Abstract
The lectin chaperone calreticulin (CRT) assists the folding and quality control of newly synthesized glycoproteins in the endoplasmic reticulum (ER). It interacts with ERp57, a thiol-disulfide oxidoreductase that promotes the formation of disulfide bonds in glycoproteins bound by CRT. Here, we investigated the interaction between CRT and ERp57 by using biochemical techniques and NMR spectroscopy. We found that ERp57 binds to the P-domain of calreticulin, an independently folding domain comprising residues 189–288. Isothermal titration calorimetry showed that the dissociation constant of the CRT(189–288)/ERp57 complex is (9.1 ± 3.0) × 10−6 M at 8°C. Transverse relaxation-optimized NMR spectroscopy provided data on the thermodynamics and kinetics of the complex formation and on the structure of this 66.5-kDa complex. The NMR measurements yielded a value of (18 ± 5) × 10−6 M at 20°C for the dissociation constant and a lower limit for the first-order exchange rate constant of koff > 1,000 s−1 at 20°C. Chemical shift mapping showed that interactions with ERp57 occur exclusively through amino acid residues in the polypeptide segment 225–251 of CRT(189–288), which forms the tip of the hairpin structure of this domain. These results are analyzed with regard to the functional mechanism of the CRT/ERp57 chaperone system.
Glycoprotein folding and quality control in the endoplasmic reticulum (ER) are assisted by two homologous molecular chaperones, calreticulin (CRT) and the membrane-bound calnexin (CNX). CRT and CNX are lectins that interact with monoglucosylated trimming intermediates of N-linked core glycans, cooperating with enzymes involved in the trimming and modification of the glycans (1–3). In vivo, both proteins also interact with ERp57 (4), a soluble luminal homologue of protein disulfide isomerase (PDI). Like PDI, ERp57 is composed of four thioredoxin-like domains with active site CXXC sequence motifs in the N- and C-terminal domains (5). During the folding of viral glycoproteins in the ER of living cells, ERp57 has been shown to form transient intermolecular disulfide bonds with glycoprotein substrates bound to CNX and CRT (6). When the association of CNX and CRT with glycoproteins is inhibited, the formation of intermolecular disulfide bonds with ERp57 is abrogated. Thus, the interaction between the glycoprotein substrates and either of the lectin chaperones seems to be required for the interaction with ERp57.
The three-dimensional structure of both the CRT P-domain, CRT(189–288) (7) and the CNX ectodomain (including the CNX P-domain) (8) recently have been solved. They show that the P-domain comprises an unusual, extended hairpin fold, which in the crystal structure of the CNX ectodomain protrudes as a long arm from a compact, globular lectin domain. To gain insights into the cooperation of CRT and CNX with ERp57 during glycoprotein folding, we have characterized the interaction between the CRT P-domain and ERp57 by using biochemical methods and transverse relaxation-optimized spectroscopy (TROSY)-NMR.
Materials and Methods
Protein Expression and Purification.
CRT(189–288) was prepared as described (9). 15N,2H labeling of the protein was obtained by expressing it in 0.5 liters of M9 minimal medium supplemented with 1 g Celtone dN powder (Martek Biosciences, Columbia, MD) and 2 g/liter glucose. Recombinant human PDI was expressed and purified as described (10), except for an additional, final purification step on a Superdex 200 gelfiltration column (Amersham Pharmacia). For the recombinant expression of human ERp57 we used Escherichia coli BL21(DE3) cells freshly transformed with the pHisERp57 expression vector, which encodes a N-terminal 22-aa affinity tag containing a deca-histidine sequence and a factor Xa cleavage site. Three liters of LB medium containing ampicillin (100 μg/ml) was inoculated with 30 ml of preculture of pHisERp57-containing E. coli BL21(DE3) cells that had been grown at 37°C for 3 h. At OD600 = 0.6 expression was induced with 0.4 mM isopropyl l-d-galactopyranoside. The cells were harvested after 4 h, and the pellet was resuspended in 30 ml buffer A (25 mM Tris⋅HCl, pH 8.0/500 mM NaCl/10 mM β-mercaptoethanol). After sonication, the cell lysate was centrifuged at 20,000 g for 30 min, and the supernatant was applied to a Ni2+-charged NTA column (Qiagen, Chatsworth, CA). The fusion protein was eluted with a linear gradient of 0–500 mM imidazole in buffer A. After dialysis against 25 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, the N-terminal fusion tail was removed by factor Xa cleavage, performed for 20 h at room temperature by using a ratio of 1:200 (wt/wt) of factor Xa to fusion protein. After cleavage, the protein was dialyzed against buffer B (100 mM KH2PO4, pH 7.0/25 mM NaCl/10 mM β-mercaptoethanol) and loaded onto a MonoQ 10/10 anion-exchange column (Amersham Pharmacia). ERp57 was eluted with a linear gradient of 0–500 mM NaCl in buffer B. Yields were about 20 mg pure ERp57 per liter of culture medium. The correct molecular weights of all recombinant proteins were verified by matrix-assisted laser desorption ionization–time of flight.
