Abstract
Cell-surface microvilli play a central role in adhesion, fusion, and signaling processes. Some adhesion and signaling receptors segregate on microvilli but the determinants of this localization remain mostly unknown. In this study, we considered CD4, a receptor involved in immune response and HIV infection, and p56Lck, a CD4-associated tyrosine kinase. Analysis of CD4 trafficking reveals that p56Lck binds tightly to CD4 independently of its activation state and inhibits CD4 internalization. Electron microscopy analysis established that p56Lck mediates CD4 association with microvilli whereas biochemical data indicate that p56Lck expression renders CD4 insoluble by the nonionic detergent Triton X-100. In addition, cytoskeleton-disrupting agent increased CD4 solubility, suggesting the involvement of cytoskeletal elements in CD4 anchoring to microvilli. This concept was supported further by the observation that the lateral mobility of CD4 within the plasma membrane was decreased in cells expressing p56Lck. Finally, isolation of detergent-resistant membranes revealed that the complex CD4-p56Lck is enriched within these domains as opposed to conditions in which CD4 does not interact with p56Lck. In conclusion, our results show that p56Lck targets CD4 to specialized lipid microdomains preferentially localized on microvilli. This localization, which prevents CD4 internalization, might facilitate CD4-mediated adhesion processes and could correspond to the signaling site of the receptor.
CD4 is a 55-kDa glycoprotein expressed at the surface of various hematopoietic cells (1). In T helper lymphocytes, CD4 plays a crucial role during antigenic stimulation by MHC class II-bearing cells. CD4 has a dual function in this process. First, it acts as an adhesion molecule that binds to nonpolymorphic regions of MHC class II. Second, CD4 acts as a signal transduction receptor by triggering the activation of the CD4-associated tyrosine kinase p56Lck, which modulates, in turn, signaling through the TCR (2). Whereas the physiological role of CD4 remains mostly unknown in p56Lck-negative cells (3), a pathological role for CD4 is well documented in all CD4-positive cells, where CD4 acts as part of the receptor complex used by HIV to infect its target cell (4).
The p56Lck kinase is a member of the Src family of nonreceptor tyrosine kinases expressed primarily in thymocytes and T lymphocytes. This kinase is associated with the cytosolic side of the plasma membrane and interacts specifically with CD4 through noncovalent bonds coordinated by a Zn2+ ion (5, 6). Although both CD4 and p56Lck possess the necessary determinants for their sorting, they associate early in the secretory pathway and reach the plasma membrane together (7).
A tight regulation of CD4 surface expression is crucial to ensure a correct immune function or efficient HIV infection (8, 9). Endocytic processes play a primordial role in the control of CD4 surface expression, and p56Lck is a key partner in these events. Indeed, p56Lck inhibits CD4 internalization by preventing CD4 incorporation into clathrin-coated pits, whereas in p56Lck-negative cells, CD4 is internalized and recycled to the surface efficiently (10). However, the exact mechanism by which p56Lck prevents CD4 recruitment in endocytic structures is unknown. One hypothesis is that the CD4-p56Lck complex behaves like some tyrosine kinase receptors (i.e., insulin/EGF receptors), which, in their inactivated state, are anchored to microvilli and therefore are kept away from the internalization gates. Endocytosis of these receptors only occurs when they are activated by their ligand, which leads to receptors' translocation to domains in which endocytosis occurs (11).
Consequently, in the present study, we examined the surface localization of CD4 in cells expressing or not expressing p56Lck as well as the role of p56Lck activation in CD4 trafficking. Our results indicate that p56Lck targets CD4 within particular microdomains of the plasma membrane associated with microvilli and that CD4 internalization is independent of the p56Lck activation state.
Materials and Methods
Reagents and Antibodies.
[γ-32P]ATP was purchased from Amersham Pharmacia, and [3H]palmitic acid was purchased from NEN. Other chemicals were of analytical grade and obtained from Fluka or Sigma. Polyclonal anti-CD4 antibody used for Western analysis was provided by the National Institutes of Health AIDS Research and Reference Reagent Program (Rockville, MD). R-phycoerythrin- and FITC-conjugated mAbs to CD4 were purchased from Dako; RPA-T4 was purchased from PharMingen; Leu-3a, from Roche Molecular Biochemicals; OKT4, from Ortho Diagnostics; mAbs to p56Lck were purchased from Santa Cruz Biotechnology; mAbs to CD71, from Zymed, and polyclonal antibodies to AlkP, from Rockland (Gilbertsville, PA).
