Abstract
The current understanding of the relationships between cell shape, intracellular forces and signaling, nuclear shape and organization, and gene expression is in its infancy. Here we introduce a method for investigating gene-specific responses in individual cells with controlled nuclear shape and projected area. The shape of the nuclei of primary osteogenic cells were controlled on microfabricated substrata with regiospecific chemistry by confining attachment and spreading of isolated cells on adhesive islands. Gene expression and protein synthesis were altered by changing nuclear shape. Collagen I synthesis correlated directly with cell shape and nuclear shape index (NSI), where intermediate values of nuclear distension (6 < NSI < 8) promoted maximum synthesis. Osteocalcin mRNA, a bone-specific differentiation marker, was observed intracellularly by using reverse transcription in situ PCR at 4 days in cells constrained by the pattern and not detected in unconstrained cells of similar projected area, but different NSI. Our data supports the concept of gene expression and protein synthesis based on optimal distortion of the nucleus, possibly altering transcription factor affinity for DNA, transport to the nucleus, or nuclear matrix organization. The combination of microfabricated surfaces, reverse transcription in situ PCR, and NSI measurement is an excellent system to study how transcription factors, the nuclear matrix, and the cytoskeleton interact to control gene expression and may be useful for studying a wide variety of other cell shape/gene expression relationships.
Keywords: patterning‖cell size‖osteocalcin‖reverse transcription–PCR‖ photolithography
Cell morphology has a profound effect on a range of cellular events, such as proliferation (1–3), differentiation (4–7), cytoskeletal organization (8), and presumably gene expression. Changes to the cytoskeleton lead to altered stress levels imparted on the nucleus (9–11) and could affect organelle and DNA organization and distribution, ultimately altering cell function. For example, the rate of albumin secretion from hepatocytes can be altered by constraining cell size on patterned culture surfaces (3). In human epidermal kareatinocytes cell shape can modulate between terminal differentiation and proliferation (4). Furthermore, cells can be forced to enter the apoptotic cascade when the area on which the cell is allowed to spread is constrained (1). One proposed mechanism for the transduction of cell shape information into gene expression is through mechanical forces transmitted by means of the direct link of the cytoskeleton to the nucleus (9, 12), and in particular to nuclear matrix proteins (NMPs) such as NMP-1 and NMP-2 (13). These architectural transcription factors, which are components of the nuclear scaffold, induce changes in DNA supercoiling and can interact directly with gene promoter sequences (14). This interaction between the nuclear architecture and regulation of transcription or DNA topography raises the question of whether gross deformation of the cell, and hence its nucleus, can modulate the NMP/DNA interactions and gene expression. However, the details in the cascade of mechanical events involving cell morphology, cytoskeletal organization, intracellular signaling, nuclear shape, nuclear matrix organization, promoter geometry, and gene expression are poorly understood.
We report a method for both precisely controlling cell and nuclear shape and measuring in situ mRNA expression in an effort to help elaborate these mechanisms. Microfabrication and fluidic techniques have been used to control cell adhesion and spreading on the μm scale and have proven useful for asking basic questions in cell biology (1, 3, 8, 15–19). Although previous studies have used surfaces with spatially resolved chemistry to study cell shape/function relationships and their effect on protein synthesis (1, 3), studies examining in situ gene expression (mRNA) and protein synthesis as a function of the cell projected area and nuclear shape have not been published. Therefore, the methods developed in this work were designed to assess cell and nuclear shape-dependent changes in gene expression and protein synthesis of individual cells. An interpenetrating polymer network of poly(acrylamide) and polyethylene glycol [p(AAm-co-EG)] grafted to silane-derivatized glass was used as a nonadhesive surface chemistry amenable to the thermal cycling of PCR (20). Combining photolithographic and photopolymerization techniques, we created islands of adhesive surface chemistry [an amine-terminated silane that preferentially adsorbs vitronectin (18)] surrounded by the p(AAm-co-EG) interpenetrating polymer network. A range of geometric shapes and sizes were created to impart different mechanical environments within different cells maintained on the same culture surface. Because of our interest in bone, we patterned primary bone-derived cells from rat calvaria and probed for type I collagen (CollI) synthesis and osteocalcin (OC) mRNA expression to determine whether constraining the projected area and nuclear shape of a bone-derived cell altered the time required for differentiation into the osteoblast phenotype. We chose CollI and OC because they are markers of osteoblast differentiation, and NMPs (e.g., Cbfa1, a transcriptional activator of osteoblast differentiation) have been shown to bind to the promoter region of genes for both OC and CollI (13).
