural model includes residues 119–198 and 205–362, and the second includes residues 123–191 and 203–362. The remaining residues in each of the structures are disordered. The Tap structure comprises two domains, i.e., a noncanonical ribonucleoprotein (RNP) domain (119–198) and a leucine-rich repeat (LRR) domain (203–362). Importantly, Tap 102–372 does not bind to the CTE, but a slightly longer fragment of Tap including residues 96–372 does bind to the CTE in vitro (16).
Previously, it has been demonstrated that the MPMV CTE displays species specificity, i.e., the CTE functions in human but not in quail cells. More importantly, the demonstration that CTE function can be rescued in quail cells by expression of human Tap in trans (15) has provided a basis for identifying residues in Tap that are critical for binding to the CTE (17). Because the CTE is the only well-defined substrate for Tap/NXF1, determining residues involved in binding to this retroviral RNA element may provide key insights into how this nuclear export factor interacts with cellular mRNAs.
To accomplish this goal, we have determined the crystal structure of a functional RNA-binding domain from Tap, comprising residues 96–372, and investigated the interactions of the CTE with surface residues of the RNP and LRR domains of Tap by using in vivo CTE function and RNA-binding assays. We have identified four residues in the LRR domain that are critical for CTE binding and cluster on a concave surface of the LRR domain [distinct from that in another report (16) near an identified critical residue, Arg-249 (17)]. We now propose that this conserved concave surface of the LRR domain, the previously identified region in the RNP domain, and the polypeptide linking the RNP and LRR domains play key roles in interactions of Tap with the CTE. Surprisingly, the CTE-interacting surfaces on both the LRR and RNP domains of Tap are entirely different from the RNA-interacting surfaces in the spliceosomal U2B′′-U2A′-RNA complex (18), which comprises structurally analogous RNP (U2B′′) and LRR (U2A′) domains as separate polypeptides.
Materials and Methods
Plasmid Construction.
A DNA sequence encoding amino acids 96–372 of Tap was PCR-amplified by using primers that introduced flanking NdeI and XhoI sites. The digested PCR product was cloned into the pET15b expression vector (Novagen). Point mutations C143S, C252S, and C328S were introduced by using the QuikChange site-directed mutagenesis kit (Stratagene) and confirmed by DNA sequencing. All vertebrate expression plasmids are based on pBC12/cytomegalovirus (CMV) (19). The following expression plasmids have been described: pBC12/CMV/β-galactosidase (β-gal) (20), pMS2-hTAP (17), pcTat-TAP (15); the indicator constructs pDM128/CTE (21) and pDM128/4XMS2 (17); and pCTE/CAT, a reporter plasmid containing the cat gene under the control of the HIV-1 long terminal repeat, where the TAR element has been replaced with the MPMV CTE (15). Tap mutants were constructed by recombinant PCR and were cloned into the NcoI/XhoI sites of pMS2 (17). Plasmids expressing Tap mutants fused to HIV-1 Tat were generated by subcloning the mutant alleles from the pMS2 vector into the NcoI/XhoI sites of pcTat-TAP. All clones were verified by DNA sequencing and Western blot analysis.
Protein Expression and Purification.
The native Tap protein was expressed in Escherichia coli BL21 (DE3) in LB medium, and the selenomethionyl (SeMet) derivative was expressed in E. coli B834 (DE3) in LeMaster medium in the presence of 1 mg/l of thiamine (Sigma) and dl-selenomethionine (Sigma) at 50 mg/liter as described (22). Cell cultures were grown at 37°C until OD600 ≈ 0.7, induced with isopropyl β-d-thiogalactoside (IPTG) overnight at 30°C, and then harvested. The soluble lysates were applied to an Ni-NTA (Qiagen) affinity column. The fractions containing Tap were then applied to an S-Sepharose ion-exchange column and eluted by an NaCl gradient. Purified protein was then digested with Thrombin (Novagen) and reapplied to a Mono-S column (Amersham Biosciences). The final purification step and buffer exchange were performed by using a Superdex 75 (Amersham Biosciences) column.
Crystallization and Data Collection.
