Abstract
Simian virus 40 large T antigen contains an amino terminal J domain that catalyzes T antigen-mediated viral DNA replication and cellular transformation. To dissect the role of the J domain in these processes, we exploited the genetic tools available only in the yeast Saccharomyces cerevisiae to isolate 14 loss-of-function point mutations in the T antigen J domain. This screen also identified mutations that, when engineered into simian virus 40, resulted in T antigen mutants that were defective for the ability to support viral growth, to transform mammalian cells in culture, to dissociate the p130–E2F4 transcription factor complex, and to stimulate ATP hydrolysis by hsc70, a hallmark of J domain-containing molecular chaperones. These data correlate the chaperone activity of the T antigen J domain with its roles in viral infection and cellular transformation and support a model by which the viral J domain recruits the cytoplasmic hsc70 molecular chaperone in the host to rearrange multiprotein complexes implicated in replication and transformation. More generally, this study presents the use of a yeast screen to identify loss-of-function mutations in a mammalian virus and can serve as a widely applicable method to uncover domain functions of mammalian proteins for which there are yeast homologues with selectable mutant phenotypes.
Hsp40–hsc70 molecular cochaperones modulate the conformations of polypeptide substrates to perform diverse functions including the folding of nascent polypeptides, the transport of proteins across organellar membranes, and the rearrangement of multiprotein complexes (1). Members of the hsp40 family are defined molecularly by the presence of an ≈70-aa motif called the J domain and biochemically by their ability to stimulate ATP hydrolysis by hsc70 partner chaperones. Hsc70s bind and release polypeptide substrates through the coordinated actions of their amino-terminal ATPase and carboxyl-terminal substrate-binding domains; transient interactions between ATP-bound hsc70 and polypeptide substrates are stabilized upon ATP hydrolysis (2–4). The J domain of hsp40 chaperones directly interacts with the ATPase domain of hsc70s and regulates the affinity of hsc70 for its substrates by stimulating ATP hydrolysis (5–7). Other domains in hsp40s may bind specific substrates and target them to hsc70 (8–11).
Structural studies on four hsp40 homologs define a common element in the J domain (12–15). J domains consist of four α-helices arranged such that the antiparallel second and third helices form a finger-like projection. An invariant HPD sequence located in the loop connecting the second and third helices is critical for the interaction with and stimulation of hsc70 (16, 17), but additional residues on the surface of helix II may also contact hsc70 (18).
The large tumor antigen of simian virus 40 (SV40) T antigen contains a J domain that is essential for most aspects of SV40 infection and contributes to the transformation of cultured rodent cells and the promotion of tumorigenesis in transgenic mice (19–23). Wild-type T antigen associates directly with hsc70 in vivo and in vitro (24, 25) and is capable of stimulating hsc70 ATPase activity in vitro (22, 26). In addition, the T antigen J domain can functionally replace the J domain of DnaJ in Escherichia coli (27) and the J domains from two human DnaJ homologs can substitute for the amino terminus of T antigen in promoting viral DNA replication (21) or enhancing Tst-1 transcription factor activity (28). Because T antigen inactivates normal cellular growth controls by altering the retinoblastoma (Rb) family of tumor suppressors (pRb, p107, and p130) and p53 (26, 29–31), we proposed that T antigen performs its diverse functions by recruiting cellular hsc70 to modulate the activities of these multiprotein complexes (32). However, validation of this “chaperone model” requires the correlation of in vivo T antigen-mediated phenotypes with biochemical measurements of T antigen chaperone activity. To accomplish this goal, we established a genetic screen in yeast from which T antigen mutants compromised for SV40 growth and the ability to promote cellular transformation were identified. Biochemical measurements of the purified wild-type and mutant T antigens indicated that these mutants were defective for the stimulation of hsc70 ATPase activity. This study also describes the random mutagenesis of a J domain toward the goal of identifying noncomplementing mutants.
Materials and Methods
Plasmid Construction and Genetic Screen.
The Saccharomyces cerevisiae strain ACY17b (MATα ade2–1 leu2–3 112, his3–11 15 trp1–1 ura3–1 can1–100 ydj1–2∷HIS3 LEU2∷ydj1–151; ref. 33) was manipulated by standard procedures (34). The chimeric T-Ydj1 fusion was created by sequential PCRs. In the first round, oligonucleotides TJ1A (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org, for oligonucleotide sequences) and TJ3B were used to amplify DNA encoding the T antigen J domain with an EcoRI site at the 5′ end and a short sequence complementary to the region downstream of the YDJ1 J domain at the 3′ end. In the second PCR step, DNA encoding residues 82–409 of YDJ1 from pAV6 (obtained from A. Caplan, Mount Sinai School of Medicine, New York) and a NotI site at the 3′ end was amplified with oligonucleotides TJ3A and TJ2B. In the final PCR, the products from the first two amplifications were fused, and the resulting fragment was digested with EcoRI and NotI and cloned into the EcoRI and NotI sites of pYes2 (Invitrogen) to create pT-Ydj1. Mutant T-Ydj1 constructs were generated similarly with dl1135 (35), P43L/K45N, or D44E/G47R (provided by Kathy Rundell, Northwestern University School of Medicine, Chicago) T antigen constructs as template DNA in the initial PCR. The mutations in the J domains in each of these constructs were confirmed by DNA sequence analysis.