Protein Concentrations.
The concentrations of all proteins used in this study were determined from their absorbance at 280 nm by using molar extinction coefficients calculated by the method of Gill and von Hippel (11).
Chemical Cross-Linking.
Solutions of CRT(189–288) alone, ERp57 alone, or both proteins together were incubated with 20 μM of the homobifunctional cross-linker disuccinimidyl glutarate (Pierce) for 30 min on ice in a volume of 10 μl. Protein solutions were at 7 μM concentration, containing 100 mM KH2PO4, 25 mM NaCl, and 10 mM β-mercaptoethanol at pH 7.0. Excess disuccinimidyl glutarate was quenched with 20 mM glycine. The reactions were supplemented with reducing SDS/PAGE loading buffer and run on a 12% SDS/PAGE gel.
Interaction Studies by ELISA.
Microtiter wells (Nunc) were coated with 50 μl of either ERp57, PDI, or one of three different negative control proteins at 15 μg/ml in 15 mM Na2CO3, 35 mM NaHCO3 by incubating overnight at 4°C. After blocking with 200 μl 5% milk powder in buffer W (PBS/0.05% Tween 20) for 1 h at 37°C in a moist chamber, the wells were washed three times with 200 μl of buffer W. All subsequent incubation steps were performed for 8–16 h at 4°C by using protein or antibody solutions diluted in buffer W. Each incubation step was followed by washing three times with 200 μl of buffer W. The coated protein was incubated with 50 μl of 3.3 μM biotinylated CRT(189–288), followed by the incubation with 50 μl of rabbit-α-biotin antibody (Bethyl Laboratories, Montgomery, TX) (1:2,000 dilution) and 50 μl of horseradish peroxidase-conjugated goat-α-rabbit antibody (Pierce) (1:10,000 dilution). Biotinylation was performed by using EZ-Link Sulfo-NHS-LC-LC-Biotin (Pierce) according to the manufacturer's recommendations. The wells were developed by the addition of 50 μl of BM Blue (3,3′–5,5′–tetramethylbenzidine) (Roche Molecular Biochemicals) for 10 min and the subsequent addition of 30 μl of 10 mM HCl. Finally, the absorbance was recorded at 450 nm versus a reference taken at 655 nm by using an ELISA plate reader.
Isothermal Titration Microcalorimetry (ITC).
ITC was performed at 8°C on a MCS instrument (MicroCal, Northampton, MA) calibrated with either electrically generated heat pulses or by measuring the heat of standard chemical reactions. Samples of CRT(189–288) and ERp57 were prepared as described above and gel-filtrated into the same batch of buffer containing 25 mM Tris⋅HCl, pH 7.0 and 10 mM β-mercaptoethanol. Concentrations were determined after gel filtration. The cell was loaded with 0.2 mM ERp57. The titration protocol consisted of 24 12-μl injections of a 2.0 mM solution of CRT(189–288). Injection duration was 10 s, and equilibration was allowed for 5 min between injections. The stirring rate was 200 rpm. After the experiment, the data were integrated, corrected for nonspecific heat effects, normalized for the concentration, and analyzed according to a 1:1 binding model.
NMR Spectroscopy.
The NMR experiments were carried out on a Bruker DRX750 spectrometer at 20°C by using protein solutions that contained 100 mM KH2PO4, 25 mM NaCl, and 10 mM β-mercaptoethanol at pH 7.0. A [15N,1H]TROSY spectrum (12) of free 15N,2H-labeled CRT(189–288) was measured at a protein concentration of 0.4 mM, with t1,max = 88 ms, t2,max = 98 ms, a data size of 200 × 1,024 complex points, and an overall recording time of 2 h. For optimal sensitivity a polarization transfer time of 5.4 ms was used. A [15N,1H]TROSY spectrum of the CRT(189–288)/ERp57 complex was measured in a solution containing 0.14 mM 15N,2H-labeled CRT(189–288) and 0.38 mM ERp57, with t1,max = 44 ms, t2,max = 98 ms, a data size of 100 × 1,024 complex points, and an overall recording time of 24 h. For optimal sensitivity a polarization transfer time of 3.4 ms was used (13). The same NMR set-ups as for the CRT(189–288)/ERp57 complex were used in titration experiments performed to investigate the thermodynamic and kinetic stability of the complex (Table 1), except that the overall recording time per experiment was 12 h. The titration experiments were started with the 1:2.7 mixture of CRT(189–288) and ERp57, and unlabeled CRT(189–288) was added stepwise as listed in Table 1. An experiment with a freshly prepared solution containing 0.14 mM 15N,2H-labeled CRT(189–288) and 0.07 mM ERp57 was used as an independent control for this titration protocol.