Both the native and SeMet Tap 96–372 were crystallized by mixing a 1:1 ratio of protein solution at ≈ 50 mg/ml and precipitant solution containing 1.1 M potassium sodium tartrate/0.05 M sodium citrate (pH 5.6) in a hanging drop at 20°C. Bipyramid-shaped crystals appeared and grew to a size of 300 × 300 × 150 μm3 (native) and 150 × 150 × 100 μm3 (SeMet derivative) in 4–5 days. Crystals were cryoprotected in 1.15 M potassium sodium tartrate/0.05 M sodium citrate, pH 5.6/0.15 M NaCl/16% ethylene glycol and flash-frozen in liquid nitrogen. The data were collected at the National Synchrotron Light Source, beamline X25, Brookhaven National Laboratory at λ = 1.1000 Å for the native crystal and the selenomethionine (SeMet) peak wavelength (λ = 0.9789 Å) for the SeMet Tap crystal, then processed with the denzo/hkl package (23). The crystals are tetragonal, space group P41212, and include four molecules in the asymmetric unit. See Table 1 for cell constants and data processing statistics.
Table 1.
Crystallographic data Tap (96–372)
| Native | SeMet derivative | |
|---|---|---|
| Data collection statistics | ||
| Space group | P41212 | P41212 |
| Cell dimensions, Å | a = b = 136.7, c = 202.9 | a = b = 138.2, c = 205.0 |
| Limiting resolution, Å | 3.8 | 3.8 |
| Completeness, % | 99.7 (99.6) | 95.5 (92.5) |
| Rsym, % | 7.2 (57.8) | 13.2 (46.8) |
| I/σ | 21.6 (1.9) | 9.8 (2.0) |
| Refinement statistics | ||
| Resolution limits | 8.0–3.8 | 8.0–3.8 |
| Rigid body (R/Rfree), % | 40.8/39.9 | 37.9/37.4 |
| Refinement (R/Rfree), % | 27.5/35.5 | 31.7/39.5 |
Rsym = {Σ| (I − < I >)|}/{ Σ (I) }, where < I > refers to the average intensity of multiple measurements of the same reflection. Rwork and Rfree = (Σ | Fobs − Fcalc |)/(Σ | Fobs |), where Rfree was calculated over 3% of the amplitudes not used in refinement. R values are reported for all data within the specified resolution range. Values in parentheses are statistics for data in the highest resolution shell 3.94–3.8 Å. The native Tap protein includes three amino acid substitutions (C143S, C252S, and C328S) and SeMet Tap includes a single substitution (C252S).
Structural Determination and Refinement.
Phasing was obtained by molecular replacement (24), using the Tap structure (Protein Data Bank ID code 1FT8) (16) as the search model for both the native and SeMet Tap. The SeMet Tap data were used to verify the molecular replacement solution. In a Se Bijvoet-difference Fourier map calculated by using the molecular replacement phases in cns (25), 14 peaks (of the 16 expected) corresponding to Se positions were well above the noise level of the map and coincided with the S position of methionine residues in the structural model of Tap. There should be three Se sites in the LRR domain and one site in the RNP domain of each Tap molecule. The two Se sites expected in the RNP domains of the B and D molecules, which are not as well-ordered as the RNP domains from the A and C molecules, are not evident. Model refinement for both crystal structures was carried out by using cns (25) and interspersed with model building with o (26). The final SeMet Tap model includes two intact molecules, A (105–362) and C (101–362). For the other two molecules, only the LRR domains have been included in the refinement (203–367 of B and 204–372 of D). The native model includes the same residues as the SeMet Tap for the A molecule, residues 201–367 for B, 105–362 for C, and 201–372 for D. For both structures, there is sufficient electron density to approximately position but not accurately model the RNP domains of the B and D molecules. areaimol (27) was used to calculate buried surface areas. Ribbon renderings of the structural models were generated by using molscript (28) and RASTER3D (29), and a molecular surface rendering was generated by using grasp (30). Coordinates for the structures have been deposited in the PDB, native (1KOH) and SeMet (1KOO).
Cell Culture and Transfections.
