Abstract
Interleukin-2 tyrosine kinase (Itk) is a nonreceptor protein tyrosine kinase of the Tec family that participates in the intracellular signaling events leading to T cell activation. Tec family members contain the conserved SH3, SH2, and catalytic domains common to many kinase families, but they are distinguished by unique sequences outside of this region. The mechanism by which Itk and related Tec kinases are regulated is not well understood. Our studies indicate that Itk catalytic activity is inhibited by the peptidyl prolyl isomerase activity of cyclophilin A (CypA). NMR structural studies combined with mutational analysis show that a proline-dependent conformational switch within the Itk SH2 domain regulates substrate recognition and mediates regulatory interactions with the active site of CypA. CypA and Itk form a stable complex in Jurkat T cells that is disrupted by treatment with cyclosporin A. Moreover, the phosphorylation levels of Itk and a downstream substrate of Itk, PLCγ1, are increased in Jurkat T cells that have been treated with cyclosporin A. These findings support a novel mode of tyrosine kinase regulation for a Tec family member and provide a molecular basis for understanding a cellular function of the ubiquitous peptidyl prolyl isomerase, CypA.
Normal cell growth depends on the precise control of protein tyrosine kinase activity (1). For certain families of kinases, the mechanism of catalytic regulation is well understood. Structures of Src tyrosine kinases (2, 3) reveal intramolecular interactions mediated by the Src homology 2 (SH2) and Src homology 3 (SH3) domains that control catalytic activity of the neighboring kinase domain. Specifically, distortion of the Src kinase active site is achieved in part by SH2 binding to a phosphorylated tyrosine residue in the C-terminal tail of Src (4). For other families of kinases, the mechanistic details of catalytic regulation remain elusive. In particular, the Tec family of nonreceptor tyrosine kinases (5) displays distinguishing characteristics that point to an alternative mode of regulation. The Tec family kinases modulate hematopoietic cellular responses to external stimuli (6). The T cell-specific Tec family member, interleukin-2 tyrosine kinase (Itk) (7, 8), plays a role in the maturation of thymocytes, is required for intracellular signaling following T cell receptor (TCR) crosslinking, and is involved in generation of second messengers that mediate cytoskeletal reorganization (9). Itk is homologous to Src in the region spanning the SH3, SH2, and kinase domain but lacks the Src C-terminal tail that contains the regulatory tyrosine. However, activation of Itk depends on SH2-mediated interactions with phosphorylated signaling partners (9) such as Slp-76 (10) and LAT (11), suggesting a regulatory role for the Itk SH2 domain despite the absence of a Src-like C-terminal regulatory tyrosine residue.
The Src regulatory mechanism highlights the role of molecular switches in controlling cellular signaling pathways. Well studied covalent modifications such as phosphorylation generate recognition sites that affect the activity and binding behavior of many signaling proteins (1, 12–14). Moreover, there are potential regulatory switches intrinsic to polypeptides such as the cis/trans isomerization of peptide bonds preceding the amino acid proline (15, 16). In native proteins, the local environment surrounding a proline residue, as well as the overall tertiary structure, influences the relative energies and therefore the relative population of the cis and trans conformers. The majority of folded proteins for which three-dimensional structural information has been gathered contain trans prolyl imide bonds. The cis conformation occurs at a frequency of ≈6% in folded proteins (17), and a small subset of proteins are conformationally heterogeneous with respect to cis/trans isomerization (18–21). Furthermore, the activation energy for interconversion between cis and trans proline is high (≈20 kcal/mol) leading to slow interconversion rates (22). This barrier is a rate-limiting step in protein folding and may serve to kinetically isolate two functionally and conformationally distinct molecules. These special properties, and the fact that many proline residues are highly conserved in homologous protein sequences, suggest that prolines may play a role in conformer-specific recognition and function.
Three distinct enzyme families have been identified that catalyze the cis/trans isomerization of prolyl imide bonds. These peptidyl-prolyl cis/trans isomerases (PPIases) (23, 24) consist of the cyclophilins (Cyp), FK506-binding proteins (FKBPs), and the parvulins (25, 26). CypA and FKBP12 are the intracellular protein targets of the immunosuppressive natural products cyclosporin A (CsA) and FK506, respectively (27). These small molecules inhibit PPIase activity and block T cell activation. However, the immunosuppressant activity of these small molecules derives not from inhibition of isomerase activity but rather from interruption of signaling events by the inhibitor–PPIase complexes themselves (28). The binary CsA–CypA and FK506–FKBP12 complexes bind to and inhibit calcineurin, the serine/threonine phosphatase responsible for dephosphorylation and subsequent nuclear translocation of the IL-2 transcription factor, nuclear factor of activated T cells (29, 30). The PPIase inhibitors, CsA and FK506, have shed considerable light on immunosuppression as well as the signal transduction pathways involved in T cell signaling, yet the normal cellular roles of the PPIases remain unclear. Although recent efforts have identified several PPIase-mediated cellular processes (31–37), the range of normal cellular functions controlled by the ubiquitous PPIases and the molecular basis for their role in regulating signaling cascades have not yet been fully elucidated.
The cellular targets of the PPIases are likely folded proteins that contain a conformationally heterogeneous proline residue. We now present a structural analysis of Itk that places this protein among those that exhibit multiple, well defined, low energy conformations in solution. A single prolyl imide bond is responsible for the observed conformational heterogeneity within Itk, and we show that this proline-dependent conformational switch directly regulates Itk substrate recognition. Moreover, we demonstrate that the PPIase, CypA, catalyzes the cis/trans isomerization of the conformationally heterogeneous prolyl imide bond within Itk. In vitro and in vivo functional data reveal a stable CypA–Itk complex in T cells and point to a role for CypA in repressing Itk kinase activity. Together, these data support a novel mode of nonreceptor tyrosine kinase regulation and provide a molecular basis for understanding a cellular function of the PPIase, CypA.
