Abstract
Earlier studies of invasive breast tumors have shown that 60–80% are aneuploid and ≈80% exhibit amplified centrosomes. In this study, we investigated the relationship of centrosome amplification with aneuploidy, chromosomal instability, p53 mutation, and loss of differentiation in human breast tumors. Twenty invasive breast tumors and seven normal breast tissues were analyzed by fluorescence in situ hybridization with centromeric probes to chromosomes 3, 7, and 17. We analyzed these tumors for both aneuploidy and unstable karyotypes as determined by chromosomal instability. The results were then tested for correlation with three measures of centrosome amplification: centrosome size, centrosome number, and centrosome microtubule nucleation capacity. Centrosome size and centrosome number both showed a positive, significant, linear correlation with aneuploidy and chromosomal instability. Microtubule nucleation capacity showed no such correlation, but did correlate significantly with loss of tissue differentiation. Centrosome amplification was detected in in situ ductal carcinomas, suggesting that centrosome amplification is an early event in these lesions. Centrosome amplification and chromosomal instability occurred independently of p53 mutation, whereas p53 mutation was associated with a significant increase in centrosome microtubule nucleation capacity. Together, these results demonstrate that independent aspects of centrosome amplification correlate with chromosomal instability and loss of tissue differentiation and may be involved in tumor development and progression. These results further suggest that aspects of centrosome amplification may have clinical diagnostic and/or prognostic value and that the centrosome may be a potential target for cancer therapy.
Mitotic fidelity and cytoplasmic organization are both consequences of normal centrosome function. Defective centrosomes, exemplified by an excess number of centrioles and pericentriolar material, are characteristic of breast tumors and solid tumors in general (reviewed in refs. 1–3). These observations have implicated the centrosome in the origin of chromosomal abnormalities in the development of malignant tumors (4–8). In this study, we investigate the relationship between centrosome amplification and aneuploidy, chromosomal instability (CIN), p53 mutation, and loss of tissue differentiation in human breast tumors.
Chromosomal abnormalities have long been recognized as a distinguishing feature of cancer cells (4). Fluorescence in situ hybridization (FISH) with centromeric probes is a sensitive technique that can detect aneusomy as numeric alterations of specific chromosomes (9–12). FISH and comparative genomic hybridization studies have shown that aneusomy may be an early event in breast tumor development (13, 14). FISH data from simultaneous detection of two or more chromosome probes can be analyzed to determine whether tissues are diploid or aneuploid, whether they are potentially polyploid, whether one or more clonal cell populations are present, and whether the chromosome number is stable (13, 14). CIN was first defined in colorectal cancers as the percent of cells with nonmodal chromosome number (15). CIN is a measure of the flux in karyotype, as opposed to aneuploidy, which describes the condition of a nondiploid karyotype. CIN is the rate of change in chromosome number, whereas aneuploidy is the state of an altered chromosome number (15).
In addition to alterations in chromosome number, most solid tumors are also characterized by centrosome amplification (1). Centrosomes nucleate and organize the cytoplasmic and mitotic spindle microtubules (MTs) in interphase and mitotic cells, respectively. It has been hypothesized that centrosome amplification affects cell and tissue architecture by altering the microtubule (MT) cytoskeleton (6, 16). Because the centrosome is actively involved in proper chromosome segregation during mitosis, it also has been hypothesized that centrosome amplification drives tumor aneuploidy by increasing the frequency of abnormal mitoses that lead to chromosome missegregation (4, 6, 16–18). Although earlier studies have suggested that centrosome amplification is a downstream consequence of p53 nullizygosity, or loss or gain of function mutations (19–21), alternative pathways not involving p53 mutation have also been demonstrated (16–18).
Three analytical measures useful in assessing centrosome amplification in tumors include centrosome size and centrosome number, which are structural measures, and centrosome MT nucleation capacity, which is a measure of function. Most often, structural centrosome amplification is measured by immunomicroscopy of cells or tissues by using antibodies against the centrosome proteins centrin, pericentrin, or γ-tubulin, followed by quantitative or subjective measurement of centrosome size and/or number (5–8). Two basic approaches to measure MT nucleation capacity have been used depending on whether cultured cells or frozen tissues are used: MT regrowth in living cultured cells or in vitro MT nucleation and growth in fresh frozen tissues (6, 7). In this study, we investigated the correlation of these three features of centrosome amplification with aneuploidy, CIN, p53 mutation, and loss of differentiation in breast tumors. We found that centrosome size and number both correlate with aneuploidy and CIN, and that centrosome MT nucleation capacity correlates with loss of differentiation. MT nucleation was significantly greater in tumors with p53 mutations. However, although the presence of p53 mutation correlated with aneuploidy, it did not correlate with CIN. Furthermore, we demonstrated that structural centrosome amplification is present in ductal carcinoma in situ (DCIS). Together, these results support the hypothesis (4, 6) that centrosome amplification is an early event in tumorigenesis that drives both chromosomal instability and loss of differentiation through independent centrosome functions.