Cell Culture, Plasmid Constructs, and Transfection.
The promyelocytic HL60 and CEM T cell lines were cultured in RPMI medium 1640 supplemented with 10% FCS (GIBCO). 293T cells were grown in DMEM (GIBCO) supplemented with 10% (vol/vol) FCS. The p56Lck alleles used in these experiments were described (12, 13). CD4, p56Lck, p56LckF505, and p56LckA273 were expressed from the cytomegalovirus immediate early promoter, in the pCMX plasmid vector (14). Transfections of 293T cells were performed by using the calcium phosphate method (15).
Internalization Assays.
Internalization was assayed by using the acid-wash technique as described (16). Briefly, cells were incubated for 2 h at 4°C with 125I-anti-CD4 (RPA-T4), washed, and shifted to 37°C to allow endocytosis. A percentage of 125I-anti-CD4 internalization was expressed as the ratio of acid-wash-resistant radioactivity to total radioactivity associated to cells at neutral pH. Alternatively, a FACS-based assay measuring the internalization of R-phycoerythrin (RPE)-conjugated anti-CD4 antibodies was used. Briefly, cells were incubated for 1 h at 4°C with RPE-conjugated anti-CD4 antibodies and washed, and internalization of the immune complex was analyzed by flow cytometry as described (17).
Immunoprecipitations.
Cells were lysed in buffer A [50 mM Tris⋅HCl, pH 7.4/50 mM NaCl/10 mM MgCl2/1 mM EGTA/2 mM VO4/2.5% glycerol/1% Triton X-100 (TX100) and mixture of protease inhibitors] for 20 min on ice. Appropriate antibodies were added to 500–800 μg of precleared lysates, and immune complexes were pulled down by using protein A-Sepharose. Immunoprecipitates were washed, resolved by SDS/PAGE, and analyzed by Western blotting by using the enhanced chemiluminescence kit from Amersham Pharmacia.
Kinase Assay.
293T cells were lysed 48 h after CD4 and p56Lck cotransfection in buffer B (50 mM Tris⋅HCl, pH 7.5/25 mM KCl/5 mM MgCl2/1 mM EGTA/1% TX100 and a mixture of phosphatase/protease inhibitors) for 20 min at 4°C. p56Lck was immunoprecipitated, and immune complexes were washed three times with buffer A and once in kinase buffer (20 mM Mops/5 mM MgCl2/5 mM MnCl2 and phosphatase inhibitors). p56Lck coupled to Sepharose beads was resuspended in 23 μl of kinase buffer containing 4 μCi (1 Ci = 37 GBq) of [γ-32P]ATP (5,000 Ci/mmol) and 5 μg of acid-denatured enolase as exogenous substrate and incubated for 20 min at 30°C. Products of the kinase reaction were resolved by SDS/PAGE and analyzed by autoradiography.
Electron Microscopy (EM) Analyses.
Surface CD4 localization by autoradiography was performed as described (16). Briefly, cells were incubated for 2 h at 4°C with 125I-anti-CD4 (RPA-T4), washed twice, and transferred at 37°C for the indicated times. Cells then were fixed, dehydrated, and processed for EM autoradiography. Surface CD4 localization by immunogold complexes was performed as described (16). Briefly, cells were incubated for 90 min at 4°C with Leu-3a anti-CD4, washed, and incubated a second time with an anti-mouse IgG coupled to 10-nm colloidal gold particles for 90 min at 4°C. Cells then were washed, fixed, dehydrated, and processed for EM analyses. The ratio of villous vs. nonvillous plasma membrane was determined as described (16).
FACS Analysis.
CD4 solubility in TX100 was assessed as described (18) with minor modifications. Briefly, cells were labeled with FITC-conjugated anti-CD4 at 4°C in buffer C (10 mM Tris⋅HCl, pH 8.0/150 mM NaCl/2 mM MgCl2/2 mM EGTA/1% BSA) for 45 min. Cells then were washed twice, incubated 15 min at 4°C with or without the addition of 1:10 (vol/vol) of buffer C containing 10% TX100, and analyzed immediately by flow cytometry. Cytochalasin D treatment (5 μg/ml, 30 min at 37°C) was performed before CD4 labeling with the FITC-conjugated antibody.