Materials and Methods
Surface Preparation.
Surfaces were prepared as described in detail by Thomas et al. (8) to generate a pattern of N-(2-aminoethyl)-3-aminopropyl-trimethoxysilane (EDS; United Chemical Technologies, Bristol, PA) islands with a p(AAm-co-EG) background (20). The spatial distribution of the chemistry on these substrata was confirmed by using time-of-flight secondary ion MS with imaging (8). Cell attachment and spreading could be confined to the adhesive islands of the slide, which ranged in area from 75 to 10,000 μm2 (8). Alternatively, some experiments were performed on patterned slides prepared in an identical manner with adhesive square EDS islands with areas of 400, 1,600, 3,600, 64,000, or 10,000 μm2. Homogeneous EDS samples were used as control surfaces for the subsequent experiments.
Cell Culture and Molecular Probes.
Primary rat bone cells were isolated from the calvaria of 10- to 12-day-old rats (Harlan–Sprague–Dawley) following methods described by Whitson et al. (21). Patterned slides were sterilized with 70% ethanol, dried, and rinsed with PBS. A 2-well chamber system (Falcon 4102) was attached to the slide to confine cells to two separate regions of the pattern. Rat bone cells were plated at a density of 5.0 × 104 cells/cm2 and maintained in culture before fixation. Patterned cultures were fixed/permeabilized (ORTHO Permea Fix, Ortho Diagnostics) (22), dehydrated with a graded series of ethanol, and rinsed with chloroform to ensure complete dehydration. F-actin staining was performed by blocking samples with BSA and incubating them for 1 h with 5 units/ml Oregon Green 488 phalloidin (Molecular Probes) in PBS. Immunostaining for CollI was performed to examine expression kinetics (2, 4, and 8 days) by using 1:3,000 (vol/vol) monoclonal anti-CollI (C-2456; Sigma). In some experiments, cells were labeled with BrdUrd followed by immunostaining to determine the percentage of cells proliferating on the patterned substrata. BrdUrd labeling was performed for 24 h after 1, 2, and 5 days in culture. Samples were viewed by using phase-contrast and epifluorescence microscopy (Nikon Eclipse TE300), and images were captured by using Oncor image software and a Sensys high-resolution, cooled charge-coupled device camera (Photometrics, Tucson, AZ).
Nuclear Morphology Measurement.
Bone cells from rat calvaria were seeded onto EDS/interpenetrating polymer network-patterned slides and EDS-modified slides. At 24 h in culture, cells were fixed with anhydrous MeOH at −20°C for 10 min, treated with RNase A (10 μg/ml in PBS, Sigma) for 1 h, and stained with ethidium homodimer-1 (4 μM in PBS for 15 min, Molecular Probes). Slides were rinsed with PBS between steps. Two to four drops of SlowFade (Molecular Probes) were placed on the slides, which were then affixed with 150-μm-thick glass spacers (to prevent cell compression) and a 1-oz coverslip. Nuclear morphology measurements were collected with a Zeiss LSM-510 confocal microscope equipped with a 543-nm HeNe laser (80-μm pinhole diameter, optical slice <1.5 μm, ×40 oil objective, numerical aperture 1.0). Gain settings were kept constant throughout all measurements to ensure measurement repeatability. Nuclear height was measured by recording the focus position at the top and bottom of the stained nuclei, and an image was captured at the focus position with maximal nuclear area in the x-y plane. Using iplab (Scanalytics, Billerica, MA), this image was then segmented and the projected nuclear area was calculated by using a photomask for scale calibration. The nuclear space index (NSI) was calculated as maximal nuclear area in the x-y plane divided by nuclear height.
Synthesis of Riboprobes.
Digoxigenin-labeled cRNA probes to rat OC were prepared by using the DIG RNA Labeling Kit (Roche Molecular Biochemicals). The 257-bp OC fragment was subcloned into pTAdv by using PCR with the primers 5′-TGC AAA GCC CAG CGA CTC TGA G-3′ and 5′-TTG GGC TCC AGG GCA ACA CAT-3′ and a rat tooth cDNA (22). The sequence was then inserted into the pSPT18 plasmid and sequenced to determine the insert orientation. The cDNA plasmid was transcribed with T7 or SP6 RNA polymerase to create the 257-bp antisense or sense riboprobes, respectively. Digoxigenin-labeled cRNA probes were synthesized in the presence of digoxigenin-dUTP by “run off” transcription, resulting in labeled cRNA probes for in situ PCR. This reaction product was elecrophoresed on an agarose gel to verify the probe length and blotted onto a nylon filter to verify digoxigenin incorporation and determine probe concentration.