Materials and Methods
Protein Expression and NMR Spectroscopy.
Full-length Itk cDNA (7) for PCR amplification was a gift from Leslie J. Berg (University of Massachusetts Medical School). Protein samples for NMR were expressed and purified as described previously (38). Unless otherwise indicated, all NMR samples were 1.5 mM. NMR spectra were recorded at 25°C on a Bruker DRX500 spectrometer operating at a 1H frequency of 499.867 MHz. Bovine calf thymus CypA (Sigma) was used for NMR experiments.
In Vitro Kinase Assays.
Cell growth conditions, baculovirus infections, and the production of high titer baculovirus stocks were as previously described (39). Sf9 cells (obtained from American Type Culture Collection, ATCC) were infected with baculovirus-expressing Itk (gift from Leslie J. Berg). Harvested cells were lysed in 1% IGEPAL (Sigma) lysis buffer, and lysates were cleared by centrifugation at 14,000 rpm for 15 min at 4°C. Itk was immunoprecipitated with anti-Itk antibody clone 2F12 (Upstate Biotechnology, Lake Placid, NY) and protein G agarose (GIBCO/BRL), washed three times in 50 mM Tris, pH 7.6, wash buffer, and then washed once in kinase buffer (50 mM Hepes buffer, pH 7/10 mM MgCl2/10 mM MnCl2) before resuspension in ATP/kinase buffer (kinase buffer + 0.5 mM Na3VO4/0.5 mM ATP). Kinase reactions were stopped after 30 min by addition of 2 × SDS loading dye and boiling samples. Samples were resolved on a 10% SDS-polyacrylamide gel and transferred to Immobilon-P membranes (Millipore); Itk autophosphorylation was detected with anti-phosphotyrosine antibody clone 4G10 (Upstate Biotechnology). Itk was detected with anti-Itk antibody. The level of Itk phosphorylation was analyzed by normalizing detected phosphotyrosine to total Itk in each lane with nih image. Recombinant CypA (1 μM, Sigma) and CsA (3 μM, Sigma) were incubated for 6 h with constant agitation at 4°C. The complex was then centrifuged at 14,000 rpm for 20 min to remove any precipitate.
Assay of Protein Phosphorylation in Jurkat T Cells.
Jurkat clone E6–1 cells were obtained from ATCC. Cells were maintained at 37°C, 5% CO2 in RPMI medium 1640 (GIBCO/BRL) supplemented with 10% FCS (HyClone) and 1 × penicillin–streptomycin–glutamine solution (GIBCO/BRL); 1.25 mg/ml stock solutions of CsA and FK-506 were prepared in DMSO, and 2 × 107 Jurkat cells were incubated for 1 h in the presence of 500 ng/ml of CsA, FK-506 (Calbiochem), or an equal volume of DMSO at 37°C, 5% CO2. Before stimulation, the cells were washed twice with ice-cold media lacking FCS and 1 × penicillin–streptomycin–glutamine solution containing CsA, FK-506, or DMSO and then resuspended in 400 μl of the same media. Stimulations were performed by incubating the cells on ice for 20 min with 10 μg/ml anti-CD3 antibody (PharMingen), quickly pelleting the cells, and resuspending in 50 μg/ml rabbit anti-mouse IgG (Sigma) at 37°C for the indicated times. Stimulations were stopped by diluting the cells 3-fold into ice-cold PBS resuspension buffer (PBS supplemented with 1 mM Na3VO4/20 mM NaF). Cells were pelleted and resuspended in ice-cold RIPA lysis buffer, and lysates were cleared by centrifugation. Cleared lysates were depleted of remaining antibodies used during the stimulation by incubation with protein G agarose for 1 h at 4°C with constant rocking. Itk, Zap-70, or PLCγ1 was immunoprecipitated from precleared lysates with anti-Itk antibody, anti-Zap-70 antibody (Upstate Biotechnology), or anti-PLCγ1 antibody clone B-6-4 (Upstate Biotechnology) and protein G or protein A agarose. Immunoprecipitates were washed three times in RIPA lysis buffer and once in PBS resuspension buffer. Samples were resolved on an 8% or a 12.5% (for CypA detection) SDS-polyacrylamide gel. Itk detection was as described above, and Zap-70 and PLCγ1 were detected with anti-Zap-70 antibody and anti-PLCγ1 antibody, respectively. CypA was detected with anti-CypA (Calbiochem), and FKBP12 was detected with anti-FKBP (N-19) (Santa Cruz Biotechnology). The identity of the CypA band was confirmed by comigration with purified protein purchased from Sigma.
Competitive Binding Assays.
Lysates and immunoprecipitated Itk were obtained as described above from unstimulated Jurkat T cells. The protein G agarose, containing immunoprecipitated Itk, was briefly centrifuged, and the supernatant was removed. The agarose was then resuspended in 100 μl of RIPA cell lysis buffer that contained 50 μg of the indicated ligand. After 1 h of incubation at 4°C with constant rocking, the agarose was washed three times in RIPA lysis buffer and once in PBS resuspension buffer before resuspension in 20 μl of 2 × SDS loading dye. Samples were resolved on a 12.5% SDS-polyacrylamide gel. Detection of CypA and Itk were as described above. Itk GST-SH3 was detected with anti-GST (Upstate Biotechnology). The level of CypA coimmunoprecipitation was analyzed by normalizing detected CypA to total Itk in each lane by using nih image. Synthesis of the ADpYEPP and QQPPVPPQRPMA peptides was carried out by Gautam Sarath at the University of Nebraska Lincoln Protein Core Facility.