Table 1.
Composition of the solutions used in the titration of CRT(189–288) with ERp57
| mol:mol ratio CRT(189–288)/ERp57 | Concentration, mM
|
||
|---|---|---|---|
| 15N,2H-labeled CRT(189–288) | Unlabeled CRT(189–288) | Unlabeled ERp57 | |
| 1:2.7 | 0.14 | 0 | 0.38 |
| 1.1:1 | 0.12 | 0.24 | 0.33 |
| 1.8:1 | 0.11 | 0.41 | 0.29 |
| 2.4:1 | 0.1 | 0.54 | 0.26 |
| 3:1 | 0.09 | 0.64 | 0.24 |
| 4:0 | 0.28 | 0 | 0 |
| 2:1* | 0.14 | 0 | 0.07 |
The protein solutions contained 100 mM KH2PO4, 25 mM NaCl, and 10 mM β-mercaptoethanol at pH 7.0 and 20°C.
As an independent control for the titration experiment, spectra were recorded of a freshly prepared solution of unlabeled ERp57 containing a 2-fold excess of labeled CRT(189–288).
The NMR data were processed with the program prosa (14) and analyzed with the program xeasy (15). Drawings of molecular models were performed by using the program molmol (16).
Results
The Calreticulin P-Domain Binds ERp57.
Evidence for intermolecular interactions between CRT(189–288) and ERp57 was independently obtained from chemical cross-linking (Fig. 1a), ELISA experiments (Fig. 1b), ITC (Fig. 1c), and chemical shift perturbation in TROSY-NMR experiments (Fig. 2).
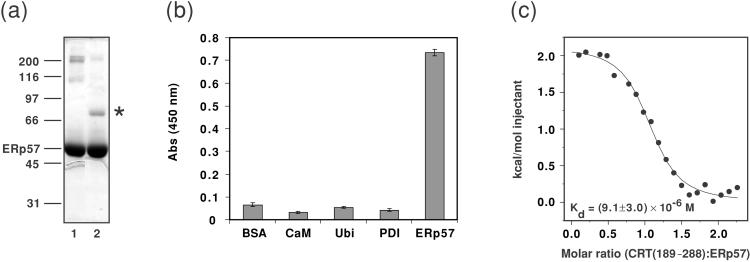
Figure 1.
(a) Chemical cross-linking of CRT(189–288) and ERp57. The homobifunctional cross-linker disuccinimidyl glutarate was added to solutions of CRT(189–288) alone (data not shown), ERp57 alone (lane 1), or an equimolar mixture of the two proteins (lane 2). After incubation for 30 min on ice, the reaction was quenched with an excess of glycine. Potential protein complexes were analyzed by 12% reducing SDS/PAGE, and the gel was stained with Coomassie blue. Molecular mass standards given in kDa are indicated on the left, along with the position of ERp57. The suggested position of the CRT(189–288)/ERp57 complex is indicated by *. (b) The interaction of CRT(189–288) and ERp57 studied by ELISA. Potential ligands for CRT(189–288) were coated in microtiter wells, and the interaction was probed with biotinylated CRT(189–288), followed by the binding of a primary antibody directed against biotin and a horseradish peroxidase-conjugated secondary antibody. Development by the addition of BM Blue (3,3′–5,5′–tetramethylbenzidine) (Roche Molecular Biochemicals) substrate gives rise to a signal at 450 nm for positive enzymatic reactions. BSA, calmodulin (CaM), and ubiquitin (Ubi) were used as control proteins. The data presented in the graph are mean values of triplicates. The experiment was repeated three times with closely similar results. Standard deviations are given by the vertical bars. Nonspecific binding of primary and secondary antibodies to wells coated with protein ligand in the absence of biotinylated CRT(189–288) was subtracted for each well. (c) Binding isotherm describing the formation of the CRT(189–288)/ERp57 complex followed by ITC at 8°C. ● represent the integrated heats at each injection after correction for nonspecific heat effects and normalization for the molar concentration. The continuous line visualizes a least-squares nonlinear fit of the data according to a 1:1 binding model defined by the following parameters: n = 1.04 ± 0.03, Kd = (9.1 ± 3.0) × 10−6 M [Ka = (1.1 ± 0.3) × 105 M−1], and ΔH = 2.1 ± 0.1 kcal mol−1.