In Situ Reverse Transcription (RT) and PCR.
To detect the low level of expressed OC mRNA (below the 0.161 pg/10,000 μm2 detection limit) in single cells isolated on patterned substrata, low copy mRNA was amplified by using RT followed by in situ PCR. Hybridization was performed by using the digoxigenin-labeled cRNA probes prepared from the 257-bp OC fragment. Briefly, experimental samples were treated with DNase overnight at 37°C in a humid chamber and reverse-transcribed by using the Gene Amp In Situ PCR System 1000 (Perkin–Elmer) with random hexamers. Each patterned array of cells (Fig. 1A) was covered with an AmpliCover disk and clip (Perkin–Elmer), allowing for experimental and control reactions to be performed on the same slide. The cycle had the following format: denature at 95°C for 1 min, anneal at 56°C for 2 min, and extend at 72°C for 1.5 min (30 cycles). See Fig. 4A for an outline of the experimental procedure and the controls used for RT in situ PCR. The positive control group received no DNase treatment and resulted in the amplification of the DNA sequence of interest, and the negative control did not receive RT or PCR. After RT-PCR, hybridization was performed at 42°C overnight in the GeneAmp 1000 by using 300 ng/ml of the cRNA probe in hybridization solution. Samples were washed and blocked before probe detection, which was performed by using antidigoxigenin mAb conjugated with rhodamine (Roche Molecular Biochemicals) diluted 1:1,000 in blocking solution. Samples were viewed by using phase-contrast and epifluorescence microscopy (Nikon Eclipse TE300), and images were captured by using Oncor image software and a Sensys high-resolution, cooled charge-coupled device camera (Photometrics). All images were taken at the same magnification and intensity scaling.
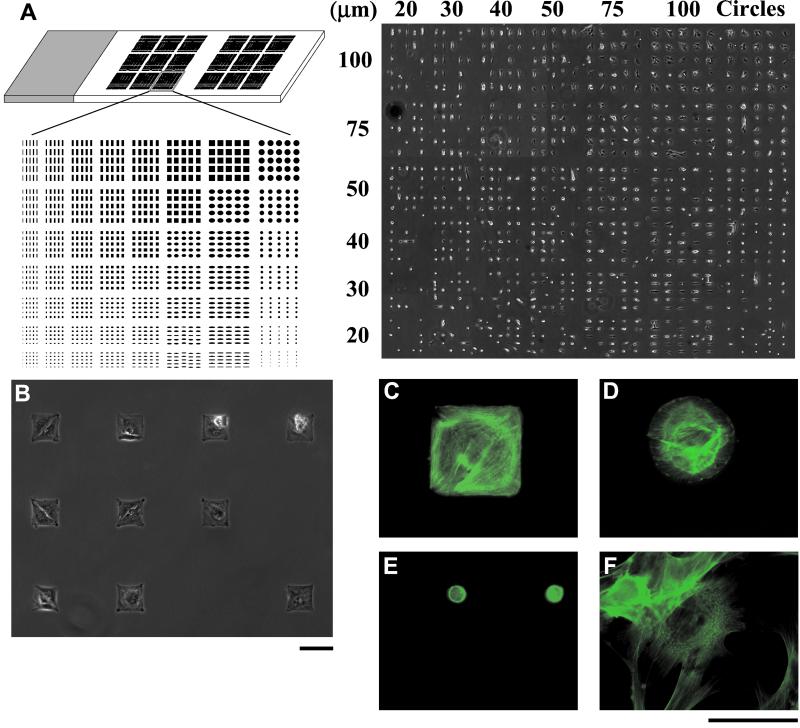
Figure 1.