Results
Proline Isomerization Induces Conformational Heterogeneity Within the Itk SH2 Domain.
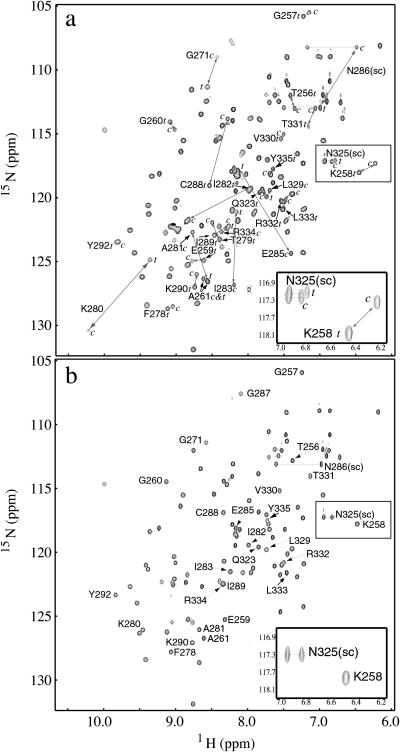
Structural analysis of the purified Itk SH2 domain shows two well defined, low energy conformations are populated in solution (Fig. 1a). NMR resonance assignments for the 15N-labeled Itk SH2 domain reveal two sets of resonances in slow exchange for 35 of the 109 SH2 residues. Characteristic NOE correlations (40) between Asn-286 and Pro-287 indicate that proline cis/trans isomerization around the Asn-286–Pro-287 imide bond is the source of the conformational heterogeneity. A heteronuclear single quantum correlation (HSQC) (41) spectrum of the Itk SH2 domain in which Pro-287 is mutated to Gly (Fig. 1b) confirmed this assessment. For the P287G mutant, in which the 286–287 amide bond adopts only the trans conformation, all of the 1H-15N cross-peaks assigned to the cis conformer are absent. For many of the SH2 residues in the wild-type protein, the differences in chemical shift values between the cis and trans conformers are large (>6 ppm in the 15N dimension and >1 ppm in the 1H dimension), reflecting significant differences in the chemical environment of nuclei in the two forms (Fig. 1a). The largest chemical shift differences between the cis and trans conformers occur for residues surrounding Pro-287 in primary amino acid sequence. However, it is notable that large chemical shift differences are also apparent in regions removed in primary sequence from Pro-287, in particular, residues located within the C terminus of the SH2 domain (residues 329–335). Together, the conformationally heterogeneous regions map to a contiguous surface of the Itk SH2 domain (Fig. 2 a and b).
Figure 1.
The Itk SH2 domain adopts two stable structures in solution. (a) 1H-15N HSQC spectrum of purified, recombinant 15N-labeled Itk SH2 domain. The resonances corresponding to the cis (c) and trans (t) conformers are connected by a gray double-headed arrow. (Inset) An expansion of the boxed region of the spectrum containing representative doubled resonances for the Asn-325 side chain (sc) NH2 and the Lys-258 backbone NH. (b) HSQC spectrum of the Itk SH2 domain in which Pro-287 is mutated to Gly. Labeled cross-peaks correspond to those that are labeled in a, and the cross-peak corresponding to Gly-287 (introduced by mutation) is assigned. (Inset) The same region as in a but showing a single resonance for Asn-325 (sc) and Lys-258.
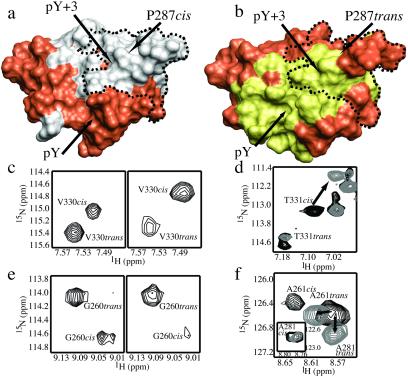
Figure 2.
Surface representations (rendered with molmol; ref. 51) of the Itk SH2 cis (a) and trans (b) structural models (R.J.M. and A.H.A., unpublished results). The SH2 residues that display conformational heterogeneity are demarcated with a dotted line on each model. Pro-287 is indicated, and the pY and pY + 3 binding pockets are labeled. Chemical shift perturbations that occur on addition of equimolar Itk SH3 domain to the Itk SH2 domain (38) are mapped onto the cis model and highlighted in gray (a). Likewise, SH2 residues that undergo chemical shift perturbations on addition of three times excess phosphopeptide (ADpYEPPPSNDE) are highlighted in yellow on the trans model (b). For both surface models, residues that do not exhibit chemical shift changes on addition of ligand are orange. (c) Select region of the HSQC spectrum of a 0.5 mM Itk SH2 sample. (Left) Itk SH2 domain alone, where the trans/cis ratio is approximately 60:40 based on the cross-peak volumes for each set of doubled resonances. (Right) The same region of the Itk SH2 HSQC spectrum after addition of 10 times excess unlabeled Itk SH3 domain (trans/cis ratio 10:90). (d) Superposition of the same region of two HSQC spectra of 15N-labeled Itk SH2 domain in the absence (black) and presence (gray) of unlabeled Itk SH3 domain. The Thr-331 cis cross-peak shifts on addition of equimolar Itk SH3 domain (arrow), whereas the Thr-331 trans cross-peak remains at the same frequency. (e) (Left) Itk SH2 domain (trans/cis 60:40). (Right) Addition of 3 times excess phosphopeptide to Itk SH2 (trans/cis 90:10). (f) Superposition of two HSQC spectra of 15N-labeled Itk SH2 domain in the absence (black) and presence (gray) of 3 times excess phosphopeptide. The Ala-261 and Ala-281 trans cross-peaks shift on addition of phosphopeptide (arrows), whereas the Ala-261 and Ala-281 cis cross-peaks resonate at the same frequency regardless of ligand present (the Ala-281 cis cross-peak is shown in the Inset because of the large difference in resonance frequencies for the cis and trans resonances of Ala-281).