Human 293T cells and quail QCl-3 cells were maintained as described (21, 31) and were transfected by using Fugene-6 (Roche Molecular Biochemicals) or DEAE-Dextran (31), respectively. All transfections were performed in 35-mm plates. Levels of DNA used in each transfection experiment are denoted in the Fig. 2 legend, with pBC12/CMV/β-gal included as an internal control. In all transfection experiments, CAT enzyme levels were determined ≈48 h after transfection, as described, and normalized to the level of β-gal activity present in the cell lysate (21).
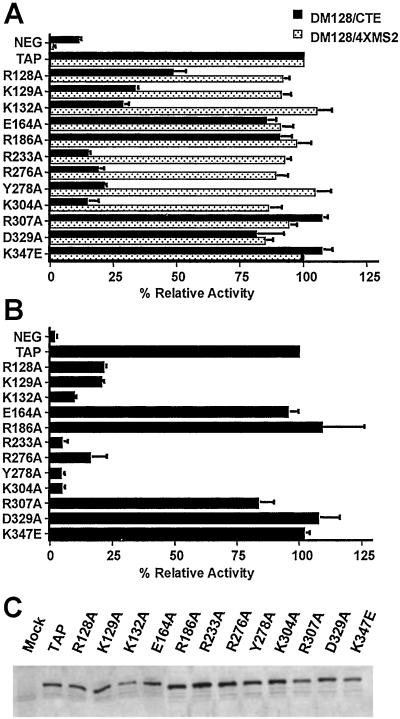
Figure 2.
Mutation of critical residues on the surface of Tap inhibits both CTE function and CTE binding. (A) Fusion proteins consisting of the MS2 coat protein linked to wild-type or mutant Tap were assayed in QCl-3 cells for their ability to induce the nuclear export of an unspliced cat mRNA encoded by pDM128/CTE. These constructs were also assayed in 293T cells for their ability to activate nuclear export when tethered via the MS2 operator RNA present in pDM128/4XMS2. Cell cultures (35-mm) were transfected with 25 ng of pDM128/CTE or pDM128/4XMS2, 50 ng of the pBC12/CMV/β-gal internal control plasmid, and 200 ng of the indicated effector plasmid. The parental plasmid pBC12/CMV served as the negative control. Induced CAT activities are expressed relative to the culture transfected with pMS2-Tap expression plasmid. The average of three transfection experiments with SD is indicated. (B) The indicated Tap mutants were fused to the HIV-1 Tat protein and assayed for their ability to bind to the MPMV CTE in vivo. 293T cells were transfected with 100 ng of pCTE/CAT, 50 ng of pBC12/CMV/β-gal and 500 ng of the indicated Tat-Tap expression plasmid. Induced CAT activities are expressed relative to the culture transfected with the wild-type pcTat-Tap plasmid. The average of three transfection experiments with SDs is indicated. (C) Western analysis of the expression of the indicated Tat–Tap fusion proteins in transfected 293T cells was performed by using a rabbit polyclonal anti-Tat antiserum (36).
Results and Discussion
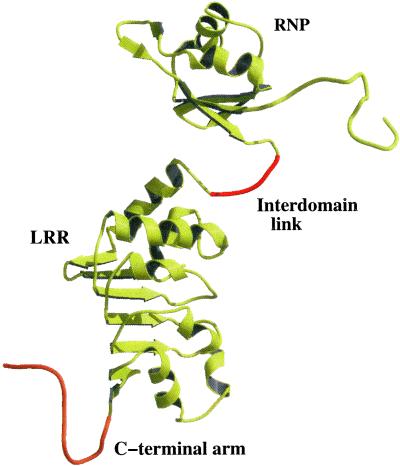
We have determined the crystal structure of a functional RNA-binding domain from the human Tap protein, comprising residues 96–372, at 3.8 Å. The crystals of the SeMet and native Tap proteins are tetragonal (space group P41212) but are not isomorphous to a reported tetragonal form space group P41212 (16). Our crystals were obtained by using a different construct and under different crystallization conditions (see Materials and Methods and Table 1). The structures were determined by molecular replacement. Se Bijvoet-difference Fourier analysis of the SeMet Tap data was used to verify the molecular replacement solution in addition to standard refinement methods. There are four molecules (A–D) in the asymmetric unit of the crystal, which are not related by 4-fold noncrystallographic symmetry (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). The interactions within the asymmetric unit are primarily between the LRR domains but are not conserved in the different interacting pairs of LRR domains, suggesting that they are not biologically relevant. The structure of the RNP domain from the crystal structure determined at 3.15 Å resolution (PDB ID code 1FT8) and that of the LRR domain from the structure determined at 2.9 Å resolution (PDB ID code 1FO1) have been reported as independent structural domains of Tap (16). An important contribution of our work is the complete structural model for the intact RNA-binding domain of Tap (see Fig. 1), establishing the relative position of the RNP domain with respect to the LRR domain.