(A Left) Multidomain patterning on a 1 × 3-inch glass slide. The array shown is repeated 18 times on the slide. Two groups of nine arrays were patterned so that control RT in situ PCR and hybridization groups could be run on the same slide as experimental groups. (A Right) A montage of phase-contrast images of bone-derived cells patterned on one multidomain array on a 1 × 3-inch glass slide. [Reproduced with permission from ref. 8 (Copyright 1999, American Society of Mechanical Engineers).] (B) Phase-contrast image of individual cells on 80-μm square islands. (C–F) Cells stained for F-actin with phalloidin-Oregon green. Cells on large patterns, 100-μm square (C) and 80-μm circle (D), arranged their cytoskeletons to conform to the patterned features, whereas cells seeded on 20-μm circles (E) showed diffuse staining indicative of a disorganized cytoskeleton. (F) Cells seeded on unpatterned EDS surfaces showed organized cytoskeletons, but the orientation was different than those on the constraining islands. (Scale bars = 100 μm.)
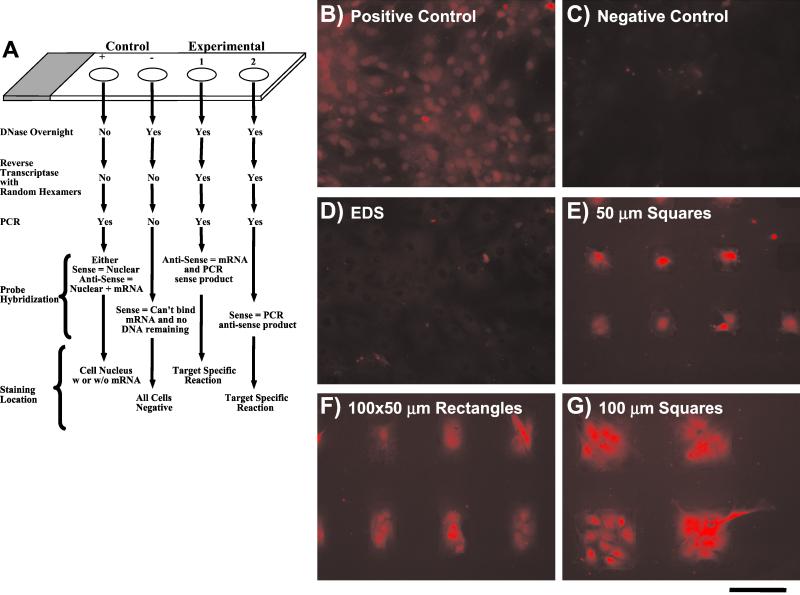
Figure 4.
(A) Schematic of the steps necessary for the RT in situ PCRs with positive and negative controls. (B–G) In situ hybridization for OC after RT-PCR for 4-day cultures. (D) Control homogeneous EDS. (B) Positive RT-PCR/hybridization control, cells on homogeneous EDS. (C) Negative RT-PCR/hybridization control, cells on homogeneous EDS. (E–G) cells on patterned surfaces: (E) 50-μm square; (F) 100 × 50-μm rectangle; and (G) 100-μm square. (Scale bar = 100 μm.)
Results
Cell Patterning.
When cells were plated on the patterned surfaces, they attached and spread on the adhesive (EDS) islands, assumed the shape of the underlying pattern (Fig. 1 A and B), and oriented their cytoskeleton to the features on which they were cultured (Fig. 1 C–F). Cells were nonadherent to the interpenetrating polymer network.
Cell Proliferation, CollI Synthesis, and Nuclear Shape.