To screen for T antigen J domain mutants, the J domain in pT-Ydj1 was mutagenized by modified error-prone PCR (36). Two sets of PCRs containing A or G at a final concentration of 20 μM and the other deoxynucleotides at 200 μM were performed with oligonucleotides SWF3 and SWF4 and TaqDNA polymerase (Roche Molecular Biochemicals). DNA from multiple reactions was purified from agarose gels and cotransformed with a gapped version of pT-Ydj1 into ACY17b. To generate the gapped pT-Ydj plasmid, a Bgl11 site was introduced just downstream of the T antigen J domain with the Quickchange Mutagenesis Kit (Stratagene) and oligonucleotides SWF1 and SWF2. The resulting plasmid was digested with EcoR1 and BglII, purified, and transformed into yeast. Cells containing plasmids repaired by homologous recombination between the mutagenized J domain fragment and the gapped vector (37) were selected by plating transformation mixtures onto synthetic complete medium lacking uracil. Transformants were subsequently screened for growth on galactose-containing medium at 25°C, 35°C, and 37°C. Of a total of 1,800 transformants screened, 115 candidates were selected. Expression of the T-Ydj1 mutants was subsequently examined by immunoblot analysis using mAb419 that recognizes the T antigen J domain (38). From this analysis, ≈50% of the mutant proteins were not expressed and were not examined further. Plasmids in the remaining temperature-sensitive candidates were rescued from yeast, amplified in bacteria, and reintroduced into ACY17b to confirm the loss-of-function phenotype, resulting in 41 mutants. DNA sequencing using the Amplicycle sequencing kit (Perkin–Elmer) identified 14 unique point mutations in the T antigen J domain.
Viral Assays.
Plaque and cellular transformation assays were performed as described (26). The Y34N, H42R, and K53R mutations were engineered into pSV40* (26) by using the Quickchange site-directed mutagenesis kit (Stratagene) and oligonucleotides SWF7 and SWF8; SWF9 and SWF10; SWF11 and SWF12, respectively.
Purification of T Antigen from Yeast.
To express T antigen in yeast, a 2.2-kb BglII–BamHI fragment containing an intronless version of large T antigen was removed from pSV40*-Xho (26) and cloned into the BamHI site in pYes2 (Invitrogen). The H42R and K53R mutations were site-directed into the pYes2-TAg plasmid by using the oligonucleotides and methods described above. The pYes2-TAg, pYes2-TagH42R, and pYes2-TagK53R plasmids were subsequently transformed into W3031b (W3031b MATα ade2–1 his3–11,15 leu2–3,112 ura3–1 trp1–1 can1–100), and 4 liters of cells was grown at 30°C in synthetic complete medium lacking uracil containing 2% raffinose to an OD at 600 nm of 0.8. The cultures were subsequently diluted into an equal volume of medium containing galactose at a final concentration of 2%. After 5 h at 30°C, the cells were harvested, converted to spheroplasts (39), and frozen overnight at −20°C. Spheroplasts were suspended in 40 ml of ice-cold lysis buffer (0.2 M LiCl/20 mM Tris, pH 8.0/1 mM EDTA, pH 8.0/1 mM DTT), supplemented with PMSF, pepstatin-A, and leupeptin at the manufacturer's recommended concentrations (Sigma), and lysed by agitation with glass beads six times with 1-min pulses on a Vortex mixer and with 1-min incubations on ice between each pulse. The resulting lysate was clarified by centrifugation 2 × 2,000 g for 10 min and 2 × 13,000 g for 10 min at 4°C. The final supernatant was loaded onto a 5-ml Q-Sepharose fast flow column (Amersham Pharmacia) that was pre-equilibrated in lysis buffer. The column was washed with 15 ml of lysis buffer containing NaCl at a final concentration of 250 mM instead of LiCl, and T antigen was eluted with 10 ml of the same buffer but containing 800 mM NaCl. This eluate was diluted with 2 vol of lysis buffer and T antigen was further purified by immunoaffinity chromatography with a protein G-419 antibody column as described (40). T antigen-containing fractions were identified by Bradford assay, confirmed by Immunoblot analysis using mAb419, and stored at −20°C. Wild type and each mutant T antigen were purified at least twice for biochemical studies. At initial time points, yeast-purified T antigen stimulated Hsc70 ATPase activity comparably to T antigen from insect cells (40), and more than 90% of the Hsc70-bound ATP was hydrolyzed after a 10-min incubation with either yeast or insect-purified T antigen (compare figure 4 to figure 7 in ref. 26).
Hsc70 ATPase and Gel-Shift Assays.
Single turnover ATPase experiments and E2F gel-shift assays were performed as described (26).
Results
The T Antigen J Domain Functionally Replaces the J Domain of YDJ1.