Methods
Tissues.
Human breast tissues were collected according to a protocol approved by the Mayo Clinic Internal Review Board. Portions of the tissues were snap-frozen in liquid nitrogen within 30 min of surgery and stored at −70°C until use. All tumors were graded according to the Nottingham grading system by a single pathologist (C.R.), using standard hematoxylin-and-eosin-stained sections from paraffin-embedded portions of the same tumor.
Centrosome Volume, Number, and Area.
Centrosome volume and number were determined as described by using confocal microscopy for volume reconstruction of centrosomes immunolabeled with a monoclonal antibody against centrin (6). Determinations in this study were based on average values for all cells in four fields of view including a minimum of 50 cells. Alternatively, the centrosome area was determined by confocal microscopy and image analysis of immunofluorescence signal by using a polyclonal antibody against pericentrin (Covance, Berkeley, CA) Similar results were obtained with antibodies against the centrosomal protein γ-tubulin (data not shown; see refs. 6 and 22). On each tissue section, the average signal from five fibroblast centrosomes was used to normalize the values of epithelial cell centrosomes from five fields of view. Centrosome area was determined for 5 normal tissues from reduction mammoplasties, 7 DCIS tumors, 15 lymph node negative invasive ductal carcinomas, and 14 lymph node positive invasive ductal carcinomas. A normalized size and number index for centrosome amplification was calculated as follows: [(tumor centrosome size/0.023 μm3) + (tumor centrosome number/1.55)]/2 = centrosome size and number index, where the average value for centrosome size in normal breast epithelial cells is 0.023 μm3 and 1.55 is the average number of centrosomes in normal breast epithelial cells. Correlative light and electron microscopy on selected tissues demonstrated that multiple anti-centrin-staining spots corresponded to supernumerary centrioles and/or excess pericentriolar material (22).
MT Nucleation.
The capacity of centrosomes to nucleate MTs was determined by using an in vitro assay on touch preparations from frozen tissues according to published methods (23). This functional assay reflects the in vitro ability of centrosomes to nucleate MTs under defined conditions where time, temperature, and tubulin concentration were maintained such that MT nucleation and growth occurred only in association with centrosomes and did not occur spontaneously. Normal and tumor preparations were assayed in parallel by using identical conditions. The number of MTs nucleated per cell was determined for 100 consecutively viewed cells. Cells with significant overlap of MT arrays were excluded from analysis, as were obviously damaged cells. A normalized MT index was calculated from these results by dividing the tumor MT number by 4.8 MT, the average number of MTs nucleated by normal breast epithelial cells.
FISH Analysis.
FISH probes to pericentromeric regions of chromosomes 3 (CEP3), 7 (CEP7), and 17 (CEP17) (Vysis, Downers Grove, IL) were hybridized to touch preparations of nuclei from frozen tissues according to published methods (9, 10). Probes were labeled with SpectrumOrange (CEP3), SpectrumGreen (CEP7), and SpectrumAqua (CEP17) for simultaneous analysis. DNA was counterstained with 4′,6-diamidino-2-phenylindole before mounting coverslips. For each tissue, 100 consecutive nuclei were scored for the number of signals for each of the three probes per nucleus with methods described (10).
Classification of tissue ploidy was a two-step process. In the first step, each of the 100 cells for a given tissue was classed as disomic if all three probes had two signals, as polysomic if two or more probes had more than two signals, as monosomic for a given probe, or otherwise as nondisomic. For the second step, the sum of the cells in each of the categories above was used to determine the tissue status as follows: (i) diploid if at least 50% of the cells were disomic, fewer than 15% were polysomic, and fewer than 50% were monosomic for the same chromosome, (ii) monosomic if 75% of the cells had the same chromosome loss and fewer than 15% were polysomic, or (iii) aneuploid if more than 15% of the cells were polysomic. Results from the seven reduction mammoplasties (assumed to be diploid) were used to calculate these cutoff values.