Figure 1.
A ribbon rendering of a composite model is shown for the RNA-binding domain of Tap 96–372. The model consists of the N-terminal arm, the RNP and LRR domains (yellow), the polypeptide linking the two domains, and the C-terminal tail of the LRR domain (orange). The structures of the RNP and LRR domains of the four intact molecules (A and C molecules for native and SeMet Tap) superimpose well with rms deviations for alpha carbons of 0.9–15 Å for individual pairs of atoms. The C-terminal tail is ordered in the D molecules of the native and SeMet Tap structures and has been included here to complete the composite model.
In contrast to the previous report, our structural model of the RNA-binding domain of Tap includes the RNP and LRR domains, interdomain linking polypeptide, and N-terminal arm resulting from contiguous electron density within a given molecule in the asymmetric unit. Despite the modest resolution of the structure, the residues linking the two domains (198–203) in Tap and the N-terminal residues were readily apparent in initial difference electron density maps resulting from molecular replacement phasing for two (A and C) of the four molecules in both structures. Our models include the residues linking the RNP and LRR domains, 198–203, and residues 105–118 for the A and C molecules in each structure (see Materials and Methods). It is apparent that there is some flexibility provided by the polypeptide linking the two domains as evidenced by the less well-ordered RNP domains from the B and D molecules. However, the flexibility is limited by the fact that the interdomain linker includes only 6 residues (all nonglycine) and is flanked by proline residues on either side (P196, P197, and P206). In addition, the surface area buried by the interdomain contacts including the linking region is ≈700 Å2, suggesting that there is a reasonable domain interface between the LRR and RNP domains.
An additional feature in our model is the structure of the C-terminal tail of the LRR domain. For one molecule (D), there was interpretable electron density for the C-terminal residues of the LRR domain 363–372. These residues interact with the same C-terminal residues from a symmetry-related D molecule. In the structural model 1FT8 (PDB ID code), a partial model of an RNP domain (designated the E molecule) has been placed in a similar position to that occupied by the C termini of the D molecules. Placement of the complete RNP domain in this position for the 1FT8 model results in atomic overlap with a symmetry-related molecule and is therefore not consistent with crystal packing constraints.
We have reported that the quail Tap protein does not bind the retroviral CTE (15) and that the inability to bind results from a single amino acid difference compared with the human Tap protein (17). The finding that Arg-249, a residue located on the exterior of the LRR domain of Tap, is critical for CTE binding suggested that other residues on this surface of the Tap protein might also be involved in CTE binding. To delineate further the surface of the Tap protein that is required for CTE binding, we introduced substitutions for a number of surface amino acid residues and measured the ability of the resultant Tap mutants to rescue CTE function in transfected quail cells (Fig. 2A). The assay used to measure CTE function utilizes the reporter construct pDM128/CTE (15, 21, 32). This plasmid contains a cat gene and the MPMV CTE flanked by 5′ and 3′ splice sites, under the control of the CMV immediate early promoter. Expression of the cat gene product requires the nuclear export and cytoplasmic translation of an unspliced cat mRNA and this occurs efficiently only when the CTE is functional. As described (15, 17, 33), transfection of quail QCl-3 cells with the pDM128/CTE indicator plasmid alone induces a very low level of CAT activity (Fig. 2A) because of the inability of the endogenous quail Tap protein to bind the CTE. However, cotransfection of a human Tap expression plasmid activates the CTE-dependent nuclear export of the unspliced cat mRNA and, hence, increased CAT enzyme activity.