To test our chaperone model for T antigen function and to initiate a structure–function analysis of the T antigen J domain, we required a pool of unique J domain mutants. Because traditional methods for isolating mutations are labor intensive or generate compound mutations (19), we developed a genetic screen that exploited a functional chimeric gene in yeast to rapidly identify chaperone-defective point mutations in the T antigen J domain. In principle, our screen should be generally applicable to investigators who want to generate a large set of random mutations in a defined domain in a mammalian gene as long as mutation of a yeast homologue displays a tractable phenotype.
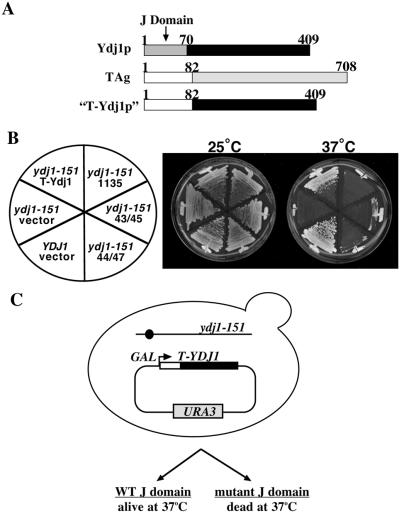
To construct this chimeric gene, we replaced the J domain from S. cerevisiae YDJ1 with that from T antigen to generate a “T-Ydj1” fusion (Fig. 1A). YDJ1 encodes an hsp40 chaperone with diverse cellular functions (33, 41–47). The first ≈70 aa of Ydj1p constitute a J domain (Fig. 1A) and purified Ydj1p stimulates the steady-state ATPase activity of a cytosolic yeast hsc70, Ssa1p, by ≈9-fold (48). Cells harboring the ydj1–151 allele grow at 25°C but do not survive at temperatures above 35°C and purified Ydj1–151p exhibits a reduced capacity to stimulate Ssa1p ATPase activity (33). When expressed in yeast with the inducible GAL1 promoter, T-Ydj1p suppressed the temperature-dependent lethality of ydj1–151 yeast at 37°C (Fig. 1B, T-Ydj1). However, mutations in or near the conserved HPD residues (double mutants P43L/K45N or D44E/G47R) or deletion of residues 17–27 in the J domain (dl1135) of T-Ydj1p abrogated the ability of this chimera to suppress the ydj1–151 thermosensitive phenotype (Fig. 1B).
Figure 1.
A T-Ydj1 chimera suppresses the conditional lethality of ydj1–151 yeast. (A) To create the T-Ydj1 chimera, the first 70 aa of Ydj1p were replaced with T antigen residues 1–82. (B) Wild-type (YDJ1) or mutant (ydj1–151) yeast transformed with different T-Ydj1 constructs or a control vector were streaked onto synthetic complete medium lacking uracil + galactose plates and incubated 3 days at the indicated temperatures. T-Ydj1 contains the wild-type T antigen J domain; 1135, 43/45, and 44/47 are T-Ydj1 chimeras that have a deletion of T antigen J domain residues 17–27, or the P43L and K45N or D44E and G47R mutations, respectively. (C) To isolate novel point mutations in the T antigen J domain, we screened for loss-of-function alleles of the T-Ydj1 chimera that failed to suppress the temperature-dependent lethality of ydj1–151 yeast at 35°C and 37°C (see Materials and Methods). WT, wild type.
Isolation of T Antigen J Domain Mutants.
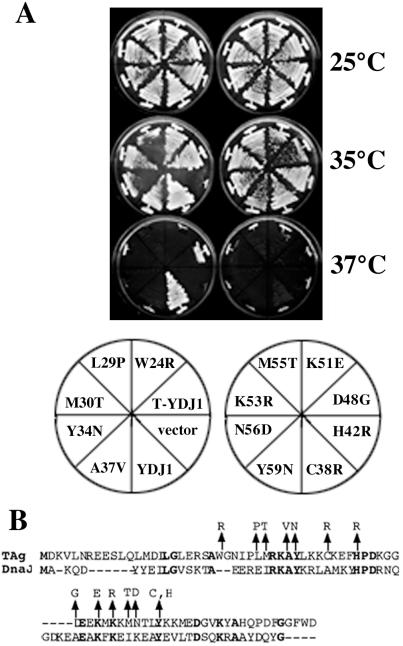
We next exploited the functionality of the T-Ydj1 chimera in yeast to isolate random mutations in the T antigen J domain by screening for loss-of-function alleles (Fig. 1C). Mutagenesis was targeted to the T antigen J domain by using error-prone PCR and gap repair by homologous recombination in yeast (see Materials and Methods), and the resulting mutant T-Ydj1p candidates were screened for their ability to suppress the temperature sensitivity of ydj1–151 yeast. The expression of T-Ydj1p in all candidates that failed to support growth at temperatures of 35°C and above was examined by immunoblot analysis to eliminate frameshift and nonsense codon-containing mutants from the pool. DNA sequence analysis of T-Ydj1 chimeras in the resulting candidates revealed 14 unique point mutations in the T antigen J domain that impaired the growth of the ydj1–151 strain at 37°C (Fig. 2).
Figure 2.