Fluorescence Recovery After Photobleaching (FRAP).
FRAP analysis was performed as described (19) with minor modification. Cells were labeled with FITC-conjugated anti-CD4 antibodies in cold KGR+ buffer (Krebs–Ringer phosphate buffer, pH 7.3/10 mM glucose/1 mM CaCl2/1 mM MgCl2), washed, and processed for fluorescence microscopy analysis. Fluorescently labeled receptors were photobleached with an argon laser at 488 nm. The ×63 oil-immersion planachromatic objective used gave an estimated bleach spot radius (w) of 0.89 mm at 1/e2 intensity. The diffusion coefficient (D, ×10−10 cm2/s), denoting the rate of receptor diffusion at 37°C, was calculated according to ref. 20, and the mobile fraction (R, %), reflecting the proportion of mobile receptors, was determined according to ref. 21.
Isolation of Detergent-Resistant Membranes (DRMs).
DRMs were isolated as described (22). Briefly, cells (25 × 106) were solubilized for 30 min on ice with 1 ml of TNE buffer (10 mM Tris⋅HCl, pH 7.5/150 mM NaCl/5 mM EDTA and a mixture of protease inhibitors) containing 1% TX100. Precleared lysates were adjusted to 40% sucrose and overlaid with 5 ml of 30% sucrose and 2 ml of 5% sucrose solution prepared in TNE. Samples were ultracentrifuged in a SW41Ti rotor (Beckman) for 18 h at 200,000 × g. Two-milliliter fractions were collected from the top and TCA-precipitated. The samples then were processed for SDS/PAGE and Western blot analysis.
Metabolic Labeling.
Labeling of cells with [3H]palmitic acid was performed as described (23) with minor modifications. Cells were labeled with 0.5 mCi/ml of [3H]palmitic acid for 5 h at 37°C in RPMI medium 1640, washed twice with PBS, and lysed with 1 ml of buffer D (10 mM Tris⋅HCl, pH 7.4/1% Nonidet P-40/0.4% deoxycholate/66 mM EDTA/10 mM 1,10-O-phenanthroline). SDS (0.1%) was added to precleared lysates, and CD4 was immunoprecipitated by using the OKT4 anti-CD4 antibody. Immunoprecipitates were analyzed by SDS/PAGE and fluorography.
Results
Inhibition of CD4 Endocytosis by p56Lck Is Independent of p56Lck Kinase Activity.
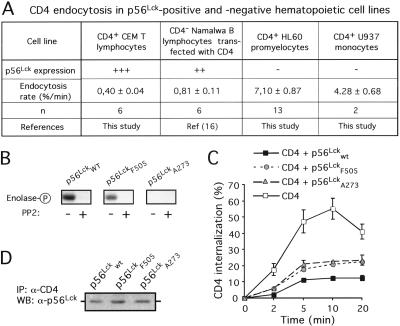
Studies in cells coexpressing p56Lck and CD4 showed that p56Lck inhibits CD4 internalization by preventing CD4 interaction with clathrin-coated endocytic structures (10). In the present study, we assessed whether a similar regulatory p56Lck function occurs in lymphoid cell lines naturally expressing p56Lck. CD4 internalization was measured in p56Lck-positive CD4+ CEM T lymphocytes and CD4− Namalwa B lymphocytes stably transfected with CD4 and in p56Lck-negative CD4+ cell lines (HL60 promyelocytes and U937 monocytes). Our results support a p56Lck-dependent inhibition of CD4 internalization in hematopoietic cell lines naturally expressing the kinase (Fig. 1A).
Figure 1.
p56Lck prevents CD4 endocytosis independently of the kinase activation state. (A) Rates of CD4 internalization were calculated based on the percentage of the total cell-associated radioactivity incorporated after 5 min at 37°C by using iodinated anti-CD4 antibodies as described in Materials and Methods. (B) Kinase activity of p56Lck mutants was assessed in a kinase assay monitoring phosphorylation of enolase as a substrate. As a control of the reaction, 2 μM PP2 was used to inhibit the reaction. Data are representative of three independent experiments. (C) Internalization of CD4 in 293T cells expressing p56Lck mutants by using a FACS-based assay as described in Materials and Methods. Data are means ± SE of four independent experiments. (D) p56Lck association with CD4 in 293T cells cotransfected with CD4 and p56Lck mutants. Data are representative of three independent experiments.