Cell clones were identified by evaluating the percentage of cells having identical probe signal patterns (24). A cell population with the same probe signal pattern was considered clonal if it represented at least 10% of the total cells. In tissues with high clonal heterogeneity, fewer than 10% of the cells had identical probe signal patterns. Tissues were considered to have low clonal heterogeneity if more than 20% of the cells were clonal, and one or two clones, each accounting for at least 10% of the total cells, could be identified.
The modal signal number of each chromosome was determined for each tissue. Bimodal values emerged in six tissues that had clones representing a significant percentage of the population. As described (15, 17), CIN was calculated for each chromosome as the percent of cells with nonmodal signal numbers. However, in this study, bimodal values were included in the calculation to minimize artificial inflation of CIN values. The average CIN values for the seven normal tissues plus and minus three standard deviations was used to identify tumors with unstable karyotypes.
p53 Mutation Analysis.
Denaturing high-performance liquid chromatography was used to screen tissues for p53 mutations according to published methods (25). DNA was extracted from frozen sections of 7 reduction mammoplasties and 20 invasive tumors (the same tissues that were analyzed by FISH). Six regions of genomic DNA, which included exons 4–9 and spanned the splice sites, were amplified in separate PCRs. Amplified DNA was separated under partial denaturing conditions by using a WAVE System denaturing high-performance liquid chromatography (Transgenomic, San Jose, CA), and the resulting curves were compared with curves for wild-type p53. DNA from curves that did not conform to the wild-type profile was sequenced to confirm and identify mutations.
Statistical Analysis.
Linear regression analysis with Kendall's τ test was used to determine that CIN for chromosomes 3, 7, and 17 each correlate with centrosome size and number (P < 0.05), but not with MT nucleation capacity. Significance of differences in centrosome size, number, and MT nucleation capacity between ploidy groups, and in MT nucleation capacity between the different grades of tumor, and in MT nucleation capacity between tumors with wild-type or mutant p53 was determined by using the Wilcox rank-sum test.
Results
FISH Analysis.
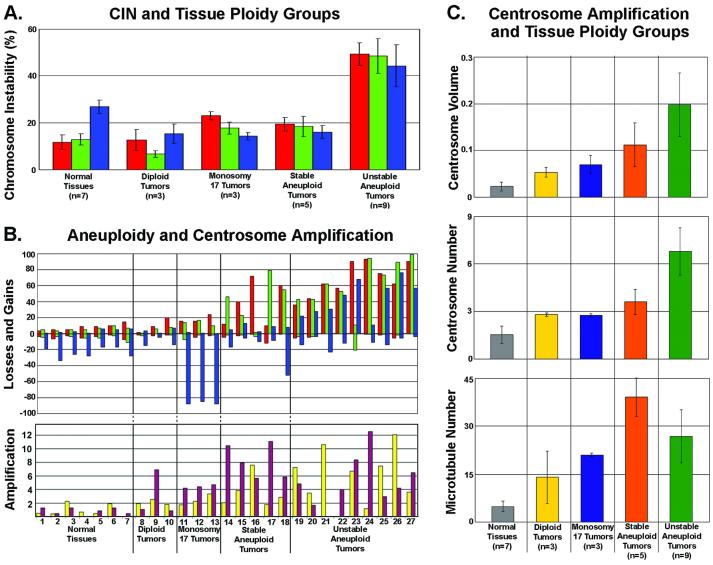
FISH data were analyzed to establish ploidy and to identify tumors with unstable karyotypes. Three of 20 invasive tumors were diploid, with chromosome gains and losses falling well within the range of normal values (Figs. 1 and 2 A and B). Three additional tumors were classified as monosomic for chromosome 17, because more than 75% of the cells had only one chromosome 17 signal with no other significant chromosome aberrations (Fig. 2B). None of the tumors were monosomic for chromosomes 3 or 7 (Fig. 2B). Fourteen of the 20 invasive tumors were aneuploid. This group of aneuploid tumors was subdivided into a group of 5 tumors with normal CIN values and another group of 9 tumors with high CIN values (Figs. 1 and 2 A and B and Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). The five aneuploid tissues with near-normal CIN values are examples of tumors with stable, but aneuploid, karyotypes. The nine tumors with high CIN are examples of tumors with karyotypes in a state of flux.
Figure 1.
FISH analysis. Flow chart of human breast tissue and tumor classification used in this study based on FISH analysis for chromosomes 3, 7, and 17. (Inset) FISH signals from two representative tumor nuclei: red signal, chromosome 3; green signal, chromosome 7; blue signal, chromosome 17.
Figure 2.