Mutations in the T antigen J domain abrogate T-Ydj1 suppression of ydj1–151 temperature sensitivity. (A) ydj1–151 yeast transformed with T-Ydj1 plasmids isolated in our genetic screen, a plasmid expressing wild-type Ydj1p (YDJ1), or a control vector (vector) were streaked onto synthetic complete medium lacking uracil + galactose plates and incubated for 3 days at the indicated temperatures. Note that the T-Ydj1 suppression of ydj1–151 temperature sensitivity was stronger at 35°C than 37°C and that the degree of suppression by different mutants at these temperatures was variable. (B) Alignment of T antigen residues 1–82 with the J domain of E. coli DnaJ is shown with identical residues depicted in bold text. The locations of the 14 point mutations are marked with arrows.
Y34N, H42R, and K53R Mutant T Antigens Are Defective in Vivo.
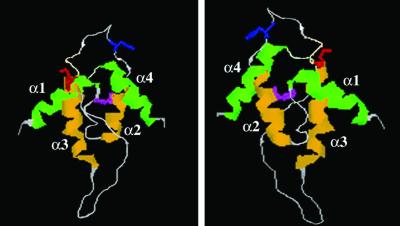
To validate our genetic screen and to assay the effects of these mutations on T antigen function, we engineered three of the mutations identified from the yeast screen, Y34N, H42R, and K53R, into the SV40 genome. Y34N and H42R were chosen because the tyrosine and histidine are conserved among all hsp40 J domains and are predicted to be buried within the J domain or to directly contact hsc70, respectively (Fig. 3; refs. 15 and 18). In contrast, the lysine at position 53 is solvent exposed and located opposite to the surface of the J domain that is thought to directly contact hsc70 (Fig. 3; refs. 15 and 18). The effects of these specific mutations in the J domain of T antigen have not been examined to date.
Figure 3.
The predicted positions of the Y34N, H42R, and K53R mutations in the T antigen J domain. Two views of the crystal structure of the T antigen J domain (15) are shown. Helices 2 and 3 are depicted in yellow, and helices 1 and 4 are shown in green. Y34 (magenta) is buried within the interhelical contacts that stabilize helices 2 and 3. H42 (blue) resides in the invariant HPD loop that connects helices 2 and 3 and contacts hsc70. K53 (red) is solvent-exposed and oriented opposite to the surface of the J domain that is thought to contact hsc70.
First, because the J domain is required for SV40 replication (19, 21, 26), we infected BSC40 cells with wild type and the three T antigen mutants. Wild-type SV40 produced visible plaques within 9 days of infection. The three mutant SV40 viruses, however, failed to produce plaques even 16 days after infection, at which time the plates infected with wild-type SV40 were completely clear (Table 1). Cotransfection of the three mutants with dl1007, an SV40 mutant with wild-type T antigen but lacking the viral coat protein genes (49), produced plaques 11 days after infection (data not shown). Thus, the failure of SV40 viruses containing the Y34N, H42R, or K53R mutations to produce plaques is caused strictly by defects in T antigen.
Table 1.
Large T antigen J domain mutants
| Plaque formation | Cellular transformation | Stable | |
|---|---|---|---|
| Wild type | + | + | + |
| Y34N | − | − | − |
| H42R | − | +/− | + |
| K53R | − | +/− | + |
| Mock | − | − | N/A |
Assays were performed at least twice for each construct. In cellular transformation assays, the number of foci/dish observed was 162 and 189 for wild-type T antigen; 60 and 71 for H42R T antigen; 92 and 77 for K53R T antigen. In plaque formation assays, no plaques were detectable 24 days after transfection with Y34N, H42R, and K53R mutant T antigen SV40 DNAs, at which time transfections with wild-type SV40 DNA cleared the plate. N/A, not available.
Next, to examine the effects of the three mutations in T antigen in a second in vivo assay, the ability of each mutant to transform a nonpermissive rodent cell line (REF52) was assessed by using a dense focus assay (50). Transformation was scored 4–6 weeks after transfection by the appearance of cells that had lost contact inhibition. We found that the H42R and K53R mutant T antigens transformed REF52 cells with ≈50% the efficiency of wild-type T antigen, whereas the Y34N mutant failed to transform these cells (Table 1). To determine whether this phenotype was related to the stability of the mutant T antigens, we examined the steady-state levels of these proteins in stably transfected cell lines. The H42R and K53R mutant T antigens accumulated to near wild-type levels; however, the Y34N mutant T antigen was barely detected (Table 1). Thus, the failure of the Y34N mutant T antigen to elicit cellular transformation likely results from poor expression and/or stability.
Mutations in the J Domain Compromise the Ability of T Antigen to Stimulate hsc70 ATP Hydrolysis.