The Cis and Trans Proline Conformers Bind Different Ligands.
We have previously demonstrated that the Itk SH2 domain mediates two distinct binding events: canonical recognition of phosphotyrosine-containing peptides and phosphotyrosine-independent dimerization via contacts to the SH3 binding pocket of another Itk molecule (38). Chemical shift mapping reveals the contiguous SH2 surface residues that are involved in binding to either the Itk SH3 domain (Fig. 2a) or phosphopeptide (Fig. 2b). The overlap between the SH3 and phosphopeptide binding surfaces is within the conformationally heterogeneous region of the Itk SH2 domain. This suggests that the conformational heterogeneity induced by proline isomerization plays a role in regulating ligand selection. Indeed, addition of excess Itk SH3 domain to the SH2 domain shifts the cis/trans equilibrium to favor the cis SH2 conformer (Fig. 2c). As well, shifts in the positions of cross-peaks corresponding to the cis proline conformer but not those of the trans SH2 conformer are observed in the HSQC spectrum of the Itk SH2 domain to which recombinant Itk SH3 domain has been added (Fig. 2d). Thus, the cis Asn-286–Pro-287 imide bond, and not the trans, is required for Itk dimerization. In contrast, a phosphotyrosine-containing peptide binds preferentially to the trans SH2 conformer. Addition of excess phosphopeptide to the Itk SH2 domain shifts the equilibrium to favor the trans conformer (Fig. 2e). Chemical shift changes are observed for cross-peaks corresponding to some residues in the trans SH2 conformer but not for those same residues in the cis (Fig. 2f). This demonstrates conformer-specific ligand recognition mediated entirely by isomerization of a single imide bond.
The Itk SH2 Domain Is a Substrate for the PPIase, CypA.
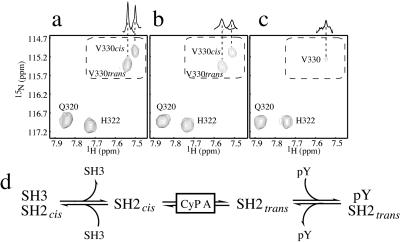
NMR linewidths (42) were examined in spectra of 15N-labeled SH2 domain in the presence of substoichiometric amounts of two different PPIases (CypA and FKBP-12). Line broadening of the conformationally heterogeneous resonances within the Itk SH2 domain was observed on addition of 3 mol% and 20 mol% CypA (Fig. 3). In contrast, the spectrum of the Itk SH2 domain incubated with 13 mol% FKBP-12 was identical to that of SH2 domain alone (data not shown). Addition of CypA to the Itk SH2 domain also caused small increases in all linewidths and specific chemical shift perturbations that were localized to residues surrounding Pro-287. The P287G mutation within the Itk SH2 domain abrogates interaction with CypA (data not shown). Together, these data provide evidence for a direct, specific, and productive interaction between CypA and a folded domain of a protein tyrosine kinase. By lowering the kinetic barrier to interconversion, CypA accelerates cis/trans isomerization and may play an integral role in a coupled equilibrium system involving conformer-specific recognition (Fig. 3d). Such a mechanism would allow Itk to switch binding partners more rapidly in response to exogenous signaling events.
Figure 3.
(a) Region of the HSQC spectrum for the 15N-labeled Itk SH2 domain (0.5 mM) that includes cross-peaks for the Gln-320, His-322, and Val-330 backbone NH resonances. Gln-320 and His-322 are not affected by cis/trans isomerization around the Asn-286–Pro-287 imide bond; therefore, each appears as a single cross-peak. In contrast, the Val-330 amide resonance is doubled as a result of slow exchange between the cis and trans forms. (b) Same region of the HSQC spectrum for the Itk SH2 domain (0.5 mM) in the presence of 3 mol% CypA. Line broadening is apparent for all of the doubled resonances in the SH2 domain spectrum on addition of CypA, whereas those peaks that correspond to residues unaffected by the isomerization event do not broaden significantly. (c) Addition of 20 mol% CypA to the Itk SH2 domain results in further line broadening and coalescence to the chemical shift value that represents the average of the cis and trans conformers. One-dimensional projections through the center of the Val-330 cross-peaks along the 1H axis illustrate the exchange mediated broadening and coalescence. (d) Equilibrium model encompassing the CypA-catalyzed Itk SH2 cis/trans interconversion, cis-mediated SH3 binding (Itk dimerization), and trans-mediated phospholigand (pY) binding.
CypA Is an Inhibitor of Itk Kinase Activity.
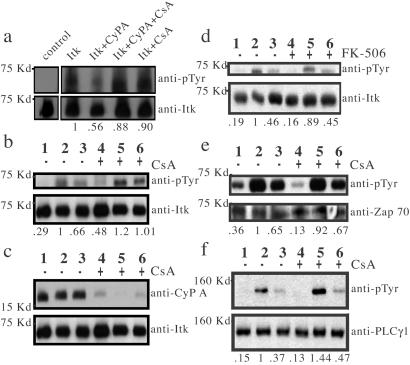
Given the fact that CypA catalyzes the cis/trans isomerization within the Itk SH2 domain, we carried out in vitro functional assays to examine the role of CypA in regulating Itk activity. Itk was expressed in Sf9 cells with the baculovirus expression system and purified by immunoprecipitation. Following incubation with ATP, in vitro Itk autophosphorylation (39) was detected by Western blotting. Itk autophosphorylation was decreased significantly in the presence of CypA (Fig. 4a). CsA reversed the effect of CypA on Itk kinase activity (Fig. 4a). Thus, CypA inhibits Itk kinase activity in vitro.