Stimulation of T lymphocytes by MHC class II-bearing cells activates p56Lck and induces CD4 internalization (24). We therefore examined whether p56Lck activation was required to allow dissociation of the CD4-p56Lck complex and CD4 internalization. CD4 endocytosis was recorded in 293T cells coexpressing CD4 and either a p56Lck wild-type allele or a constitutively activated form of the kinase (p56LckF505) (12) or an inactive mutant of p56Lck (p56LckA273) (13). As shown in Fig. 1B, the p56Lck mutants exhibited the expected kinase activity defects in an in vitro assay monitoring the phosphorylation of enolase as exogenous substrate. As a control of the in vitro reaction specificity, the addition of a specific inhibitor of src kinases (PP2) prevented phosphorylation of enolase. As expected, in cells expressing wild-type p56Lck, CD4 internalization was inhibited as compared with the rapid internalization of CD4 in the absence of p56Lck. Interestingly, each p56Lck kinase mutant was still capable to associate with CD4 (Fig. 1D) and to prevent CD4 internalization (Fig. 1C), suggesting that p56Lck interferes with CD4 endocytosis independently of its state of activity.
In summary, both active and inactive forms of p56Lck bind to CD4 and prevent its internalization, suggesting that additional signaling events other than p56Lck activation are required to dissociate the CD4-p56Lck complex and allow CD4 endocytosis.
p56Lck Anchors CD4 to Microvilli.
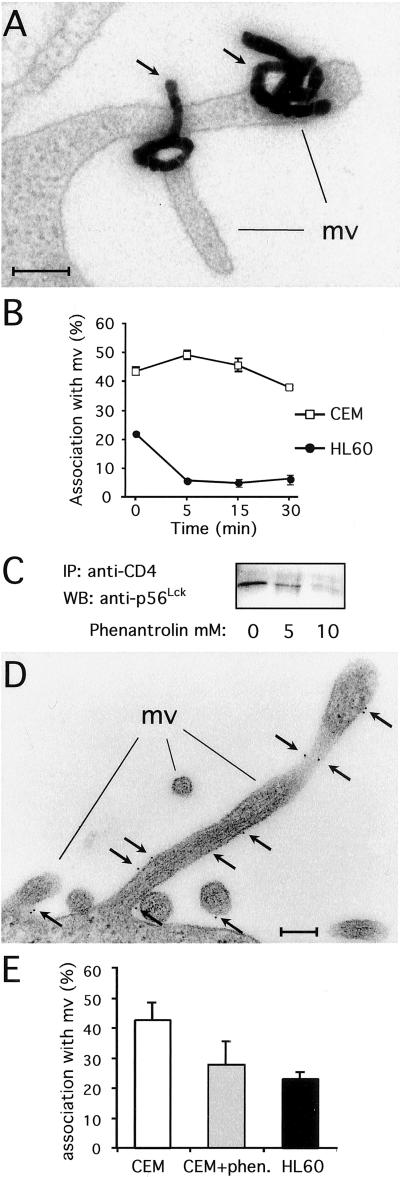
Previous studies suggested that p56Lck does not simply mask an endocytosis signal but has additional effects (10). One hypothesis is that p56Lck anchors CD4 to microvilli, where endocytosis does not occur. We thus investigated the native distribution of CD4 at the ultrastructural level in CEM (p56Lck+) and HL60 (p56Lck−) cells, which represent the most extreme phenotypes in terms of constitutive CD4 internalization (Fig. 1A). CD4 was tagged with 125I-labeled anti-CD4 antibodies and localized by quantitative EM autoradiography (Fig. 2A). At 4°C, approximately 45% and 20% of the labeling was found associated with microvilli in CEM and HL60 cells, respectively (Fig. 2B). At 37°C, CD4 remains associated with microvilli on CEM cells whereas it redistributes almost exclusively to the nonvillous surface in HL60 cells (Fig. 2B). Because the ratio of villous membranes vs. planar membranes is similar in CEM and HL60 cells (34.2 ± 1.6% and 29.6 ± 2.1% of villous membranes, respectively), CD4 concentrates (up to 1.5-fold) on microvilli in p56Lck-positive cells whereas it preferentially associates with planar domains in p56Lck-negative cells.