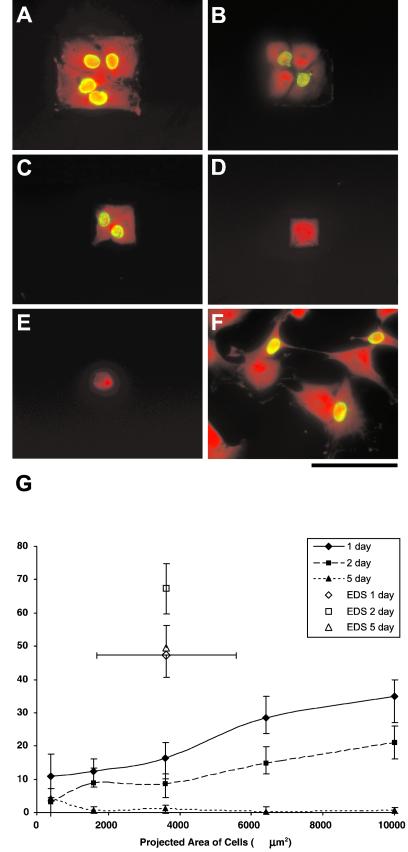
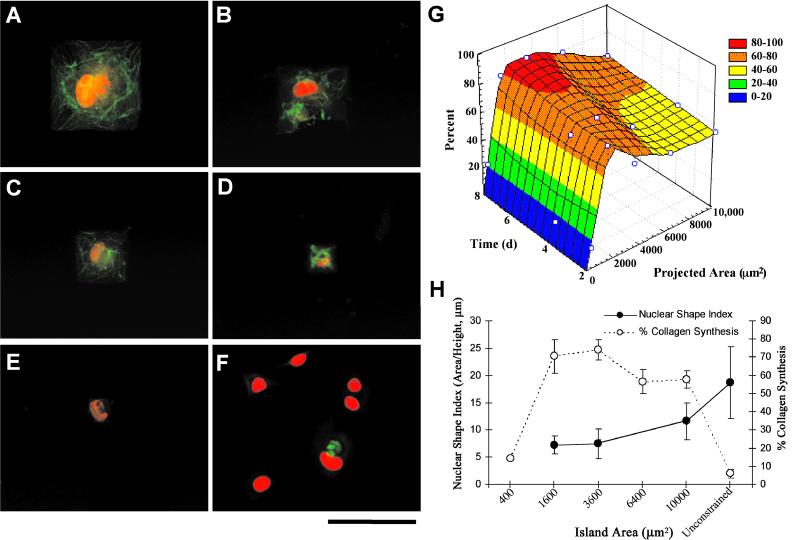
Fluorescent images of cells proliferating on islands of various dimensions taken after day 5 of exposure are shown in Fig. 2 A–F. Proliferation was greatly restricted for cells confined to smaller islands compared with unpatterned EDS (Fig. 2 A–G). Independent of the specific cell projected area, proliferation rates decreased significantly at 1-, 2-, and 5-day time points when compared with unpatterned EDS surfaces that did not limit cell spreading (P < 0.5, multiple ANOVA, Newman–Keuls post hoc test). After 5 days, proliferation essentially stopped, and the smallest island sizes (400, 1,600 and 3,600 μm2) demonstrated the lowest proliferation rates at each time point (Fig. 2G) (P < 0.5). CollI synthesis was also affected by the island size. CollI synthesis was strongly coupled to cell projected area, with a significantly higher percentage of cells synthesizing CollI when the island projected area was ≥1,600 μm2 compared with 400 μm2 EDS islands or control homogeneous EDS surfaces (Fig. 3 A–G). The temporal variation in CollI synthesis as a function of cell projected area is given in Fig. 3G as a three-dimensional plot. Maximum CollI synthesis occurred between 1,600 and 3,600 μm2 at each time point and increases with time. Examination of the proliferation (BrdUrd) and the CollI production results provided a kinetic profile of the transition from cell growth and division to phenotypic expression of extracellular matrix production. Islands that reduced the percentage of proliferating cells also decreased the activation time for CollI synthesis (Figs. 2G and 3G). However, most significantly the level of CollI synthesis correlated to a metric of nuclear shape (area/height) (Fig. 3H), where intermediate values of NSI (6–8) and projected cell area (1,600 and 3,600 μm2) demonstrated maximum CollI synthesis. Embedded in these data are the fact that unconstrained cells on EDS had mean projected areas (3,626 ± 1,948 μm2; mean ± SD) in the range for maximum CollI synthesis for cells on patterned substrata (1,600–3,600 μm2); however, these unconstrained cells had very low values of CollI synthesis and a high number of cells (≈50%) proliferating (Figs. 2G and 3G). The major difference between these unconstrained and constrained cells of similar area was that the NSI was significantly different (Fig. 3H).
Figure 2.
Images of cells cultured for 5 days on substrates with square islands with side lengths of (A) 100 μm, (B) 80 μm, (C) 60 μm, (D) 40 μm, and (E) 20 μm, and (F) homogeneous EDS surfaces (unconstrained). Cells were labeled with BrdUrd (green nuclei) to indicate DNA synthesis and counterstained with ethidium homodimer (red) for determination of the number of cells in any given island. Counterstained images were enhanced to illustrate the shape of cells. (Scale bar = 100 μm.) (G) The percentage of islands with only one cell that was actively proliferating versus projected area. Cells were exposed to BrdUrd for 24 h, labeling all cells that proliferated during that exposure period.