We constructed 38 alanine scanning mutants dispersed throughout the human Tap protein by using PCR, a subset of which is shown in Fig. 2A. Surface residues were selected for substitution based on the crystal structure. The following alanine mutations were introduced for residues within the RNP domain (119–198): R128A, K129A, K132A, S141A, K142A, T148A, E151A, E164A, D165A, S167A, S170A, K178A, R182A, E183A, R186A, S193A, and H198A. Three of the alanine substitutions of residues in the RNP domain (R128A, K129A, and K132A) affected the ability of Tap to rescue CTE function, which was consistent with another report (16). Three mutations, R100A, N118A, and T199A, were introduced within the N-terminal arm or linking region and had no effect on the ability of Tap to rescue CTE function. Of 19 mutations introduced within the LRR domain, 15 (E203A, K205A, K218A, S223A, N242A, E266A, N274A, R279A, R307A, K313A, K316A, D329A, T330A, R332A, and K347E) had no effect on the ability of Tap to rescue CTE function. Four single amino acid changes in the LRR domain (R233A, R276A, Y278A, or K304A) completely blocked the ability of the Tap protein to rescue MPMV CTE function in quail cells (Figs. 2A and 3 A and B, data not shown).
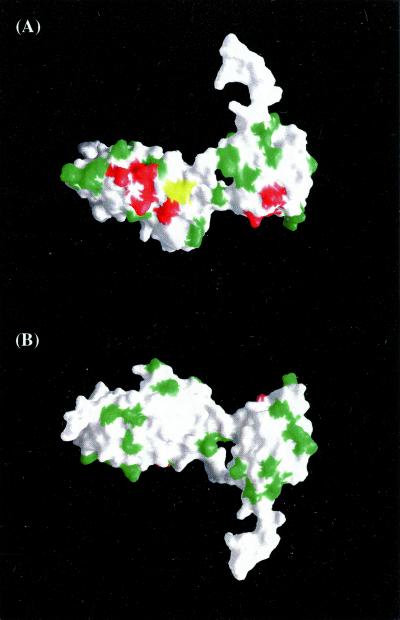
Figure 3.
The residues required for interactions of Tap with the CTE cluster on the surface of the RNP and LRR domains. (A) A surface representation is shown of the RNA-binding domain of Tap (96–372). Residues that do not affect binding are shown in green (see Results and Discussion). Residues that are important for binding are shown in red including R128, K129, K132, R233, R276, Y278, and K304. R249, which was reported (17) to be critical in binding, is shown in yellow. (B) The same surface representation of Tap as in A is shown rotated by approximately 180°.
These results suggested that the loss of CTE function might be the result of a selective loss in the ability of these Tap mutants to bind to the CTE RNA target. The data, therefore, predict that these Tap mutants would be fully functional in mediating nuclear mRNA export, if they were recruited by means of a heterologous RNA target. To test this hypothesis, we used a previously described tethering assay. The indicator construct pDM128/4XMS2 (17) is similar to pDM128/CTE, except that it contains four tandem MS2 RNA-binding sites within the intron instead of a copy of the CTE. In both nuclear mRNA export assays, Tap mutants were expressed as fusion proteins linked to the MS2 coat protein. Thus, Tap mutants that fail to rescue CTE function in quail cells, yet retain the ability to activate nuclear mRNA export when recruited by means of the heterologous MS2 RNA target, are likely defective for CTE binding.
As shown (17), nuclear export of the unspliced cat mRNA bearing four MS2-binding sites was activated by a fusion protein consisting of Tap linked to the MS2 coat protein. All of the Tap mutants that failed to export mRNAs bearing the CTE were capable of inducing the nuclear export of a similar cat mRNA when tethered by means of the MS2 RNA-binding domain (Fig. 2A). We conclude from these results that the mutant Tap proteins are fully functional mRNA export factors when recruited to a heterologous mRNA target. Therefore, the inability of the Tap mutants to rescue CTE function in quail cells must be caused by a defect in CTE binding rather than by general misfolding of these mutant proteins.