Analysis of chromosomal instability, aneuploidy, and centrosome amplification for the five tissue ploidy groups. (A) Plot of CIN (% cells showing chromosome number differing from the modal value for that particular chromosome) for the various tissue ploidy groups. (B) Plots of aneuploidy and centrosome amplification. (Upper) Plot of chromosome losses and gains for each sample in each tissue ploidy group. For both A and B: red bars, chromosome 3; green bars, chromosome 7; blue bars, chromosome 17. (Lower) Plot of centrosome amplification for each sample in each tissue ploidy group. Yellow bars, normalized centrosome size and number index; maroon bars, normalized microtubule nucleation index. (C) Plots of three different measures of centrosome amplification (centrosome volume in μm3, centrosome number, and microtubule nucleation) for each tissue ploidy group. Bars in A and C indicate standard deviation.
Centrosome Amplification.
In Fig. 2B, centrosome amplification (Lower) and chromosome gains and losses (Upper) for each individual tissue are shown. A clear positive association was apparent between increasing centrosome amplification and chromosomal gains and losses (Fig. 2B). Centrosome volume and centrosome number in normal tissues averaged 0.023 μm3 and 1.55, respectively (Table 1, Figs. 2 B and C and 3A). Two centrosome amplification indices for each tissue are presented in Fig. 2B Lower: the size and number index (yellow bars) reflects the combined normalized contributions of centrosome size and centrosome number, and the MT nucleation index (maroon bars) reflects the normalized MT nucleation capacity. Average centrosome volume of the stable aneuploid tumors was 0.112 μm3 and average volume of unstable aneuploid tumors was 0.198 μm3, more than 5- and 8-fold greater than normal values (0.023 μm3), respectively (Fig. 2C Upper). Immunofluorescence of centrosome size and number for each of the ploidy groups are presented in Fig. 3 A–E. Centrosome volume in the 14 aneuploid tumors spanned a wide range, indicating a high degree of variability among tumors. Monosomy 17 and diploid tumors had centrosome volumes intermediate between normal and aneuploid tumors (Figs. 2C Top and 3 A–C).
Figure 3.
Examples of immunofluorescence of centrosomes stained for centrin (green) in normal breast tissue (A) and centrosome amplification in breast tumors (B–E) and for microtubule nucleation (F–H) (green, anti-tubulin). (A) Region of a normal breast duct (lumen, center right) showing nuclei (red) located in the basal region of epithelial cells and pairs of centrioles (green, anti-centrin) located apically. (B–E) Examples of breast tumors showing the range of centrosome amplification in tumor tissue: (B) Diploid tumor. (C) Monosomy 17 tumor. (D) Stable aneuploid tumor. (E) Unstable aneuploid tumor. (F–H) Three examples of microtubule nucleation in touch preparations of breast tumor cells: (F) Diploid tumor. (G) Monosomy 17 tumor. (H) Stable aneuploid tumor.
Centrosome number was significantly greater in the unstable aneuploid tumors than in normal tissues (P < 0.02) (Figs. 2C Middle and 3 A–E). Unstable aneuploid tumors had an average centrosome number of 6.8 centrosomes per cell compared with 3.6 (stable aneuploid tumors), 2.7 (monosomy 17 tumors), 2.8 (diploid tumors), and 1.5 (normal tissues). Centrosome numbers in diploid and monosomy 17 tumors were not significantly different from each other, but were nearly 2-fold greater than in normal tissues.
Centrosomes of two of the three diploid tumors had MT nucleation capacity indistinguishable from normal tissues, whereas one diploid tumor nucleated significantly more MTs (Fig. 2B Lower). All other tumors had a higher MT nucleation capacity than did normal tissues (Figs. 2B Bottom and 3 F and G). Normal tissues showed significantly lower MT nucleation than monosomy 17 tumors (P < 0.02), stable aneuploid tumors (P < 0.005), and unstable aneuploid tumors (P < 0.01). On average, however, MT nucleation capacity was not significantly different between diploid tumors, monosomy 17 tumors, and unstable aneuploid tumors (Fig. 2C Bottom). Only stable aneuploid tumors had a MT nucleation capacity significantly higher than monosomy 17 tumors (P < 0.04) (Fig. 2C Bottom). Although stable aneuploid tumors nucleated more MTs than unstable aneuploid tumors, the difference was not statistically significant (Fig. 2C Bottom).
Centrosome Amplification in Preinvasive and Invasive Lesions.