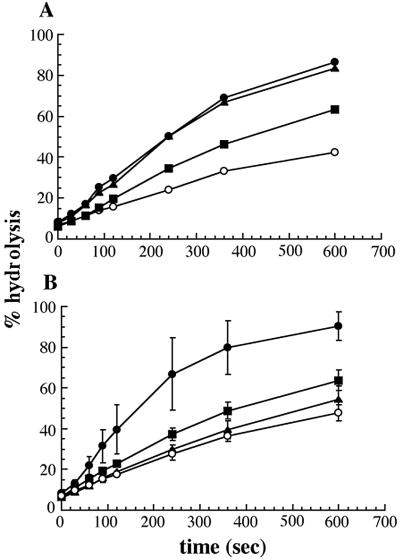
To determine whether the in vivo defects of the H42R and K53R mutant T antigens resulted from impaired chaperone activity, we purified wild-type T antigen and the H42R and K53R mutants and examined their ability to stimulate the ATPase activity of hsc70 in single turnover experiments, which specifically measure the hydrolysis of hsc70-bound ATP. When 1.5 μg of wild-type T antigen was added to the hsc70–ATP complex, the hydrolysis of ATP increased 3- to 4-fold; addition of 3.0 μg of T antigen failed to increase the rate of hydrolysis further (Fig. 4A). In this experiment, 1.5 μg of T antigen represents an approximately equimolar ratio of T antigen to hsc70. Using this amount of purified T antigen, we found that the H42R and K53R mutants increased hsc70-mediated ATP hydrolysis by 1.5- or 1.2-fold, respectively (Fig. 4B), suggesting that their inability to support SV40 viral replication and T antigen-mediated cellular transformation arises from compromised chaperone activity. We previously demonstrated that T antigen lacking the entire J domain stimulated hsc70 ATP hydrolysis by 0.4-fold (26). Thus, the H42R and K53R mutants are partially active in this assay.
Figure 4.
H42R and K53R mutant T antigens are defective for the stimulation of hsc70 ATP hydrolysis. (A) The concentration of purified wild-type T antigen required to maximally stimulate the hydrolysis of hsc70-bound ATP was assayed. hsc70, ○; hsc70 + 0.5 μg T antigen, ■; hsc70 + 1.5 μg T antigen, ▴; hsc70 + 3 μg T antigen, ●. (B) Single turnover measurements of hsc70, ○; hsc70 and 1.5 μg wild-type T antigen, ●; hsc70 and 1.5 μg H42R T antigen, ■; or hsc70 and 1.5 μg K53R T antigen, ▴. Data represent the means from experiments using at least two independent purifications of each protein, ± SD.
H42R and K53R Mutant T Antigens Are Defective in E2F Activation.
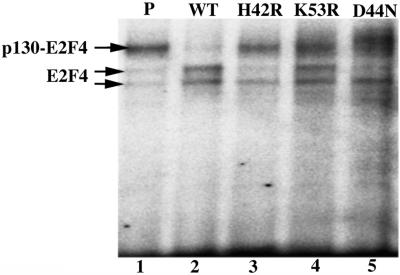
One mechanism by which T antigen disrupts cellular growth controls is by inactivating members of the retinoblastoma (Rb) protein family of tumor suppressors, which include pRb, p107, and p130 (29–32). When bound by Rb proteins, the E2F family of transcriptional activators is unable to transactivate the genes required for progression into S phase (51, 52). To measure the level of E2F-activated transcription in wild-type and H42R, K53R, and D44N T antigen-transfected cells, we first assayed the expression of a luciferase reporter gene fused to a consensus E2F promoter. D44N was included in this experiment because it was previously shown to transform REF52 cells with approximately the same efficiency as the H42R and K53R mutants described here (ref. 22; Table 1) and is defective for enhancing Hsc70 ATPase activity (26). We found that there was an ≈15-fold increase in the level of activity in T antigen-transformed cells compared with parental cells and that luciferase activity was slightly reduced in cells transfected with the three J domain mutants as compared with those containing wild-type T antigen (see Fig. 6, which is published as supporting information on the PNAS web site). More specifically, because we previously demonstrated that a p130–E2F4 DNA binding complexes could be disrupted by T antigen in REF52 cells (26), we examined E2F4–DNA complexes in lysates from stable, postconfluent REF52 cells by using a gel-shift assay. The parental REF52 cells contained E2F4 and p130–E2F4 DNA complexes but cells transfected with wild-type T antigen showed only E2F4 complexes (Fig. 5, lanes 1–2). In contrast, both H42R and K53R mutant T antigens appeared unable to dissociate fully the p130–E2F4 complex (Fig. 5, lanes 3–4). Similar results were observed with the D44N mutant T antigen (Fig. 5, lane 5), as was shown previously (30, 31). These data indicate that the J domain mutant T antigens are at least partially defective for T antigen-mediated activities that might promote entry into S phase.
Figure 5.
H42R and K53R mutant T antigens fail to disrupt the E2F4–p130 complexes in transfected REF52 cells. Lysates from parental REF52 cells (P; lane 1) and REF52 cells transfected with wild-type T antigen (WT; lane 2), H42R T antigen (lane 3), K53R T antigen (lane 4), or D44N T antigen (lane 5) were incubated with a radiolabeled E2F consensus binding site probe and subjected to gel-shift analysis. The specificity, migration, and identities of the p130–E2F4- and E2F4-containing bands in this assay have been described (26).