To directly demonstrate that these Tap mutants indeed fail to bind the MPMV CTE, we used a described in vivo RNA-binding assay that makes use of the Tat transcriptional activator encoded by HIV-1 (15, 17). Tat normally activates transcription from the HIV-1 long terminal repeat (LTR) promoter by binding to a promoter-proximal RNA element termed TAR (for a review see ref. 34). However, Tat can also effectively activate transcription when recruited by means of a heterologous RNA target, substituted in place of TAR, when expressed as a fusion protein displaying the appropriate RNA-binding specificity. In this case, Tat acts as a transcriptional activation domain whereas the fused protein functions exclusively as an RNA-binding domain.
As demonstrated (15, 17), a fusion protein consisting of Tat fused to Tap activates the expression of the cat reporter gene linked to an HIV-1 long terminal repeat (LTR) bearing the MPMV CTE in place of TAR (Fig. 2B). Consistent with our hypothesis that the four LRR domain Tap mutants are defective for CTE binding, R233A, R276A, Y278A, and K304A were unable to effectively recruit Tat to the HIV-1 LTR bearing the MPMV CTE (Fig. 2B). Three RNP mutants, R128A, K129A, and K132A, also showed reduced activity in this same assay. In contrast, E164A, R186A, R307A, D329A, and a reverse charge mutation, K347E, reported to affect CTE RNA binding in vitro (16), had no effect on CTE function (Fig. 2A) or CTE binding (Fig. 2B) in vivo. All Tat–Tap fusion proteins were expressed at similar levels, as determined by Western blot analysis and shown in Fig. 2C. We conclude from these data that the Tap mutants that failed to rescue CTE function in quail cells are defective because they are unable to bind to the MPMV CTE in vivo. In addition, the finding that CTE RNA binding requires residues located in both the LRR and RNP domains of Tap (Fig. 3 A and B) suggests that CTE binding is cooperative.
To assess the interactions of the CTE with Tap, the following relevant mutational and structural analyses are summarized below. We and others (16) have shown that the minimal fragment of Tap that retains RNA-binding activity includes residues 96–372. Remarkably, a fragment including residues 102–372 does not bind the CTE (16), suggesting that even a small deletion of the N-terminal arm renders Tap incapable of binding the CTE. A cis arrangement of the RNP and LRR domains is required for binding to the CTE, i.e., no binding to the CTE was observed in the presence of competitor for the RNP or LRR domains expressed as separate polypeptides (16). This finding is consistent with a direct interaction of the RNA with the residues linking the two domains. Alternatively, the conformation of the RNP and N-terminal arm relative to the LRR domain for productive binding to the CTE may require the interdomain linking region. In each of the eight Tap molecules found in our crystal structures, the RNP domain is in approximately the same position relative to the LRR. The Tap residues implicated in direct interaction with the CTE are clustered, clearly defining the interacting surfaces of the RNP and LRR domains (Fig. 3 A and B). The remaining surfaces of the LRR and RNP subdomains have been sufficiently well-sampled by point mutagenesis to suggest that they in fact are not involved in CTE binding.
The CTE includes two very similar internal loops that are potentially related by approximate 2-fold symmetry within the CTE molecule. Our data suggest that each loop of the CTE will bind to a Tap molecule (i.e., a second Tap molecule will bind to the second loop) based on structural considerations and modeling of an RNA hairpin in the binding site of Tap (data not shown). In addition, our mutational data, although limited, also suggest that the interactions with the N-terminal arm and the polypeptide linking the RNP and LRR domains are likely to be with the polypeptide backbone. Taken together, the mutational data and our intact structural model for the RNA-binding domain of Tap provide compelling evidence for direct interactions of the CTE with a concave surface of the LRR domain and a surface loop within the RNP domain. In addition, the proximity of the interdomain linking region may result in direct interactions with the CTE as well.