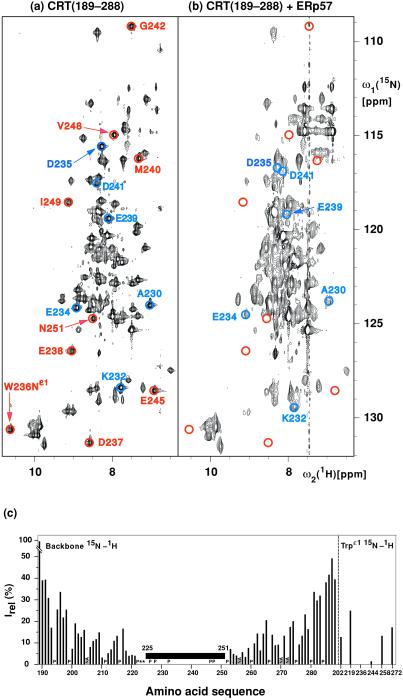
Figure 2.
(a and b) 15N–1H correlation NMR spectra used to delineate the surface areas of CRT(189–288) that interact with ERp57. These spectra contain peaks corresponding to backbone amide 15N–1H groups and, in the lower left, indole 15N–1H groups of tryptophyl residues. Color-coded peaks indicate outstandingly large effects from ERp57 interactions: Blue circles identify cross peaks with chemical shift differences of more than 0.2 ppm along the 15N dimension between free and ERp57-bound CRT(189–288). Red circles identify cross-peak positions in free CRT(189–288) for which no counterpart could be observed in the spectrum of the complex. The color-coded cross peaks are labeled with the sequence-specific resonance assignments. (a) [15N,1H]TROSY spectrum of free 15N,2H-labeled CRT(189–288) (protein concentration = 0.4 mM, polarization transfer time = 5.4 ms, data size 200 × 1,024 complex points, zero-filling to 512 × 2,048 points, t1,max = 88 ms, t2,max = 98 ms, total measuring time 2 h). (b) [15N,1H]TROSY spectrum of a solution of 15N,2H-labeled CRT(189–288) and unlabeled ERp57 [protein concentrations of 0.14 mM for 15N,2H-labeled CRT(189–288) and 0.38 mM for ERp57, polarization transfer time = 3.4 ms, data size 100 × 1,024 complex points, zero-filling to 512 × 2,048 points, t1,max = 44 ms, t2,max = 98 ms, total measuring time 24 h]. (c) Plot of the relative peak intensities, Irel, of the [15N,1H]TROSY cross peaks in the CRT(189–288)/ERp57 complex and free CRT(189–288) versus the amino acid sequence of CRT(189–288). The Trp indole cross peaks are listed separately on the right. The black horizontal bar spanning the polypeptide segment 225–251 indicates that for these residues the cross peaks in the complex have either large chemical shift differences (> 0.2 ppm along the 15N dimension) or very small signal intensities [< 3% of the intensity in free CRT(189–288)] (see text). P designates proline residues, NA stands for residues for which no resonances could be assigned, and * indicates residues for which no reliable data could be measured because of spectral overlap.
In the cross-linking experiments, purified recombinant proteins were mixed and treated with the cross-linker disuccinimidyl glutarate (see Materials and Methods). Fig. 1a shows that in mixtures of CRT(189–288) and ERp57 a band appeared at an approximate molecular mass of 75 kDa. Because the free 12-kDa CRT(189–288) migrates as a protein of approximately 18 kDa, the mobility of this band is compatible with a 1:1 complex of CRT(189–288) and the 54.5-kDa ERp57.
In the ELISA experiments, ERp57 showed efficient binding to CRT(189–288), whereas three control proteins (BSA, calmodulin, and ubiquitin) did not show appreciable binding. Furthermore, it is noteworthy that PDI, which is the closest known homologue of ERp57, did not interact with CRT(189–288) (Fig. 1b). The specific association of the calreticulin P-domain with ERp57 is consistent with in vivo (4) and in vitro (17) data that demonstrated the presence of functional complexes between CNX or CRT and ERp57, but not between CNX or CRT and PDI.
ITC performed at 8°C detected a measurable heat effect on mixing of the two proteins, which reached saturation with increasing molar ratio of CRT(189–288) to ERp57. The resulting titration curve followed the shape of a typical binding isotherm (Fig. 1c) with 1:1 stoichiometry. The best fit of the data were obtained with a dissociation constant of (9.1 ± 3.0) × 10−6 M.