Figure 4.
(a) CypA, CypA/CsA complex, or CsA was added to baculovirus-expressed Itk immediately before resuspension in ATP/kinase buffer. A mock kinase assay of Itk (left lane) was performed in which ATP was excluded from the kinase reaction buffer. Itk phosphorylation is normalized to Itk protein levels, and net changes in phosphorylation are indicated below each lane. (b–f) Lanes 1, 2, and 3 are mock stimulation, 40 s and 2 min, respectively, following TCR stimulation of Jurkat cells in the absence (−) of drug treatment, whereas lanes 4, 5, and 6 represent the same conditions in the presence (+) of drug. (b) Itk phosphorylation following TCR stimulation of Jurkat cells in the presence and absence of CsA. (c) CypA binding to Itk was monitored by detection of Itk immunoprecipitates with an anti-CypA antibody. (d) Itk phosphorylation in the presence and absence of FK-506. (e) Zap-70 phosphorylation and (f) PLCγ1 phosphorylation in the presence and absence of CsA. Phosphorylation levels of Itk, Zap-70, and PLCγ1 are normalized to total protein in each lane. Net changes in phosphorylation relative to phosphorylation levels at 40 s following stimulation in the absence of drug (lane 2) are indicated below each lane.
To assess the role of cyclophilin in regulating Itk activity in vivo, the level of Itk phosphorylation was monitored following TCR/CD3-induced stimulation of Jurkat T cells that were either untreated or treated with CsA. Itk tyrosine phosphorylation, as monitored by Western blotting, was apparent at early time points following TCR stimulation. The level of Itk phosphorylation in untreated cells increased after a 40-s stimulation and then decreased toward basal levels after a 2-min stimulation (Fig. 4b). In the CsA-treated cells, the level of Itk phosphorylation was higher than the untreated cells at both time points (Fig. 4b). Additionally, endogenous CypA was detected in Itk immunoprecipitates prepared from Jurkat T cells that have not been treated with CsA, indicating the presence of a stable Itk–CypA complex in T cells (Fig. 4c). In Itk immunoprecipitates of cells pretreated with CsA, the amount of CypA was less than in untreated cells (Fig. 4c). The effect of CypA on Itk phosphorylation is specific because FK-506 treatment of Jurkat cells had no detectable effect on Itk phosphorylation (Fig. 4d), and FKBP-12 was not detected in Itk immunoprecipitates of untreated cells or cells treated with FK-506 (data not shown). This observed specificity is consistent with NMR experiments that reveal that CypA, but not FKBP, affects the cis/trans isomerization within the Itk SH2 domain (Fig. 3). Because both CsA and FK-506 treatment lead to inhibition of calcineurin and subsequent immunosuppression (29, 30), the absence of an FK-506-mediated effect on Itk phosphorylation levels indicates that the increase in Itk phosphorylation on treatment with CsA (Fig. 4b) is the direct result of inhibition of CypA activity and is not related to the immunosuppressive action of CsA.
CsA Treatment Affects the Phosphorylation State of PLCγ1 but Not Zap-70 in Jurkat T Cells.
Phosphorylation of the upstream and downstream signaling partners of Itk, Zap-70, and PLCγ1, respectively (43–45) was monitored following TCR/CD3-induced stimulation of Jurkat T cells carried out in the presence and absence of CsA. Zap-70 phosphorylation levels at both 40-s and 2-min stimulations are unchanged by treatment with CsA (Fig. 4e), consistent with the placement of Zap-70 upstream of Itk. In contrast, an increase in the level of PLCγ1 phosphorylation in a manner that parallels Itk is observed following CsA treatment of Jurkat cells. PLCγ1 phosphorylation is higher after both the 40-s and 2-min stimulations in CsA-treated cells as compared with the same time points from untreated cells (Fig. 4f). Therefore, Zap-70, a signaling molecule upstream of Itk, is not affected by CsA treatment, whereas PLCγ1, a signaling molecule downstream of Itk, is up-regulated by CsA treatment. These data suggest that an increase in Itk activity resulting from inhibition of CypA leads to a concomitant increase in downstream PLCγ1 activity.
The Itk SH2 Domain Mediates CypA Binding in T Cells.
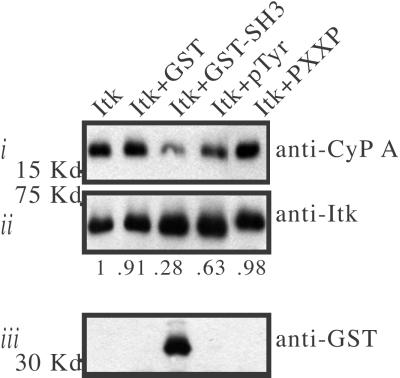
Competitive binding assays with ligands that specifically target the Itk SH2 domain provide evidence that the Itk–CypA interaction in Jurkat T cells is mediated by the Itk SH2 domain. The presence of two different SH2 ligands in Itk immunoprecipitates from Jurkat T cells, Itk SH3 domain (38) or the phosphopeptide (ADpYEPP, denoted pTyr), decreased the amount of coimmunoprecipitated CypA (Fig. 5 Top). In contrast, the presence of a peptide ligand that binds specifically to the Itk SH3 domain (QQPPVPPQRPMA, denoted PXXP) caused no detectable change in the amount of coimmunoprecipitated CypA (Fig. 5 Top). Consistent with our previous structural analysis of Itk dimerization (38), a significant amount of Itk GST-SH3 was detected following washing of Itk immunoprecipitates that have been incubated Itk GST-SH3 (Fig. 5 Bottom). This observation is consistent with the presence of the cis SH2 conformer in full-length Itk from Jurkat T cells. Taken together, in vitro, in vivo, and NMR data implicate CypA as a regulator of Itk activity in T cells and indicate that Pro-287 within the Itk SH2 domain mediates critical regulatory interactions with CypA.