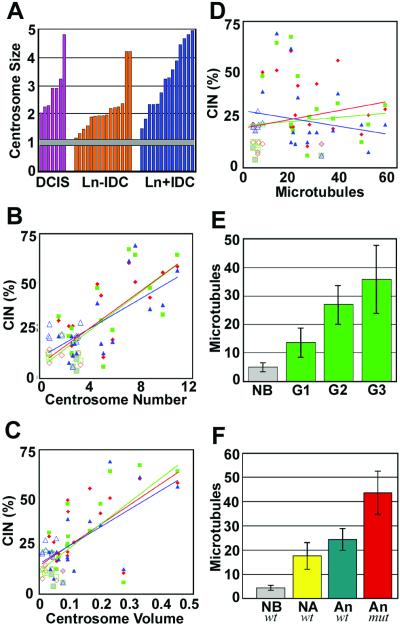
Centrosomes in normal breast epithelial tissues showed a consistent and narrow range of size with a standard deviation less than 7% of the average value, indicated by the horizontal bar in Fig. 4A. The centrosomes in noninvasive DCIS tumors displayed significant amplification in size, similar in range to that of both lymph negative and lymph node positive invasive tumors (Fig. 4A). FISH was not performed on these tissues.
Figure 4.
(A) Analysis of centrosome amplification, CIN, Nottingham grade, and p53 mutations. Bar graph of centrosome amplification normalized against fibroblast centrosomes for individual DCIS, lymph node negative invasive ductal carcinoma (Ln-IDC), and lymph node positive invasive ductal carcinoma (Ln+IDC). Gray horizontal bar indicates average for five normal breast epithelial tissues including standard error. (B–D) Plots of CIN and centrosome amplification. CIN (%) for each tissue and each chromosome is plotted against centrosome number (B), centrosome volume (μm3) (C), and microtubule nucleation (D). Open symbols are values for normal breast tissue, gray-filled symbols are values for diploid tumors, and closed symbols are values for aneuploid tumor tissue. Red symbols, chromosome 3; green symbols, chromosome 7; blue symbols, chromosome 17. (E) Bar graph of microtubule nucleation for normal breast tissue (NB) and tumors of Nottingham grades G1, G2, and G3. (F) Bar graph of microtubule nucleation and p53 status (wild type, wt; mutant, mut) for normal breast tissue (NB), nonaneuploid (diploid + monosomy 17) tumors (NA), and aneuploid tumors (An). Bars in E and F indicate standard deviation.
p53 Mutation Analysis.
The seven normal tissues from reduction mammoplasties had wild-type p53 (Table 1). Of 20 invasive tumors, we identified 5 with mutant p53. These mutations were all in the DNA-binding domain. All 5 tumors with p53 mutations were aneuploid, 2 had stable karyotypes (low CIN), and 3 had unstable karyotypes (high CIN). None of the diploid or monosomy 17 tumors had p53 mutations. Only tumors with sequence-confirmed p53 mutation were positive for p53 by immunohistochemistry, and only those positive by immunohistochemistry had sequence-confirmed mutations (data not shown).
Statistical Correlations.
When plotted against the CIN values for each of the three chromosomes, both centrosome number and centrosome volume of individual tumors showed significant, positive, linear correlations (P < 0.04) (Fig. 4 B and C). MT nucleation capacity did not correlate with CIN values for any of the three chromosomes (Fig. 4D). However, MT nucleation capacity did correlate with loss of tissue differentiation for Nottingham grades 2 and 3 compared with normal tissue (P < 0.01) as illustrated in Fig. 4E. Average MT nucleation was significantly greater in p53 mutant tumors than in all other tissue groups, including aneuploid p53 wild-type tumors, nondiploid p53 wild-type tumors, and normal tissues (Fig. 4A, Table 1).
Discussion
Studies in cell lines have implicated centrosome defects in abnormal mitoses leading to genomic instability in breast (17), pancreas (8), colon (26), and prostate (16) cancers. Here we demonstrate by using primary breast tumors that two aspects of centrosome amplification correlate independently with distinct features of breast cancer. Increased centrosome size and centrosome number correlate with CIN. We demonstrated that increased centrosome size is present in most in situ lesions, supporting the hypothesis that centrosome abnormalities drive chromosomal aberrations as an early event in DCIS. In addition, increased MT nucleation capacity of centrosomes correlated with loss of tissue differentiation. Loss of differentiation as indicated by high histologic grade is an indicator of poor prognosis (27), probably because of the increased metastatic potential of cells with altered cytoskeletons and adherent properties (28).