Figure 3.
After 4 days in culture, immunostaining for CollI (green) was performed on substrates with square islands with side lengths of (A) 100 μm, (B) 80 μm, (C) 60 μm, (D) 40 μm, and (E) 20 μm, and (F) homogeneous EDS surfaces (unconstrained). Ethidium homodimer was used as a nuclear counterstain (red). (Scale bar = 100 μm.) (G) A three-dimensional plot of the percentages of single cells producing CollI are shown versus projected area and time. Cell exposure to patterned adhesive islands between 1,600 and 3,600 μm2 resulted in the highest CollI expression levels. (H) A dual ordinate plot of NSI (area/height), a metric of nuclear spreading, and CollI synthesis indicate an inverse relationship.
RT in Situ PCR and Hybridization.
In situ hybridization without RT-PCR resulted in no OC signal detection at 4 days in culture; therefore, RT-PCR was necessary for signal amplification and was performed as described in Fig. 4A. Cells cultured on homogeneous EDS samples did not stain positively for OC mRNA (Fig. 4D). However, cells maintained on the EDS/p(AAM-co-EG)-patterned surface stained strongly for OC mRNA (Fig. 4 E–G), particularly in their nuclei, indicating active OC mRNA synthesis. The positive control (Fig. 4B) shows random spatially distributed nuclear staining; the result of amplification of the OC sequence found in the native DNA and the negative control resulted in low background staining (Fig. 4C). The drawback of performing PCR amplification of OC mRNA was the loss of quantitative information, resulting in only the determination of cells either producing or not producing OC mRNA. However, comparison of cells with similar projected areas and different NSI, >15 for unconstrained cells on EDS (Fig. 4D) and ≈6–8 for cells patterned on 50-μm square islands (Fig. 4E), the mRNA for OC is clearly detected only in the latter.
Discussion
Insight into the impact of cell shape on cell function is key for the understanding of the regulation of gene expression in all cell types, including those in the disease state (12). For example, loss of cell growth dependence on cell shape and surface contact in anchorage-dependent cells is a hallmark of tumor cells in vivo and has been induced in vitro (23). However, methods for the examination of how the mechanical and chemical factors of the cytoskeleton and the nuclear matrix integrate into a higher-order system are just beginning to emerge. Therefore, the creation and improvement of tools that aid in this research are critical to the success of these efforts.
The micropatterned surfaces used in these studies significantly affected the proliferative and protein synthetic activities of bone-derived cells. A bone-derived cell population was cultured on EDS/p(AAm-co-EG)-patterned surfaces with island areas ranging from 75 to 10,000 μm2. Differentiation into the osteoblast phenotype was determined by a decrease in cell proliferation, CollI synthesis, and the initiation of OC mRNA expression detected by using in situ hybridization procedures. Cytoskeletal staining of cells cultured on these patterned surfaces revealed that cells on islands <400 μm2 did not form stress fibers, whereas cells on larger islands organized their F-actin filaments preferentially at the periphery of the cell. The island domains also had a marked effect on cell proliferation, essentially abolishing cell division on islands of all sizes by day 5. This diminished proliferative capacity was concurrently accompanied by a marked increase in bone-specific protein expression (e.g., CollI) on the patterned surfaces. However, the NSI was the critical metric in distinguishing the amount of protein synthesis of cells spread to the same degree.
Constraining cell spreading and nuclear shape can effectively decrease the time required for phenotypic expression and prompt osteoblasts to begin expressing OC after only 4 days in culture. OC typically is expressed in cultures isolated from rat calvaria after the mineralization process has begun, with mRNA expression detectable after ≈10 days in culture and protein secretion detectable after ≈15 days in culture (24). Regardless of the adhesive region geometry and cell projected area, OC mRNA was observed in cells cultured on all patterned surfaces that limited cell spreading. The staining was observed in the cytoplasm and localized to the nuclear regions of the cells, which is indicative of active transcription of the OC gene. Cells cultured on control homogeneous EDS surfaces did not exhibit staining above negative control background levels, indicating that OC mRNA was not being expressed in cells cultured on unpatterned surfaces after 4 days. For cells with similar projected area, but different NSI, only cells constrained on the patterned substrata with NSI between 6 and 12 stained positive for OC mRNA.