Chemical Shift Mapping with TROSY-NMR.
The NMR structure and the complete sequence-specific NMR assignments of CRT(189–288) (7, 9) provided a basis for structural studies of the interactions between CRT(189–288) and ERp57. Because the CRT(189–288)/ERp57 complex was reconstituted in vitro from the isolated components, uniform isotope labeling of CRT(189–288) with 15N and 2H enabled selective observation of the 15N–1H fingerprint of this protein in the 66.5-kDa complex with the use of [15N,1H]TROSY (18).
From the amino acid sequence of CRT(189–288) one expects a “fingerprint” containing backbone 15N–1H peaks of 84 residues, of which 81 have previously been assigned in the free protein (9). Under the conditions of the present experiments, the 15N–1H fingerprint of free CRT(189–288) contains 80 of these cross peaks (Fig. 2a). With the TROSY experimental scheme used here, the 15N–1H cross peaks of the Gln and Asn side chains, which would appear in the spectral region 112–115 ppm along ω1(15N) and 6.7–8.7 ppm along ω2(1H), are nearly completely suppressed (19). In the fingerprint of 15N,2H-labeled CRT(189–288) in a solution containing a 2.7-fold excess of unlabeled ERp57 (Fig. 2b), 72 backbone 15N–1H cross peaks and five Trp indole peaks could be identified.
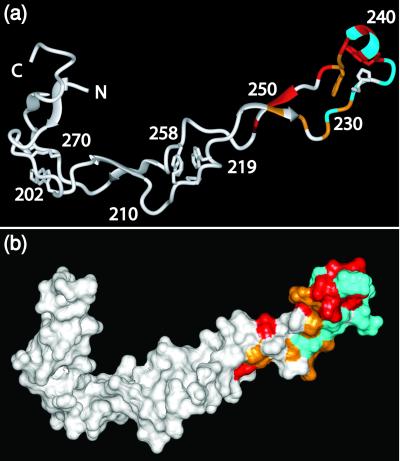
Superposition of the [15N,1H]TROSY spectra of free CRT(189–288) and the CRT(189–288)/ERp57 complex revealed that a large number of peaks in the complex superimposed exactly with peaks in free CRT(189–288). However, Fig. 2 also shows that the relative intensities as well as the peak positions of numerous corresponding, well-resolved peaks in free and complexed CRT(189–288) were significantly different. Titration experiments (see below) enabled transfer of sequence-specific resonance assignments in free CRT(189–288) to 15N,2H-labeled CRT(189–288) bound to ERp57. We thus found that for the backbone 15N–1H-moieties of D237, E238, M240, G242, E245, V248, I249, and N251, and the indole15N–1H of Trp-236 no peaks were observed in the complex, and that for the residues I225, D227, D229, K231, W236, E243, and W244 the peak intensities in the complex were weaker than 3% of the intensities in free CRT(189–288) (Fig. 2 a and b). In addition, the six backbone amide cross peaks of A230, K232, E234, D235, E239, and D241 exhibited chemical shift changes relative to free CRT(189–288) of 0.2 to 1.0 ppm along the 15N dimension and 0.0 to 0.5 ppm along the 1H dimension in the 1:2.7 complex (Fig. 2). A systematic evaluation of the signal intensities by measurement of the peak heights in the two spectra yielded the data shown in Fig. 2c. Whereas the residues near both chain ends of CRT(189–288) had similar intensities in the free and bound protein, there was a continuous loss of peak intensity from the chain ends toward the center of the polypeptide chain. All 21 residues mentioned above with either a sizeable chemical shift change, complete absence of a cross peak, or very weak cross-peak intensity in the bound form are located in the contiguous structural region of residues 225–251, indicating that the ERp57-binding site on CRT(189–288) is at the tip of the hairpin structure of this protein, as illustrated in Fig. 3.
Figure 3.
Visual display of the molecular regions of CRT(189–288) that are in direct contact with ERp57. (a) Ribbon representation. (b) Space-filling model displaying the molecular surface of the NMR structure. The residues for which the 15N–1H-moieties show chemical shift changes caused by interactions with ERp57 in excess of 0.2 ppm along the 15N dimension are colored blue, those for which no cross peak is observed in the [15N,1H]TROSY spectrum of the complex are red, and those for which the peak intensity relative to free CRT(189–288) is weaker than 3% are orange. In a the positions of selected residues areindicated.
NMR Characterization of Thermodynamics and Kinetics of the CRT(189–288)/ERp57 Interaction.