That the native quail Tap protein is fully functional in nuclear export of endogenous mRNA but does not bind to the CTE (15) suggests that the required interactions of Tap with the CTE vs. cellular mRNAs are in fact distinct or at least not directly overlapping. It has been suggested that nuclear export of mRNA may require formation of a larger complex involving additional proteins and is not likely to involve sequence-specific RNA interactions. In contrast, binding of the CTE to Tap is highly sequence-specific, as shown in an analysis of RNA-binding specificity of mutant CTEs (35).
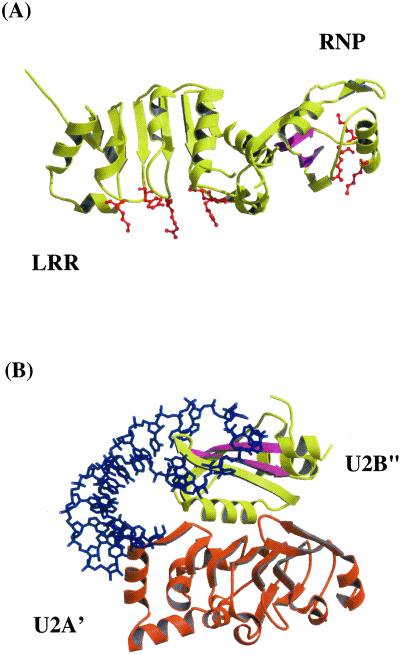
The critical interactions of the CTE with Tap are clearly distinct from RNA interactions in the spliceosomal U2B′′- U2A′-U2 hairpin-loop RNA fragment complex, currently the only other example of RNA binding requiring RNP and LRR domains, albeit in trans (See Fig. 4). In the spliceosomal complex, the RNA interacts directly with residues in the RNP1 and RNP2 motifs of the RNP domain (U2B′′) in strands β3 and β1, respectively, and also with the C terminus of the LRR domain (U2A′) (18) as shown in Fig. 4B. The residues within the RNP motifs found in U2B′′ are not conserved in Tap (16). In addition, the residues in the RNP domain of Tap that are involved in binding the CTE are located in loop 1 and the start of α1 and not in the preceding β1-strand. Further, these CTE-interacting residues in Tap do not superimpose directly with residues in the U2B′′ RNP structure (i.e., they are structurally unique). The CTE-interacting residues in the LRR domain in Tap are located primarily in loops and define a fairly extensive concave surface that is novel and clearly distinct from the small C-terminal RNA-interacting surface in the LRR domain (U2A′) of the spliceosomal complex. It has been proposed that the interactions of Tap with the CTE are similar to those found in the spliceosomal complex (ref. 16; see Fig. 4). As noted above, this proposal is not consistent with our data defining the extensive CTE-interacting surface in the LRR domain.
Figure 4.
Comparison of the RNA-interacting regions of Tap with those found in the U2B′′-U2A′-RNA complex. (A) The Tap protein (96–372) is shown as a yellow ribbon rendering. The β1- and β3-strands of the RNP fold are shown as magenta arrows for comparison with the spliceosomal complex shown in B. Residues in the RNP and LRR domains of Tap that are critical for CTE binding (R128, K129, K132, R233, R249, R276, Y278, and K304) are shown as red balls and sticks. (B) A ribbon rendering of the structurally analogous spliceosomal complex including an RNP domain (U2B′′) in yellow and LRR domain (U2A′) in orange and a blue stick model of the hairpin fragment of U2 RNA as reported in the crystal structure (PDB ID code 1A9N) is shown. The same strands, β1 and β3, which include the residues of the RNP motifs (RNP2 and RNP1), are shown in magenta for comparison with Tap. The RNA-interacting surfaces of U2B′′ and U2A′ are clearly distinct from the CTE-interacting surfaces in Tap identified in our mutational analysis.
The relative position of the RNP domain with respect to the LRR domain in the Tap structure is completely different from that in the spliceosomal complex, consistent with the novel interactions of Tap with the CTE that we have proposed based on our structural and mutational analysis (see Fig. 4). In addition, although the folds of the RNP and LRR domains are similar, the structures of these domains in Tap and the spliceosomal proteins differ significantly in detail. We suggest that type D retroviruses have evolved through an iterative molecular selection process to take advantage of an existing binding site in Tap. This is a remarkable example of a nonnative RNA element binding to a conserved surface of a protein, in this case the nuclear RNA export factor Tap, to facilitate nuclear export of unspliced retroviral RNA.