Collectively, the decreased cell proliferation, the synthesis CollI and mRNA for OC, and nuclear shape data signify a shift from cell growth to differentiation. Patterned cells are forced to cease dividing and differentiate at a significantly earlier time point than unpatterned cells. One hypothesis to explain the mechanism of this effect involves the continuous physical linkage between cytoskeletal elements and the nuclear matrix, by means of extracellular matrix-integrin engagement, which can alter DNA topography and transcription (14). The osteoblast phenotype has been influenced by morphology, external forces, cytoskeletal organization, the extracellular matrix, and nuclear matrix architecture (14, 25). Additionally, several nuclear matrix-specific proteins such as NMP-2 (also identified as Cbfa1 or Osf2) have been shown to bind to the promoter region of the OC gene (13). Therefore, changes in the nuclear matrix architecture could affect the degree of promoter supercoiling and bending and result in altered transcriptional activity of OC. Similarly, NMP-1 (or YY1), which is not osteoblast-specific but binds the OC promoter, is also found in the nuclear matrix fractions of osteoblasts and is recognized as a complex DNA binding/bending protein with multiple roles (13). Both NMP-1 and NMP-2 have been characterized as DNA binding and DNA bending proteins whose involvement in OC expression has been identified but not fully elaborated. However, the continuous link between the extracellular matrix, cytoskeleton, nuclear matrix, and promoter region of the OC gene indicates the importance of cell/nuclear morphology and intracellular signaling in OC gene activation (13, 26). Potentially, engagement of α2 integrins with CollI and subsequent activation of the mitogen-activated protein kinase pathway (MAPK)/MAPK kinase leads to NMP-2 activation and expression of the OC gene (26). Because NMP-2 binds to the OCN promoter region, optimal NSI may represent the appropriate spatial organization of the promoter amenable to the activated NMP-2. This linkage represents a possible mechanism by which OC expression could be prematurely initiated through alteration of the nuclear morphology as defined by the NSI metric.
These phenomena linking cell shape with nuclear architecture are not osteoblast-specific. Investigations of NMPs and how they are linked to the cytoskeleton have already led to a better understanding of nuclear pores, matrix-associated regions of DNA, and how molecular components of the nucleus are integrated into the machinery of replication, transcription, and posttranscriptional events (7, 27).
These culture surfaces have a number of conceivable applications in the fields of biotechnology and medicine. First, because they allow cell shape and area to be controlled as an independent variable, they are invaluable as experimental platforms for the study of cell shape/gene expression hierarchies. Second, because these surfaces enable the fundamental control of cell differentiation and proliferation, we hope to map the micropatterning technique to the surface modification of biomaterials to control cell behavior at the interface. For example, a more rapid differentiation of bone cells contacting the biomaterial may lead to stronger and quicker integration of the implant. One also can exploit these microfabricated surfaces to create cell-based sensors or cell culture systems that are spatially addressed on a temporal basis for evaluation of lead drug formulations on cells with a more reproducibly defined phenotype.
Acknowledgments
This research was supported by The Whitaker Foundation, National Institutes of Health/National Institute on Dental Research Grant T32 DE07042, and National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant R01 AR43187.