Figure 5.
For the indicated samples, glutathione S-transferase (GST), Itk GST-SH3, phosphotyrosine-containing peptide (pTyr), or a proline-rich peptide (PXXP) was incubated with full-length Itk following immunoprecipitation from Jurkat T-cell lysates. (Top, i) Following extensive washing, the amount of CypA that coimmunoprecipitated with Itk was determined by immunoblotting. The amount of CypA in Itk immunoprecipitates is normalized to total Itk in each lane. Net changes in the amount of coimmunoprecipitated CypA relative to the amount that coimmunoprecipitates with Itk in the absence of exogenous factors are indicated. (Bottom, iii) Detection of GST-SH3 in Itk immunoprecipitates following competitive binding assay.
Discussion
Our data suggest that displacement of CypA activates Itk. Phospholigand binding and Itk dimerization both compete with CypA binding to the Itk SH2 domain (Fig. 5). A mechanistic model for Itk activation (Fig. 6) can be proposed based on the well understood structural details of SH2-phospholigand recognition (46). The SH2 domain consists of two adjacent binding pockets: the “pY” pocket that contacts the phosphotyrosine residue itself and the “pY + 3” pocket that confers ligand-binding specificity by contacting amino acids flanking the phosphotyrosine. Unlike the Itk pY + 3 pocket, the pY binding site is not affected by proline-induced conformational heterogeneity (Fig. 2 a and b), nor does it appear, based on the lack of chemical shift perturbations, to be sterically occluded by CypA binding. Itk activation by a phospholigand may therefore be initiated by binding of the phosphotyrosine itself to the accessible pY pocket in the SH2 domain. Subsequent pY + 3 engagement would displace CypA and stabilize the active, trans SH2 conformer (Fig. 6). In fact, phospholigand binding to the SH2 domain has previously been shown to activate Itk (45, 47). Our data indicate that Itk dimerization also causes displacement of CypA (Fig. 5) but results in stabilization of the cis SH2 conformer, for which functional data have not yet been obtained. Nevertheless, the activity of Itk seems to correlate with the identity of SH2 bound ligand and therefore the configuration of the Asn-286–Pro-287 imide bond.
Figure 6.
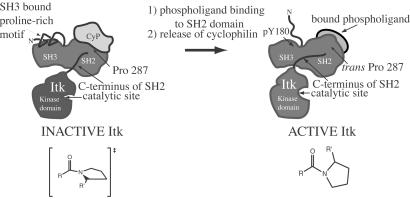
Model for Itk regulation. Itk is dark gray and includes the proline-rich region, SH3, SH2, and kinase domains (the PH domain and part of the TH domain are not shown for clarity). Two “states” of Itk are depicted: inactive, CypA-bound Itk; and active, phospholigand-bound Itk. CypA is light gray and is shown bound to the Itk SH2 domain in the region of Pro-287. The proposed configuration of the Asn-286–Pro-287 imide bond (resembling the transition state for cis/trans interconversion) is illustrated for the inactive, CypA-bound form of Itk. NMR spectroscopic data suggest that cyclophilin-bound Itk is monomeric (K.N.B. and A.H.A., unpublished results), and the proline-rich region may contact the SH3 binding pocket in an intramolecular fashion (52). Activation may be accompanied by phosphorylation of Tyr-180 in the SH3 domain expelling bound proline (53). Cis/trans isomerization around Asn-286–Pro-287 causes pronounced conformational heterogeneity in the C terminus of the Itk SH2 domain leading directly into the Itk kinase domain (see Fig. 1). The C terminus of the Itk SH2 domain may adopt a conformation in the presence of cyclophilin that structurally perturbs the kinase catalytic site rendering it inactive. Release of cyclophilin, either by Itk dimerization to favor the cis conformer (not shown) or by binding of a phosphotyrosine-containing ligand to favor the trans conformer, may lead to reorganization of the C-terminal SH2 residues and subsequent restructuring of the kinase active site.
At this time, the relative affinities of the Itk–CypA interaction, SH3/SH2-mediated Itk dimerization, and Itk SH2-phospholigand binding in a cellular context are not known. Certainly, the presence of CypA in immunoprecipitates of Itk suggests that it is a stable binding partner of Itk in T cells. NMR data show that CypA binds to the Itk SH2 domain and catalyzes the cis/trans isomerization of the Asn-286–Pro-287 imide bond. These two observations suggest two possible modes of action in vivo. CypA (as a binding partner) may act as a repressor of Itk activity by stabilizing a transition-state conformation (48) around the Asn-286–Pro-287 imide bond within Itk (Fig. 6). Alternatively, CypA (as a catalyst) may facilitate rapid interconversion between the trans and cis Itk conformers and therefore between the phospholigand bound and dimerized states, respectively (Fig. 3d). In any case, structural rearrangements in the SH2 domain because of dimerization, CypA, or phospholigand binding could allosterically regulate the neighboring kinase domain in a manner analogous to the established conformational regulation of the Src kinases (2, 3). The specific interactions mediated by the regulatory domains in the Tec family kinases are different from the Src family kinases, yet the ability of these domains to control the catalytic activity through conformational changes in the kinase domain may be similar.