We also demonstrated that centrosome amplification and aneuploidy occur independently of p53 mutation. Of 14 aneuploid tumors, all of which had structurally amplified centrosomes, 9 had wild-type p53 and 5 had mutant p53. The frequency of CIN was the same in aneuploid tumors with wild-type (6 of 10) or mutant p53 (3 of 5), therefore mutant p53 did not correlate with CIN in aneuploid tumors, indicating that although aneuploidy can arise in the absence of p53 mutation, aneuploidy occurred only in the presence of centrosome amplification, in the tissues studied here, regardless of p53 mutation status. Therefore, molecular alterations other than p53 mutation may induce centrosome amplification with the potential to drive CIN.
The frequencies of aneusomy presented here are similar to published FISH data in breast tissues (10, 14). Studies have shown that the use of probes for just two chromosomes was sufficient to segregate diploid from aneuploid tumors (29, 30). However, an advantage to the use of more than two probes is that clonal populations can be identified with greater certainty (24). High clonal heterogeneity is the likely result of aneuploidy originating directly through chromosomal instability generating multiple unrelated clones, rather than through a linear model in which endoreduplication is followed by gains and losses of chromosomes (24, 31). Here, the simultaneous use of three probes allowed us to identify six tumors that contained two clones, each of which comprised greater than 25% of the tumor cell population. We adjusted the CIN values of these tumors with low clonal heterogeneity to reflect bimodal chromosome values, thus avoiding artificial inflation of CIN values and enabling us to identify accurately five aneuploid tumors with low clonal heterogeneity and stable karyotypes (i.e., low CIN) separately from nine aneuploid tumors with high clonal heterogeneity and unstable karyotypes (high CIN). These two groups of tumors are significantly different from each other with regard to their centrosome characteristics. The unstable aneuploid tumors (high CIN) had significantly larger centrosomes and more numerous centrosomes than did the stable aneuploid tumors (low CIN). The stable aneuploid tumors described here may have regained normal centrosome function by coalescence of supernumerary centrosomes (1, 22) and thereby acquired a growth advantage, because their normal bipolar mitotic spindles faithfully segregate the successful aneuploid karyotype. That the aneuploidy found in these stable aneuploid tumors originated through centrosome amplification, and not some other mechanism, is evidenced by their retention of increased MT nucleation capacity as an independent aspect of centrosome amplification. Likewise, in an experimental cell culture system, Chiba and coworkers (32) found that chromosomal instability and centrosome amplification underwent a “convergence” to stable aneuploidy and normal centrosome numbers with continued passage in culture. Therefore, we suggest that the stable aneuploid tumors identified here may have evolved through convergence in tumors originally having high clonal heterogeneity, and that both groups of aneuploid tumors were initiated by chromosomal instability caused by amplified centrosomes.
Our results demonstrate that increased MT nucleation capacity is a feature of centrosome amplification that is independent of centrosome number and centrosome size in breast tumors. Although MT nucleation capacity did not correlate with CIN or aneuploidy, it was significantly greater in p53 mutant aneuploid tumors than in those with wild-type p53. MT nucleation capacity did correlate with increased Nottingham grade, suggesting a relationship between defects in the MT cytoskeleton and loss of tissue differentiation. In single- and multicenter studies, Nottingham grade predicted clinical outcome, with increasing grade being associated with shorter disease-free survival and overall survival (33, 34). Likewise, centrosome amplification correlates with loss of differentiation as defined by increased Gleason score in prostate tumors (16). Together these observations implicate alterations in functional properties of centrosomes in maintaining the morphological changes associated with tumor development.
Although p53 mutation has been implicated as a cause for CIN in breast cancer (35), our results, and results from other studies (36, 37), demonstrate that aneuploidy and CIN occur more often in the absence of p53 mutation. However, they do not occur in the absence of structural centrosome amplification. Although mutant p53 is present in a significant portion of breast tumors, its occurrence is not a prerequisite to the development of aneuploidy. In the cases where mutant p53 is a factor, it is likely that p53 mutation affects centrosome number to promote abnormal mitoses (5, 20, 32). Our studies further demonstrate that p53 mutations correlate with a significant increase in the MT nucleation capacity of centrosomes.