Discussion
We describe here a rapid genetic screen in yeast to isolate chaperone-defective mutations in the J domain of SV40 large T antigen. By analyzing the in vivo and in vitro phenotypes associated with the corresponding mutants, we correlated the requirement for the J domain of T antigen to stimulate the ATPase activity of hsc70 with its capacity to support viral infection and elicit maximal levels of cellular transformation in cultured cells. The results from this screen also support the efficacy of using a genetic attack in a simple eukaryote to discover fundamental principles governing the intrinsic activities of a mammalian virus.
The strongest in vivo defect observed for the three T antigen J domain mutants we constructed was for viral replication (Table 1). Mutations in the T antigen J domain have been shown to affect the replication of SV40 DNA (19, 21, 22) and the assembly of SV40 virions (20). A role for hsp40–hsc70 molecular chaperones in DNA replication is not unique to SV40 (53); binding of the human papillomarvirus-11 E1 protein to its origin of DNA replication is enhanced by cellular hsc70 and hsp40, probably by facilitating the formation of E1 dihexamers (54). In addition, replication of bacteriophage λ DNA in E. coli requires cellular DnaJ and DnaK (hsc70) to activate preinitiation complexes on the viral origin (reviewed in ref. 55).
More intriguing is the fact that the two stable mutants, H42R and K53R, were only partially defective for T antigen-mediated cellular transformation. One mechanism by which large T antigen promotes cell cycle progression is to inactivate the growth-inhibitory functions of the Rb protein family of tumor suppressors (32). We and others have demonstrated that the J domains of SV40 and polyomavirus large T antigens are required for Rb protein inactivation but do not affect the association of these tumor suppressors with T antigen (21, 22, 30, 56, 57), and a functional J domain is necessary to free E2F4 from Rb protein family members (this work and refs. 26 and 31). Our work builds on these studies by demonstrating that mutants unable to interact productively with hsc70 are compromised in an assay for E2F activity, which might provide an explanation for their reduced ability to transform REF52 cells. Another possibility that we favor is that other regions of T antigen may compensate for mutations in the J domain. This hypothesis is supported by several observations. First, the D44N mutation in the context of full-length T antigen reduces the transformation efficiency of REF52 cells by approximately 50%, but in the context of TN136 (containing only the first 136 aa of T antigen) the mutation completely abolishes the ability of this construct to transform C3H10T1/2 cells (22). Second, whereas full-length T antigen forms a stable, ATP-dependent association with hsc70, TN136 interacts only transiently with hsc70 in vitro (25). However, TN136 is capable of fully stimulating the ATPase activity of hsc70 in vitro, suggesting that a transient interaction is sufficient for J domain function (22). Finally, there is weak homology in the C-terminal region of T antigen (residues 501–520) to GrpE (25), a bacterial nucleotide exchange factor that binds to and modulates the activity of the bacterial hsc70 homolog, DnaK (58). Combined with the fact that p53 and Rb protein associate with hsc70 (59, 60), our data suggest that other domains in T antigen and/or cellular factors may compensate for the chaperone defects of these J domain T antigen mutants to support cellular transformation.
The spectrum of mutations identified in this genetic screen reveals several important features of the T antigen J domain. First, as expected, our screen uncovered mutations in helices 2 and 3 and in the HPD motif of the J domain (Fig. 2B), regions that interact directly with hsc70 or that may play a role in stabilizing this domain (refs. 16 and 18; reviewed in ref. 61; also see below). In contrast, we did not isolate T-Ydj1 plasmids with single-point mutations in helix 1, although mutations in this region combined with mutations in helices 2 or 3 were identified in some constructs (data not shown). Helix 1 is ≈2-fold longer in T antigen than in other J domain-containing proteins (14, 15, 62) and is implicated in viral DNA replication (57); thus, mutations in this domain are unlikely to affect Ydj1-specific activities in yeast. Similarly, mutations in helix 4 were not uncovered. Recent structural analysis of the T antigen J domain reveals an extended loop 3 (residues 68–90) connecting the third and forth helices, resulting in a longer J domain (residues 1–102) than that predicted by sequence comparisons (15); thus, part of loop 3 and all of helix 4 are absent in the T-Ydj1 chimera.
Second, our analysis of the K53R mutation demonstrates that a solvent-exposed residue that does not form stabilizing interhelix contacts and is oriented opposite from the hsc70-interacting face of helix 2 (refs. 14 and 15; see Fig. 3) is important for chaperone function. Either this residue contacts hsc70 that is wrapped around the J domain and is unperturbed by this interaction (18), or this face of the J domain contacts other cellular factors or regions of T antigen to support viral activities. Further analysis of this face of the T antigen J domain by site-directed mutagenesis will serve to test these hypotheses.
Finally, our data provide biochemical evidence to support previous studies implicating the requirement for a functional J domain in viral replication and cellular transformation (21, 22. 57). Whereas the failure of T antigen with mutations in conserved J domain residues to stably associate with hsc70 has been well documented (21, 25, 26, 56), we recently reported that hsc70 association is insufficient for J domain function (26). Instead, proficient J domain-mediated stimulation of hsc70 ATPase activity is a more accurate assessment of chaperone function with respect to in vivo phenotypes. This point is particularly salient as several proteins besides hsc70 have been shown to associate with the T antigen J domain (28, 63); thus, in the absence of measurements of J domain-stimulated hsc70 ATPase activity, one cannot ascertain the basis of the molecular defect(s) that lead to J domain mutant phenotypes in vivo. Therefore, one future research challenge is to isolate and biochemically characterize additional mutations in this and other domains of T antigen by using genetic screens in yeast and to identify genetic suppressors of characterized mutants in an attempt to isolate additional T antigen-interacting factors.