To date, detailed mechanistic studies of PPIase function have been limited to analysis of peptidyl-prolyl isomerization within small peptide model systems. We have identified a folded protein target for CypA that provides the opportunity to study the PPIase catalytic mechanism in the context of a physiological protein substrate. An analysis of the extent to which the tertiary structure of a physiological substrate affects CypA activity is now tractable, and comparison with structures such as the CypA–HIV capsid protein complex (49) should shed additional light on the mode of action of CypA. Moreover, the past decade has witnessed an enormous effort to elucidate the mode of action of the immunosuppressive agents, CsA and FK-506. This work has advanced our understanding of T cell signaling and our ability to control the immune response. Although these natural products target the active sites of cyclophilin and FKBP, the fact that they inhibit PPIase activity is apparently irrelevant to their immunosuppressive effects. The discovery of a nonreceptor protein tyrosine kinase as a physiological target of CypA in T cells raises the possibility that other conformationally heterogeneous kinases exist that are regulated by PPIases in a similar manner. These findings are provocative in light of the high incidence of cancer that accompanies the use of CsA in organ transplant patients (50). Hyperactivation of protein tyrosine kinases, including Itk, during clinical use of PPIase inhibitors may contribute to unregulated cellular proliferation and ultimately formation of malignancies in immunosuppressed patients. Certainly, further exploration of the physiological roles of the PPIases is warranted and may provide a framework on which more selective immunosuppressive therapies may be developed.
Acknowledgments
We are grateful to J. A. Thomas for the use of equipment for cell culture experiments. In addition, we thank J. E. Buss, L. J. Berg, S. C. Bunnell, G. Culver, K. M. Shokat, T. J. Wandless, and K. Cimprich for useful discussions and helpful comments on the manuscript. This work was funded by National Institutes of Health Grant RO1-AI43957 (to A.H.A.).
Abbreviations
- Itk
interleukin-2 tyrosine kinase
- TCR
T cell receptor
- SH2
Src homology 2
- SH3
Src homology 3
- PPIase
peptidyl-prolyl cis/trans isomerase
- CypA
cyclophilin A
- CsA
cyclosporin A
- FKBP
FK506-binding protein
- HSQC
heteronuclear single quantum correlation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hubbard S R, Till J H. Annu Rev Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- 2.Xu W, Harrison S C, Eck M J. Nature (London) 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 3.Sicheri F, Moarefi I, Kuriyan J. Nature (London) 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 4.Willams J C, Wierenga R K, Saraste M. Trends Biochem Sci. 1998;23:179–184. doi: 10.1016/s0968-0004(98)01202-x. [DOI] [PubMed] [Google Scholar]
- 5.Yang W-C, Collette Y, Nunès J A, Olive D. Immunity. 2000;12:373–382. doi: 10.1016/s1074-7613(00)80189-2. [DOI] [PubMed] [Google Scholar]
- 6.Mano H. Cytokine Growth Factor Rev. 1999;10:267–280. doi: 10.1016/s1359-6101(99)00019-2. [DOI] [PubMed] [Google Scholar]
- 7.Heyeck S D, Berg L J. Proc Natl Acad Sci USA. 1993;90:669–273. doi: 10.1073/pnas.90.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siliciano J D, Morrow T A, Desiderio S V. Proc Natl Acad Sci USA. 1992;89:11194–11198. doi: 10.1073/pnas.89.23.11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsoukas C D, Grasis J A, Ching K A, Kawakami Y, Kawakami T. Trends Immunol. 2001;22:17–20. doi: 10.1016/s1471-4906(00)01795-6. [DOI] [PubMed] [Google Scholar]
- 10.Wardenburg J B, Fu C, Jackman J K, Flotow H, Wilkinson S E, Williams D H, Johnson R, Kong G, Chan A C, Findell P R. J Biol Chem. 1996;271:19641–19644. doi: 10.1074/jbc.271.33.19641. [DOI] [PubMed] [Google Scholar]
- 11.Zhang W, Sloan-Lancaster J, Kitchen J, Trible R P, Samelson L E. Cell. 1998;92:83–92. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- 12.Scott J D, Pawson T. Sci Am. 2000;282:72–79. doi: 10.1038/scientificamerican0600-72. [DOI] [PubMed] [Google Scholar]
- 13.Pawson T, Nash P. Genes Dev. 2000;14:1027–1047. [PubMed] [Google Scholar]
- 14.Volkman B F, Lipson D, Wemmer D E, Kern D. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 15.Cheng H N, Bovey F A. Biopolymers. 1977;16:1465–1472. doi: 10.1002/bip.1977.360160707. [DOI] [PubMed] [Google Scholar]
- 16.Grathwohl C, Wüthrich K. Biopolymers. 1981;20:2623–2633. [Google Scholar]
- 17.Stewart D E, Sakar A, Wampler J E. J Mol Biol. 1990;214:253–260. doi: 10.1016/0022-2836(90)90159-J. [DOI] [PubMed] [Google Scholar]