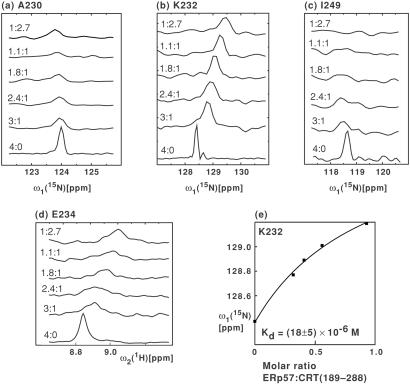
The 1:2.7 CRT(189–288)/ERp57 solution of Fig. 2b was titrated by stepwise addition of unlabeled CRT(189–288), as detailed in Table 1. In Fig. 4 a-d the change in chemical shift and peak intensity upon titration is shown for four selected residues. When following the chemical shift changes at different titration points we found that for the residues E238, I249, and N251, which showed no peak intensity in the 1:2.7 CRT(189–288)/ERp57 solution, weak cross peaks could be observed in [15N,1H]TROSY spectra measured at excess molar ratios of CRT(189–288). Specifically, at CRT(189–288)/ERp57 molar ratios of 3:1, 2.4:1, and 1.8:1, these cross peaks displayed sizeable chemical shift changes when compared with free CRT(189–288), as illustrated for I249 (Fig. 4c).
Figure 4.
Titration of the 15N,2H-labeled CRT(189–288)/ERp57 complex with unlabeled CRT(189–288). (a-c), Cross sections from [15N,1H]TROSY spectra along the ω1(15N) dimension for the residues A230, K232, and I249 of 15N,2H-labeled CRT(189–288). The numbers on the left of each trace refer to the solution conditions given in Table 1. (d) Cross sections from [15N,1H]TROSY spectra along ω2(1H) for residue E234. (e) Kd determination based on the titration data for residue K232. ■ represent the experimental 15N chemical shifts of K232 at discrete values of the ERp57/CRT(189–288) molar ratio (Table 1). The curve visualizes the fit from which the 15N chemical shift of the CRT(189–288)/ERp57 complex [δ(15Ncomplex)] was determined, using the relation δ(15N) = δ(15Nfree) + [δ(15Ncomplex) − δ(15Nfree)] ([ERp57]/[CRT(189–288)])/(([ERp57]/[CRT(189–288)]) + Kratio) where δ is the chemical shift and Kratio the Kd expressed as molar ratio, and the brackets indicate molar concentrations of the respective compounds. Subsequently, the Kd value was calculated at each experimental point by using the relation Kd = [CRT(189–288)][ERp57]/[CRT(189–288)/ERp57], yielding an average value of Kd = (18 ± 5) × 10−6 M.
The continuous chemical shift changes (Fig. 4 a-d) demonstrated that the exchange rate between bound and free CRT(189–288) was fast on the chemical shift time scale, with an estimated lower limit for the first-order exchange rate constant of koff > 1,000 s−1 at 20°C. Using the data for residue K232 (Fig. 4b), a fit of the dependence of the chemical shifts on the ERp57/CRT(189–288) molar ratio as described in the legend to Fig. 4e yielded a dissociation constant of Kd = (18 ± 5) × 10−6 M for the complex of CRT(189–288) and ERp57 (Fig. 4e).
Discussion
In the present investigation the ability to record high-resolution NMR spectra of big structures in solution with the use of TROSY (18) enabled us to supplement the evidence for CRT(189–288) binding to ERp57 obtained from biochemical experiments with a structural characterization of the complex. Thus, we obtained an identification of the surface residues in the distal end of the calreticulin P-domain that comprise the contact area with ERp57. Further NMR measurements also enabled the determination of the thermodynamic stability of the CRT(189–288)/ERp57 complex. The two Kd values of (9.1 ± 3.0) × 10−6 M at 8°C and (18 ± 5) × 10−6 M at 20°C determined independently by ITC and TROSY-NMR, respectively, are in good agreement with each other. The relatively low affinity of the CRT/ERp57 complex is comparable to that reported recently for the complex between CRT and the glycoprotein IgG in vitro, i.e., Kd = 1.9 × 10−6 M (20). However, whereas an exchange rate of koff = 0.1 s−1 was reported for the CRT/glycoprotein complex, the NMR experiment reveals a much faster rate of koff > 1,000 s−1 for the CRT(189–288)/ERp57 complex, which is thus very short-lived. The additional presence of a substrate protein might, of course, result in formation of a more stable ternary complex, which could be further stabilized upon formation of an intermolecular disulfide bond between ERp57 and the glycoprotein (see Fig. 5). The fast off-rate for the binary CRT/ERp57 complex could be of functional significance because it might allow ERp57 to “screen” for pre-existing complexes of CRT and glycoprotein.