Abbreviations
- NMP
nuclear matrix protein
- p(AAm-co-EG)
poly(acrylamide-co-ethylene glycol)
- CollI
type I collagen
- OC
osteocalcin
- EDS
N-(2-aminoethyl)-3-aminopropyl-trimethoxysilane
- NSI
nuclear space index
- RT
reverse transcription
References
- 1.Chen C S, Mrksich M, Huang S, Whitesides G M, Ingber D E. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J, Moscona A. Nature (London) 1978;273:345–349. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]
- 3.Singhvi R, Kumar A, Lopez G P, Stephanopoulos G N, Wang D I C, Whitesides G M, Ingbar D E. Science. 1994;264:696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 4.Watt F M, Jordan P W, O'Neil C H. Proc Natl Acad Sci USA. 1988;85:5576–5580. doi: 10.1073/pnas.85.15.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aulthouse A L. Anat Rec. 1994;238:31–37. doi: 10.1002/ar.1092380105. [DOI] [PubMed] [Google Scholar]
- 6.Aubin J E, Liu F, Malaval L, Gupta A K. Bone. 1995;17:77s–83s. doi: 10.1016/8756-3282(95)00183-e. [DOI] [PubMed] [Google Scholar]
- 7.Lelievre S A, Weaver V M, Nickerson J A, Larabell C A, Bhaumik A, Petersen O W, Bissell M J. Proc Natl Acad Sci USA. 1998;95:14711–14716. doi: 10.1073/pnas.95.25.14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas C H, Lhoest J-B, Castner D G, McFarland C D, Healy K E. J Biomech Eng. 1999;121:40–48. doi: 10.1115/1.2798041. [DOI] [PubMed] [Google Scholar]
- 9.Maniotis A J, Chen C S, Ingber D E. Proc Natl Acad Sci USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldherr C M, Akin D. Exp Cell Res. 1993;205:179–186. doi: 10.1006/excr.1993.1073. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez M, Long H, Onyia J, Hock J, Xu W, Bidwell J. Endocrinology. 1997;138:482–489. doi: 10.1210/endo.138.1.4852. [DOI] [PubMed] [Google Scholar]
- 12.Ruoslahti E. Science. 1997;276:1345–1346. doi: 10.1126/science.276.5317.1345. [DOI] [PubMed] [Google Scholar]
- 13.Bidwell J P, Van Wijnen A J, Fey E G, Dworetzky S, Penman S, Stein J L, Lian J B, Stein G S. Proc Natl Acad Sci USA. 1993;90:3162–3166. doi: 10.1073/pnas.90.8.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bidwell J P, Alvarez M, Feister H, Onyia J, Hock J. J Bone Miner Res. 1998;13:155–167. doi: 10.1359/jbmr.1998.13.2.155. [DOI] [PubMed] [Google Scholar]
- 15.Kleinfeld D, Kahler K H, Hockberger P E. J Neurosci. 1988;8:4098–4120. doi: 10.1523/JNEUROSCI.08-11-04098.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lom B, Healy K E, Hockberger P E. J Neurosci Methods. 1993;50:385–397. doi: 10.1016/0165-0270(93)90044-r. [DOI] [PubMed] [Google Scholar]
- 17.Healy K E, Thomas C H, Rezania A, Kim J E, McKeown P J, Lom B, Hockberger P E. Biomaterials. 1996;17:95–108. doi: 10.1016/0142-9612(96)85764-4. [DOI] [PubMed] [Google Scholar]
- 18.Thomas C H, McFarland C D, Jenkins M L, Rezania A, Steele J G, Healy K E. J Biomed Mater Res. 1997;37:81–90. doi: 10.1002/(sici)1097-4636(199710)37:1<81::aid-jbm10>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 19.Folch A, Toner M. Annu Rev Biomed Eng. 2000;2:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 20.Bearinger J P, Castner D G, Golledge S L, Rezania A, Hubchak S, Healy K E. Langmuir. 1997;13:5175–5183. [Google Scholar]
- 21.Whitson S W, Whitson M A, Bowers D E, Falk M C. J Bone Min Res. 1992;7:727–741. doi: 10.1002/jbmr.5650070703. [DOI] [PubMed] [Google Scholar]
- 22.George A, Bannon L, Sabsay B, Dillon J W, Malone J, Veis A, Jenkins D, Gilbert D J, Copeland N G. J Cell Biochem. 1996;271:32869–32873. doi: 10.1074/jbc.271.51.32869. [DOI] [PubMed] [Google Scholar]
- 23.Wittelsberger S C, Kleene K, Penman S. Cell. 1981;24:859–866. doi: 10.1016/0092-8674(81)90111-2. [DOI] [PubMed] [Google Scholar]
- 24.Wada Y, Kataoka H, Yokose S, Ishizuya T, Miyazono K, Gao Y H, Shibasaki Y, Yamaguchi A. Bone. 1998;22:479–485. doi: 10.1016/s8756-3282(98)00039-8. [DOI] [PubMed] [Google Scholar]
- 25.Xiao G, Wang D, Benson D M, Karsenty G, Franceschi R T. J Biol Chem. 1998;273:32988–32994. doi: 10.1074/jbc.273.49.32988. [DOI] [PubMed] [Google Scholar]
- 26.Xiao G, Jiang D, Thomas P, Benson M D, Guan K, Karsenty G, Franceschi R. J Biol Chem. 2000;275:4453–4459. doi: 10.1074/jbc.275.6.4453. [DOI] [PubMed] [Google Scholar]
- 27.Berezney R, Mortillaro M J, Ma H, Wei X, Samarabandu J. Int Rev Cytol. 1995;162:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]