- 18.Feng Y, Hood W F, Forgey R W, Abegg A L, Caparon M H, Thiele B R, Leimgruber R M, McWherter C A. Protein Sci. 1997;6:1777–1782. doi: 10.1002/pro.5560060821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grochulski P, Li Y, Schrag J D, Cygler M. Protein Sci. 1994;3:82–91. doi: 10.1002/pro.5560030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H J, Sheng X R, Niu W D, Pan X M, Zhou J M. J Biol Chem. 1998;273:7448–7456. doi: 10.1074/jbc.273.13.7448. [DOI] [PubMed] [Google Scholar]
- 21.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 22.Schmid F X, Baldwin R L. Proc Natl Acad Sci USA. 1978;75:4764–4768. doi: 10.1073/pnas.75.10.4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer G, Wittman-Liebold B, Lang K, Kiefhaber T, Schmid F X. Nature (London) 1989;337:476–478. doi: 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]
- 24.Takanashi N, Hayano T, Suzuki M. Nature (London) 1989;337:473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- 25.Schiene-Fischer C, Yu C. FEBS Lett. 2001;495:1–6. doi: 10.1016/s0014-5793(01)02326-2. [DOI] [PubMed] [Google Scholar]
- 26.Fischer G. Angew Chem Int Ed Engl. 1994;33:1415–1436. [Google Scholar]
- 27.Rosen M K, Schreiber S L. Angew Chem Int Ed Engl. 1992;31:384–400. [Google Scholar]
- 28.Walsh C T, Zydowsky L D, McKeon F D. J Biol Chem. 1992;267:13115–13118. [PubMed] [Google Scholar]
- 29.Liu J, Farmer J D, Lane W S, Freidman J, Weissman I, Schreiber S L. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 30.O'Keefe S J, Tamura J, Kincaid R F, Tocci M J, O'Neill M A. Nature (London) 1992;357:692–694. doi: 10.1038/357692a0. [DOI] [PubMed] [Google Scholar]
- 31.Schmid F X, Mayr L M, Mücke M, Schönbrunner E R. Adv Protein Chem. 1993;44:25–66. doi: 10.1016/s0065-3233(08)60563-x. [DOI] [PubMed] [Google Scholar]
- 32.Jayaraman T, Brillantes A M, Timerman A P, Fleischer S, Erdjument-Bromage H, Tempst P, Marks A R. J Biol Chem. 1992;267:9474–9477. [PubMed] [Google Scholar]
- 33.Bram R J, Crabtree G R. Nature (London) 1994;371:355–358. doi: 10.1038/371355a0. [DOI] [PubMed] [Google Scholar]
- 34.Arevalo-Rodriguez M, Cardenas M E, Wu X, Hanes S D, Heitman J. EMBO J. 2000;19:3739–3749. doi: 10.1093/emboj/19.14.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huse M, Chen Y G, Massagué J, Kuriyan J. Cell. 1999;96:425–436. doi: 10.1016/s0092-8674(00)80555-3. [DOI] [PubMed] [Google Scholar]
- 36.Lu K P, Hanes S D, Hunter T. Nature (London) 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 37.Yaffe M B, Schutkowski M, Shen M, Zhou X Z, Stukenberg P T, Rahfeld J U, Xu J, Kuang J, Kirschner M W, Fischer G, et al. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 38.Brazin K N, Fulton D B, Andreotti A H. J Mol Biol. 2000;302:607–623. doi: 10.1006/jmbi.2000.4091. [DOI] [PubMed] [Google Scholar]
- 39.Heyeck S D, Wilcox H M, Bunnell S C, Berg L J. J Biol Chem. 1997;272:25401–25408. doi: 10.1074/jbc.272.40.25401. [DOI] [PubMed] [Google Scholar]
- 40.Wüthrich K, Billeter M, Braun W. J Mol Biol. 1984;180:715–740. doi: 10.1016/0022-2836(84)90034-2. [DOI] [PubMed] [Google Scholar]
- 41.Mori S, Abeygunawardana C, O'Neil Johnson M, van Zijl P C M. J Magn Reson B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 42.Hsu V L, Handschumacher R E, Armitage I M. J Am Chem Soc. 1990;112:6745–6747. [Google Scholar]
- 43.Wange R L, Malek S N, Desiderio S, Samelson L E. J Biol Chem. 1993;268:19797–19801. [PubMed] [Google Scholar]
- 44.Shan X, Wange R L. J Biol Chem. 1999;274:29323–29330. doi: 10.1074/jbc.274.41.29323. [DOI] [PubMed] [Google Scholar]
- 45.Bunnell S C, Diehn M, Yaffe M B, Findell P R, Cantley L C, Berg L J. J Biol Chem. 2000;275:2219–2230. doi: 10.1074/jbc.275.3.2219. [DOI] [PubMed] [Google Scholar]
- 46.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, et al. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 47.Ching K A, Grasis J A, Tailor P, Kawakami Y, Kawakami T, Tsoukas C D. J Immunol. 2000;165:256–262. doi: 10.4049/jimmunol.165.1.256. [DOI] [PubMed] [Google Scholar]
- 48.Harrison R K, Stein R L. Biochemistry. 1990;29:1684–1689. doi: 10.1021/bi00459a003. [DOI] [PubMed] [Google Scholar]
- 49.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 50.Hojo M, Morimoto T, Maluccio M, Asano T, Morimoto K, Lagman M, Shimbo T, Suthanthiran M. Nature (London) 1999;397:530–534. doi: 10.1038/17401. [DOI] [PubMed] [Google Scholar]
- 51.Koradi R, Billeter M, Wuthrich K. J Mol Graphics. 1996;14:29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 52.Andreotti A H, Bunnell S C, Feng S, Berg L J, Schreiber S L. Nature (London) 1997;385:93–97. doi: 10.1038/385093a0. [DOI] [PubMed] [Google Scholar]
- 53.Park H, Wahl M I, Afar D E, Turck C W, Rawlings D J, Tam C, Scharenberg A M, Kinet J P, Witte O N. Immunity. 1996;4:515–525. doi: 10.1016/s1074-7613(00)80417-3. [DOI] [PubMed] [Google Scholar]