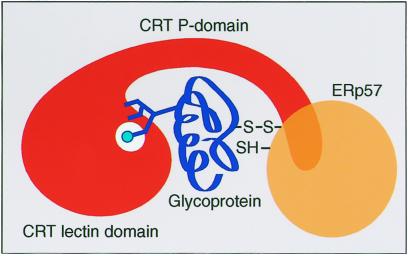
Figure 5.
Scheme for a possible mode of cooperative interaction between CRT and ERp57 in assisting the folding of a glycoprotein based on currently available structural, biochemical, and cell-biological data (2, 7, 8). The substrate glycoprotein (blue) binds to the CRT lectin domain (red) by means of a branched oligosaccharide. CRT and CNX both interact specifically with the Glc1Man9GlaNAc2 form of the oligosaccharide (26–28). The interaction of the terminal glucose (light blue circle) with the binding site (white circle) would place the glycoprotein polypeptide chain in the partially solvent-shielded cavity bounded by the CRT lectin domain, the P-domain, and ERp57 (orange) bound to the distal end of the protruding P-domain. It is further hypothesized that the CRT-ERp57 interaction places the thiol-disulfide oxidoreductase favorably for the formation of intermolecular disulfide bonds (S—S) with the glycoprotein.
Based on their close sequence similarity, CRT and CNX are likely not only to possess similar P-domains but also structurally homologous lectin domains. In CNX, the lectin domain contains a single binding site for the oligosaccharide of the substrate glycoprotein and is located close to the site from which the P-domain emerges as an elongated hairpin loop (8). The P-domain forms a slightly curved arm of about 110 Å in CRT (7) and 140 Å in CNX (8). Binding of ERp57 to the distal end of this arm is likely to generate a partially solvent-shielded cavity surrounded by the lectin domain, the P-domain, and ERp57 (see Fig. 5). Given the apparent plasticity of the P-domain (7), the size of this cavity might be variable so as to accommodate substrates of variable size.
The role of CNX and CRT as molecular chaperones seems to rely on three specific features. First, these proteins bind the substrate glycoprotein through the glycan binding site. By binding to the oligosaccharide moiety, they may tether the substrate with minimal constraints on the conformational freedom of the substrate polypeptide chain. Second, they provide a partially solvent-shielded environment for folding (Fig. 5), where access for other ER chaperones and protein folding intermediates is likely to be restricted, so that aggregation and formation of non-native intermolecular disulfides with other newly synthesized proteins is suppressed (21). Third, they provide a strategically placed binding site for ERp57, which supports productive interactions between the thiol-disulfide oxidoreductase and cysteines in the folding glycoprotein. Such interactions have previously been shown to involve the formation of a series of transient mixed disulfides between the substrate glycoprotein and ERp57 (6).
The presently described insights into the structural basis of the CRT/ERp57 chaperone system emphasize the importance of determining whether ERp57 functions as a thiol oxidase and/or as a disulfide isomerase in vivo. It will also be of interest to study in closer detail how the apparent plasticity of the P-domain might affect the function of the bound ERp57 molecule. The accessibility of the glycan to processing enzymes such as glucosidase II and the UDP-glucose:glycoprotein glucosyltransferase will also need further investigation. The available data already indicate, however, that CNX and CRT function by a different mechanism from that of previously analyzed molecular chaperones. Distinctive features are that binding of the substrate molecule occurs by means of oligosaccharide branches (22–24), that there is close structural and functional cooperation between the chaperone and a thiol-disulfide oxidoreductase (4, 25), and that the on-and-off cycle of the glycoprotein substrate is driven by independently acting enzymes that regulate covalent modification of the N-linked glycan (2).
Acknowledgments
We thank the following for kind gifts of expression vectors and purified protein: S. High, University of Manchester, Manchester, U.K. (pHisERp57), J. Winther, Carlsberg Laboratory, Copenhagen (pET12a-PDI2), and P. Gazzotti, Eidgenössische Technische Hochschule, Zürich (calmodulin). Financial support by the Schweizerischer Nationalfonds [Projects 31.51054.97 (A.H.) and 31.49047.96 (K.W.)] is gratefully acknowledged.