Supplementary Material
Acknowledgments
We thank Yunje Cho for providing the coordinates of the T antigen J domain crystal structure, Timothy D. Kierstead for construction of the T-Ydj1 expression plasmid, and Christopher S. Sullivan, Paul Cantalupo, James D. Tremblay, Sean Hasso, and other members of the Brodsky and Pipas laboratories for technical assistance. We thank Thomas Harper for assistance with the figures. This work was supported by a grant from the American Cancer Society (RPG-99–267-01-MBC) to J.L.B and by Grant CA40586 to J.M.P from the National Institutes of Health. S.W.F. was supported by a National Research Service Award (1 F32 CA 83270–01) from the National Institutes of Health and an award from the University of Pittsburgh Cancer Institute.
Abbreviations
- SV40
simian virus 40
- Rb
retinoblastoma
References
- 1.Hartl F U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 2.Schmid D, Baici A, Gehring H, Christen P. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 3.Greene L E, Zinner R, Naficy S, Eisenberg E. J Biol Chem. 1995;270:2967–2973. doi: 10.1074/jbc.270.7.2967. [DOI] [PubMed] [Google Scholar]
- 4.McCarty J S, Buchberger A, Reinstein J, Bukau B. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 5.Gässler C S, Buchberger A, Laufen T, Mayer M P, Schröder H, Valencia A, Bukau B. Proc Natl Acad Sci USA. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suh W C, Burkholder W F, Lu C Z, Zhao W, Gottesman M E, Gross C A. Proc Natl Acad Sci USA. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis J E, Voisine C, Craig E A. Proc Natl Acad Sci USA. 1999;96:9269–9276. doi: 10.1073/pnas.96.16.9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickner S, Hoskins J, McKenny K. Nature (London) 1991;350:165–167. doi: 10.1038/350165a0. [DOI] [PubMed] [Google Scholar]
- 9.Langer T, Lu C, Echols H, Flanagan J, Hayer M K, Hartl F U. Nature (London) 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 10.Szabo A, Korszun R, Hartl F U, Flanagan J. EMBO J. 1996;15:408–407. [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson J L, Craig E A. J Cell Biol. 2001;152:851–856. doi: 10.1083/jcb.152.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellecchia M, Szyperski T, Wall D, Georgeopolous C, Wüthrich K. J Mol Biol. 1996;260:236–250. doi: 10.1006/jmbi.1996.0395. [DOI] [PubMed] [Google Scholar]
- 13.Qian Y Q, Patel D, Hartl F U, McColl D J. J Mol Biol. 1996;260:224–235. doi: 10.1006/jmbi.1996.0394. [DOI] [PubMed] [Google Scholar]
- 14.Berjanskii M V, Riley M I, Xie A, Semenchenko V, Folk W R, Van Doren S R. J Biol Chem. 2000;275:36094–36103. doi: 10.1074/jbc.M006572200. [DOI] [PubMed] [Google Scholar]
- 15.Kim H-Y, Ahn B-Y, Cho Y. EMBO J. 2001;20:295–304. doi: 10.1093/emboj/20.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai J, Douglas M G. J Biol Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 17.Wall D, Zylic M, Georgopolous C. J Biol Chem. 1994;269:5446–5451. [PubMed] [Google Scholar]
- 18.Greene M K, Maskos K, Landry S J. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peden K W C, Pipas J M. Virus Genes. 1992;6:107–118. doi: 10.1007/BF01703060. [DOI] [PubMed] [Google Scholar]
- 20.Spence S L, Pipas J M. Virology. 1994;204:200–209. doi: 10.1006/viro.1994.1524. [DOI] [PubMed] [Google Scholar]
- 21.Campbell K S, Mullane K P, Aksoy I A, Stubdal H, Zalvide J, Pipas J M, Silver P A, Roberts T M, Schaffhausen B S, DeCaprio J A. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- 22.Srinivasan A, McClellan A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky J L, Pipas J M. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slinskey A, Barnes D, Pipas J M. J Virol. 1999;73:6791–6799. doi: 10.1128/jvi.73.8.6791-6799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawai E T, Butel J S. J Virol. 1989;63:3961–3973. doi: 10.1128/jvi.63.9.3961-3973.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan C S, Gilbert S P, Pipas J M. J Virol. 2001;75:1601–1610. doi: 10.1128/JVI.75.4.1601-1610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan C S, Tremblay J D, Fewell S W, Lewis J A, Brodsky J L, Pipas J M. Mol Cell Biol. 2000;20:5749–5757. doi: 10.1128/mcb.20.15.5749-5757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelley W L, Georgopoulos C. Proc Natl Acad Sci USA. 1997;94:3679–3684. doi: 10.1073/pnas.94.8.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sock E, Enderich J, Wegner M. Mol Cell Biol. 1999;19:2455–2464. doi: 10.1128/mcb.19.4.