Supplementary Material
Acknowledgments
We thank the members of the Georgiadis and Cullen laboratories for helpful discussions; Attilio De Falco and Greg Listner for systems support; and Mike Becker, Lonnie Berman, and Bill Nolan from NSLS X25. Data were collected at the National Synchrotron Light Source, Brookhaven National Laboratory, which is supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences. This work was funded by National Institutes of Health Grant GM 55026 (to M.M.G.) and by a grant from the Howard Hughes Medical Institute (to B.R.C.).
Abbreviations
- CTE
constitutive transport element
- MPMV
Mason–Pfizer monkey virus
- RNP
ribonucleoprotein
- LRR
leucine-rich repeat
- SeMet
selenomethionyl or selenomethionine
- β-gal
β-galactosidase
- CMV
cytomegalovirus
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1KOH and 1KOO).
References
- 1.Cullen B R. Mol Cell Biol. 2000;20:4181–4187. doi: 10.1128/mcb.20.12.4181-4187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malim M H, Hauber J, Le S-Y, Cullen B R. Nature (London) 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 3.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 4.Neville M, Robash M. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 6.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K-T, Rekosh D, Hammarskjold M-L. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 8.Pasquinelli A E, Ernst R K, Lund E, Brimm C, Zapp M L, Rekosh D, Hammarskjold M-L, Dahlberg J E. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saavedra C, Felber B K, Izaurralde E. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 10.Katahira J, Straber K, Podtelejnikov A, Mann M, Jung J U, Hurt E. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan W, Zolotukhin A S, Bear J, Patenaude D J, Felber B K. RNA. 2000;6:1762–1772. doi: 10.1017/s1355838200000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiegand H L, Coburn G A, Zeng Y, Kang Y, Bogerd H P, Cullen B R. Mol Cell Biol. 2002;22:245–256. doi: 10.1128/MCB.22.1.245-256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fribourg S, Braun I C, Izaurralde E, Conti E. Mol Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- 14.Braun I C, Rohrbach E, Schmitt C, Izaurralde E. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang Y, Cullen B R. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liker E, Fernandez E, Izaurralde E, Conti E. EMBO J. 2000;19:5587–5598. doi: 10.1093/emboj/19.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coburn G A, Wiegand H L, Kang Y, Ho D N, Georgiadis M M, Cullen B R. Genes Dev. 2001;15:1194–1205. doi: 10.1101/gad.888201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price S R, Evans P R, Nagai K. Nature (London) 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 19.Cullen B R. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 20.Tiley L S, Madore S J, Malim M H, Cullen B R. Genes Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]
- 21.Bogerd H P, Echarri A, Ross T M, Cullen B R. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrickson W A, Horton H R, LeMaster D M. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend. Sawyer L, Isaacs N, Bailey S, editors. Daresbury, England: Sci. Eng. Res. Counc.; 1993. pp. 56–62. [Google Scholar]
- 24.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 25.Brunger A T, Adams P A, Clore G M, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges N, Pannu N S, Read R J, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 29.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 30.Nicholls A, Sharp K, Honig B. Proteins Struct Funct. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 31.Cullen B R, Skalka A M, Ju G. Proc Natl Acad Sci USA. 1983;80:2946–2950. doi: 10.1073/pnas.80.10.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hope T J, Huang X, McDonald D, Parslow T G. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang Y, Bogerd H P, Cullen B R. J Virol. 2000;74:5863–5871. doi: 10.1128/jvi.74.13.5863-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cullen B R. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 35.Kang Y, Bogerd H P, Yang J, Cullen B R. Virol. 1999;262:200–209. doi: 10.1006/viro.1999.9906. [DOI] [PubMed] [Google Scholar]
- 36.Hauber J, Perkins A, Heimer E P, Cullen B R. Proc Natl Acad Sci USA. 1987;84:6364–6368. doi: 10.1073/pnas.84.18.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.