Abbreviations
- CNX
calnexin
- CRT
calreticulin
- ER
endoplasmic reticulum
- ITC
isothermal titration microcalorimetry
- PDI
protein disulfide isomerase
- TROSY
transverse relaxation-optimized spectroscopy
References
- 1.Ellgaard L, Molinari M, Helenius A. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- 2.Parodi A J. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- 3.Chevet E, Cameron P H, Pelletier M F, Thomas D Y, Bergeron J J. Curr Opin Struct Biol. 2001;11:120–124. doi: 10.1016/s0959-440x(00)00168-8. [DOI] [PubMed] [Google Scholar]
- 4.Oliver J D, Roderick H L, Llewellyn D H, High S. Mol Biol Cell. 1999;10:2573–2582. doi: 10.1091/mbc.10.8.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrari D M, Söling H D. Biochem J. 1999;339:1–10. [PMC free article] [PubMed] [Google Scholar]
- 6.Molinari M, Helenius A. Nature (London) 1999;402:90–93. doi: 10.1038/47062. [DOI] [PubMed] [Google Scholar]
- 7.Ellgaard L, Riek R, Herrmann T, Güntert P, Braun D, Helenius A, Wüthrich K. Proc Natl Acad Sci USA. 2001;98:3133–3138. doi: 10.1073/pnas.051630098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schrag J D, Bergeron J J, Li Y, Borisova S, Hahn M, Thomas D Y, Cygler M. Mol Cell. 2001;8:633–644. doi: 10.1016/s1097-2765(01)00318-5. [DOI] [PubMed] [Google Scholar]
- 9.Ellgaard L, Riek R, Braun D, Herrmann T, Helenius A, Wüthrich K. FEBS Lett. 2001;488:69–73. doi: 10.1016/s0014-5793(00)02382-6. [DOI] [PubMed] [Google Scholar]
- 10.Westphal V, Spetzler J C, Meldal M, Christensen U, Winther J R. J Biol Chem. 1998;273:24992–24999. doi: 10.1074/jbc.273.39.24992. [DOI] [PubMed] [Google Scholar]
- 11.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 12.Pervushin K V, Wider G, Wüthrich K. J Biomol NMR. 1998;12:345–348. doi: 10.1023/A:1008268930690. [DOI] [PubMed] [Google Scholar]
- 13.Riek R, Wider G, Pervushin K, Wüthrich K. Proc Natl Acad Sci USA. 1999;96:4918–4923. doi: 10.1073/pnas.96.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Güntert P, Dötsch V, Wider G, Wüthrich K. J Biomol NMR. 1992;2:619–629. [Google Scholar]
- 15.Bartels C, Xia T H, Billeter M, Güntert P, Wüthrich K. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 16.Koradi R, Billeter M, Wüthrich K. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 17.Zapun A, Darby N J, Tessier D C, Michalak M, Bergeron J J, Thomas D Y. J Biol Chem. 1998;273:6009–6012. doi: 10.1074/jbc.273.11.6009. [DOI] [PubMed] [Google Scholar]
- 18.Pervushin K, Riek R, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pervushin K, Braun D, Fernandez C, Wüthrich K. J Biomol NMR. 2000;17:195–202. doi: 10.1023/a:1008399320576. [DOI] [PubMed] [Google Scholar]
- 20.Patil A R, Thomas C J, Surolia A. J Biol Chem. 2000;275:24348–24356. doi: 10.1074/jbc.M003102200. [DOI] [PubMed] [Google Scholar]
- 21.Helenius A, Trombetta E S, Hebert D N, Simons J F. Trends Cell Biol. 1997;7:193–200. [Google Scholar]
- 22.Ou W J, Cameron P H, Thomas D Y, Bergeron J J. Nature (London) 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- 23.Rodan A R, Simons J F, Trombetta E S, Helenius A. EMBO J. 1996;15:6921–6930. [PMC free article] [PubMed] [Google Scholar]
- 24.Zapun A, Petrescu S M, Rudd P M, Dwek R A, Thomas D Y, Bergeron J J. Cell. 1997;88:29–38. doi: 10.1016/s0092-8674(00)81855-3. [DOI] [PubMed] [Google Scholar]
- 25.Oliver J D, van der Wal F J, Bulleid N J, High S. Science. 1997;275:86–88. doi: 10.1126/science.275.5296.86. [DOI] [PubMed] [Google Scholar]
- 26.Hammond C, Braakman I, Helenius A. Proc Natl Acad Sci USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiro R G, Zhu Q, Bhoyroo V, Soling H D. J Biol Chem. 1996;271:11588–11594. doi: 10.1074/jbc.271.19.11588. [DOI] [PubMed] [Google Scholar]
- 28.Vassilakos A, Michalak M, Lehrman M A, Williams D B. Biochemistry. 1998;37:3480–3490. doi: 10.1021/bi972465g. [DOI] [PubMed] [Google Scholar]