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J A. Mol Cell Biol. 1997;9:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zalvide J, Stubdal H, DeCaprio J A. Mol Cell Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan C S, Cantalupo P, Pipas J M. Mol Cell Biol. 2000;20:6233–6243. doi: 10.1128/mcb.20.17.6233-6243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brodsky J L, Pipas J M. J Virol. 1998;72:5329–5334. doi: 10.1128/jvi.72.7.5329-5334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caplan A J, Cyr D M, Douglas M G. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- 34.Rose M D, Winston F, Heiter P. Methods in Yeast Genetics: A Lab Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 35.Collins B S, Pipas J M. J Biol Chem. 1995;270:15377–15384. doi: 10.1074/jbc.270.25.15377. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Y, Zhang X, Ebright R H. Nucleic Acids Res. 1991;19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma H, Kunes S, Schatz P J, Botstein D. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 38.Linder K, Mole S E, Lane D P, Kenny M K. Intervirology. 1998;41:10–16. doi: 10.1159/000024910. [DOI] [PubMed] [Google Scholar]
- 39.Deshaies R J, Schekman R. J Cell Biol. 1989;109:2653–2664. doi: 10.1083/jcb.109.6.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cantalupo P, Sáenz-Robles M T, Pipas J M. Methods Enzymol. 1999;306:297–307. doi: 10.1016/s0076-6879(99)06019-x. [DOI] [PubMed] [Google Scholar]
- 41.Atencio D P, Yaffe M P. Mol Cell Biol. 1992;12:283–291. doi: 10.1128/mcb.12.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caplan A J, Douglas M G. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura Y, Yahara I, Lindquist S. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 44.Dey B, Caplan A J, Boschelli F. Mol Biol Cell. 1996;7:91–100. doi: 10.1091/mbc.7.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee D H, Sherman M Y, Goldberg A L. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brodsky J L, Lawrence J G, Caplan A J. Biochemistry. 1998;37:18045–18055. doi: 10.1021/bi980900g. [DOI] [PubMed] [Google Scholar]
- 47.Oka M, Nakai M, Endo T, Lim C R, Kimata Y, Kohno K. J Biol Chem. 1998;45:29727–29737. doi: 10.1074/jbc.273.45.29727. [DOI] [PubMed] [Google Scholar]
- 48.Cyr D M, Lu X, Douglas M G. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- 49.Lai C J, Nathans D. J Mol Biol. 1974;89:179–193. doi: 10.1016/0022-2836(74)90169-7. [DOI] [PubMed] [Google Scholar]
- 50.Peden K W C, Srinivasan A, Farber J M, Pipas J M. Virology. 1989;168:13–21. doi: 10.1016/0042-6822(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 51.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 52.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan C S, Pipas J M. Virology. 2001;287:1–8. doi: 10.1006/viro.2001.1038. [DOI] [PubMed] [Google Scholar]
- 54.Lui J-S, Kuo S-R, Makhov A M, Cyr D M, Griffith J D, Broker T R, Chow L T. J Biol Chem. 1998;273:3074–3072. doi: 10.1074/jbc.273.46.30704. [DOI] [PubMed] [Google Scholar]
- 55.Wawrzynow A, Zylicz M. In: Guidebook to Molecular Chaperones and Protein Folding Catalysts. Gething M-J, editor. New York: Oxford Univ. Press; 1997. pp. 481–488. [Google Scholar]
- 56.Sheng Q, Denis D, Ratnofsky M, Roberts T M, DeCaprio J A, Schaffhausen B. J Virol. 1997;71:9410–9416. doi: 10.1128/jvi.71.12.9410-9416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Soderbärg K, Houshmand H, You Z-Y, Magnusson G. J Virol. 2001;75:2253–2261. doi: 10.1128/JVI.75.5.2253-2261.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liberek K, Marzalek J, Ang D, Georgopoulos C, Zylicz M. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue A, Torigoe T, Sogahata K, Kamiguchi K, Takahashi S, Sawada Y, Saijo M, Taya Y, Ishii S, Sato N, Kikuchi K. J Biol Chem. 1995;270:22571–22576. doi: 10.1074/jbc.270.38.22571. [DOI] [PubMed] [Google Scholar]
- 60.Fourie A M, Hupp T R, Lane D P, Sang B C, Barbosa M S, Sambrook J F, Gething M J. J Biol Chem. 1997;272:19471–19479. doi: 10.1074/jbc.272.31.19471. [DOI] [PubMed] [Google Scholar]
- 61.Kelley W L. Trends Biol Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 62.Kelley W L, Landry S J. Trends Biochem Sci. 1994;19:227–278. doi: 10.1016/0968-0004(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 63.Tsai S-C, Pasumarthi K B S, Pajak L, Franklin M, Patton B, Wang H, Henzel W J, Stults J T, Field L J. J Biol Chem. 2000;275:3239–3246. doi: 10.1074/jbc.275.5.3239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.