In summary, our studies demonstrate that centrosome amplification is an early event in the development of breast cancer, and amplification of centrosome size and number correlate with CIN. Furthermore, centrosome amplification and CIN occur independently of p53 mutation in aneuploid tumors. Finally, MT nucleation capacity is an independent feature of centrosome amplification that correlates with loss of differentiation and is also increased significantly in tumors with p53 mutations. Because centrosome amplification precedes nuclear changes associated with aneuploidy in experimental systems (38) and is present in breast DCIS, it is possible that centrosome amplification drives CIN in breast tumor development. We suggest that centrosome amplification may increase metastatic potential through cytoskeletal alterations that affect tissue architecture in breast tumors. Although p53 mutations may exacerbate centrosome amplification, in this study they were not associated with an increased frequency of CIN. Centrosome amplification may be an indicator of CIN and unstable karyotypes in breast cancer that could be used to identify a subset of patients who would benefit from initial aggressive treatment. Finally, the centrosome presents a potential target for therapies against breast cancer through regulation of centrosome duplication and separation and through suppression of MT nucleation function.
Supplementary Material
Acknowledgments
We thank Drs. R. B. Jenkins, S. N. Thibodeau, N. J. Maihle, D. J. O'Kane, and D. Farrugia for critical review of the manuscript, Dr. S. J. Iturria for statistical analyses, and Mr. J. Tarara of the Mayo Optical Morphology Core Facility for his advice and technical assistance. This work was supported by DAMD 17-98-1-8122 from the Department of Defense Breast Cancer Research Program (to W.L.L.), by Grant CA72836 from the National Cancer Institute (to J.L.S.), and by the Mayo Foundation.
Abbreviations
- CIN
chromosomal instability
- DCIS
ductal carcinoma in situ
- FISH
fluorescence in situ hybridization
- MT
microtubule
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brinkley B R. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- 2.Lingle W L, Salisbury J L. Curr Top Dev Biol. 2000;49:313–329. doi: 10.1016/s0070-2153(99)49015-5. [DOI] [PubMed] [Google Scholar]
- 3.Salisbury J L. J Mammary Gland Biol Neoplasia. 2001;6:203–212. doi: 10.1023/a:1011312808421. [DOI] [PubMed] [Google Scholar]
- 4.Boveri T. Zur Frage der Entstehung Maligner Tumoren. Jena: Fischer; 1914. ; trans. Boveri, M. (1929) The Origin of Malignant Tumors (Williams and Wilkins, Baltimore) (English). [Google Scholar]
- 5.Carroll P E, Okuda M, Horn H F, Biddinger P, Stambrook P J, Gleich L L, Li Y Q, Tarapore P, Fukasawa K. Oncogene. 1999;18:1935–1944. doi: 10.1038/sj.onc.1202515. [DOI] [PubMed] [Google Scholar]
- 6.Lingle W L, Lutz W H, Ingle J N, Maihle N J, Salisbury J L. Proc Natl Acad Sci USA. 1998;95:2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pihan G A, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, Doxsey S J. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- 8.Sato N, Mizumoto K, Nakamura M, Maehara N, Minamishima Y A, Nishio S, Nagai E, Tanaka M. Cancer Genet Cytogenet. 2001;126:13–19. doi: 10.1016/s0165-4608(00)00384-8. [DOI] [PubMed] [Google Scholar]
- 9.Halling K C, King W, Sokolova I A, Meyer R G, Burkhardt H M, Halling A C, Cheville J C, Sebo T J, Ramakumar S, Stewart C S, et al. J Urol. 2000;164:1768–1775. [PubMed] [Google Scholar]
- 10.Persons D L, Robinson R A, Hsu P H, Seelig S A, Borell T J, Hartmann L C, Jenkins R B. Clin Cancer Res. 1996;2:883–888. [PubMed] [Google Scholar]
- 11.Pinkel D, Straume T, Gray J W. Proc Natl Acad Sci USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rennstam K, Baldetorp B, Kytola S, Tanner M, Isola J. Cancer Res. 2001;61:1214–1219. [PubMed] [Google Scholar]
- 13.Mendelin J, Grayson M, Wallis T, Visscher D W. Lab Invest. 1999;79:387–393. [PubMed] [Google Scholar]
- 14.Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi O P. Genes Chromosomes Cancer. 1998;21:177–184. [PubMed] [Google Scholar]
- 15.Lengauer C, Kinzler K, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 16.Pihan G A, Purohit A, Wallace J, Malhotra R, Liotta L, Doxsey S J. Cancer Res. 2001;61:2212–2219. [PubMed] [Google Scholar]
- 17.Miyoshi Y, Iwao K, Egawa C, Noguchi S. Int J Cancer. 2001;92:370–373. doi: 10.1002/ijc.1200. [DOI] [PubMed] [Google Scholar]
- 18.Zhou H, Kuang J, Zhong L, Kuo W L, Gray J W, Sahin A, Brinkley B R, Sen S. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 19.Wang X J, Greenhalgh D A, Jiang A, He D, Zhong L, Medina D, Brinkley B R, Roop D R. Oncogene. 1998;17:35–45. doi: 10.1038/sj.onc.1201890. [DOI] [PubMed] [Google Scholar]
- 20.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude G F. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 21.Weber R G, Bridger J M, Benner A, Weisenberger D, Ehemann V, Reifenberger G, Lichter P. Cytogenet Cell Genet. 1998;83:266–269. doi: 10.1159/000015168. [DOI] [PubMed] [Google Scholar]
- 22.Lingle W L, Salisbury J L. Am J Pathol. 1999;155:1941–1951. doi: 10.1016/S0002-9440(10)65513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingle W L, Salisbury J L. Methods Cell Biol. 2001;67:325–336. doi: 10.1016/s0091-679x(01)67022-5. [DOI] [PubMed] [Google Scholar]
- 24.Farabegoli F, Santini D, Ceccarelli C, Taffurelli M, Marrano D, Baldini N. Cytometry. 2001;46:50–56. doi: 10.1002/1097-0320(20010215)46:1<50::aid-cyto1037>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Smith D I, Rechtzigel K J, Thibodeau S N, James C D. Nucleic Acids Res. 1998;26:1396–1400. doi: 10.1093/nar/26.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghadimi B M, Sackett D L, Difilippantonio M J, Schrock E, Neumann T, Jauho A, Auer G, Ried T. Genes Chromosomes Cancer. 2000;27:183–190. [PMC free article] [PubMed] [Google Scholar]
- 27.Fitzgibbons P L, Page D L, Weaver D, Thor A D, Allred D C, Clark G M, Ruby S G, O'Malley F, Simpson J F, Connolly J L, et al. Arch Pathol Lab Med. 2000;124:966–978. doi: 10.5858/2000-124-0966-PFIBC. [DOI] [PubMed] [Google Scholar]
- 28.Engers R, Gabbert H E. J Cancer Res Clin Oncol. 2000;126:682–692. doi: 10.1007/s004320000148. [DOI] [PubMed] [Google Scholar]
- 29.Fiegl M, Kaufmann H, Zojer N, Schuster R, Wiener H, Mullauer L, Roka S, Huber H, Drach J. Hum Pathol. 2000;31:448–455. doi: 10.1053/hp.2000.6550. [DOI] [PubMed] [Google Scholar]
- 30.Takami S, Kawasome C, Kinoshita M, Koyama H, Noguchi S. Clin Chim Acta. 2001;308:127–131. doi: 10.1016/s0009-8981(01)00473-9. [DOI] [PubMed] [Google Scholar]
- 31.Kuukasjarvi T, Karhu R, Tanner M, Kahkonen M, Schaffer A, Nupponen N, Pennanen S, Kallioniemi A, Kallioniemi O P, Isola J. Cancer Res. 1997;57:1597–1604. [PubMed] [Google Scholar]
- 32.Chiba S, Okuda M, Mussman J G, Fukasawa K. Exp Cell Res. 2000;258:310–321. doi: 10.1006/excr.2000.4916. [DOI] [PubMed] [Google Scholar]
- 33.Elston C W, Ellis I O. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 34.Page D L, Gray R, Allred D C, Dressler L G, Hatfield A K, Martino S, Robert N J, Wood W C. Am J Clin Oncol. 2001;24:10–18. doi: 10.1097/00000421-200102000-00002. [DOI] [PubMed] [Google Scholar]
- 35.Sigurdsson S, Bodvarsdottir S K, Anamthawat-Jonsson K, Steinarsdottir M, Jonasson J G, Ogmundsdottir H M, Eyfjord J E. Cancer Genet Cytogenet. 2000;121:150–155. doi: 10.1016/s0165-4608(00)00260-0. [DOI] [PubMed] [Google Scholar]
- 36.Lavarino C, Corletto V, Mezzelani A, Della Torre G, Bartoli C, Riva C, Pierotti M A, Rilke F, Pilotti S. Br J Cancer. 1998;77:125–130. doi: 10.1038/bjc.1998.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauer T, Beraki K, Jebsen P W, Ormerod E, Naess O. APMIS. 1998;106:921–927. doi: 10.1111/j.1699-0463.1998.tb00240.x. [DOI] [PubMed] [Google Scholar]
- 38.Duensing S, Duensing A, Crum C P, Munger K. Cancer Res. 2001;61:2356–2360. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.