Figure 2.
p56Lck binding to CD4 triggers CD4 anchoring to microvilli. (A) Electron micrograph showing CD4 radiolabeling on microvilli in CEM cells. (Bar = 0.2 μm.) (B) Kinetics of radiolabeled CD4 association with microvilli in CEM and HL60 cells. Data are means ± SE from two to three separate experiments totaling 900–1,200 autoradiographic grains for each time point. (C) p56Lck association with CD4 in CEM cells treated with 1,10-O-phenanthroline for 30 min at 37°C before cell lysis. Data are representative of three independent experiments. (D) Electron micrograph showing CD4 gold labeling on microvilli in CEM cells. (Bar = 0.2 μm.) (E) Quantitation of gold-labeled CD4 association with microvilli at 4°C in CEM cells treated or not treated with 10 mM 1,10-O-phenanthroline and HL60 cells. Data are means ± SE from three to four separate experiments totaling 78 cells per 2,483 gold particles, 40 cells per 1,119 gold particles, and 81 cells per 1,929 gold particles counted for CEM cells, CEM cells treated with 1,10-O-phenanthroline, and HL60 cells, respectively.
The role of p56Lck in anchoring CD4 to microvilli was assessed further by using 1,10-O-phenanthroline, a membrane-permeable zinc chelator that disrupts the CD4-p56Lck complex (25). As judged by coimmunoprecipitation of p56Lck with CD4 (Fig. 2C) and CD4 immunogold labeling on ultrathin sections (Fig. 2E) from CEM cells, 10 mM 1,10-O-phenanthroline was sufficient to dissociate p56Lck from CD4 and to decrease CD4 association with microvilli to the same extent as what is observed in p56Lck-negative HL60 cells (Fig. 2E).
In conclusion, these morphological analyses demonstrate that p56Lck binding to CD4 targets this receptor to specialized plasma membrane structures, i.e., the microvilli.
The CD4-p56Lck Complex Interacts with Cytoskeletal Elements.
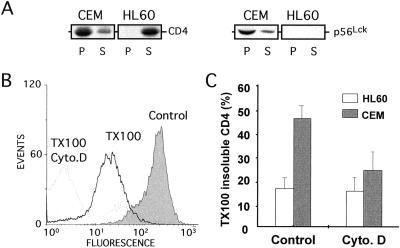
Microvilli are characterized by a dense cytoskeleton core made of a bundle of actin and its associated proteins (26). This suggests that molecules associating with microvilli might interact with cytoskeletal elements and, thus, be insoluble by nonionic detergent such as TX100 at 4°C. We thus investigated CD4 and p56Lck TX100 solubility and observed that CD4 and p56Lck are mostly insoluble by TX100 in CEM cells whereas, in p56Lck-negative HL60 cells, CD4 displays an opposite pattern of solubility (Fig. 3A). Similar results were obtained by using a FACS-based assay to quantitate the extent of CD4 solubility (Fig. 3B). In CEM cells, 45% of the initial CD4 labeling remained associated with cells after solubilization but this value dropped to 15% in HL60 cells (Fig. 3C). Of note, the percentage of CD4 insolubility correlates closely to the percentage of CD4 association with microvilli (see Fig. 2B). In addition, cytochalasin D, which prevents actin polymerization but does not disrupt the p56Lck-CD4 complex, increases to ≈80–85% of the TX100-soluble CD4 fraction in both cell types, indicating that disruption of the cytoskeleton renders CD4 soluble to a similar extent in p56Lck-positive and p56Lck-negative cells (Fig. 3C).
Figure 3.
Disruption of the cytoskeleton increases CD4 TX100 solubility in p56Lck-expressing cells. (A) Western analysis of CD4 and p56Lck solubility by TX100. Cells were lysed in buffer containing 1% TX100 for 20 min at 4°C and then centrifuged at 100,000 × g for 1 h at 4°C to fractionate the lysates in a TX100-soluble (S) and TX100-insoluble (P) fraction. Data are representative of three independent experiments. (B) Typical FACS profile of CD4-associated fluorescence in CEM cells ± 1% TX100 and cytochalasin D (Cyto. D). (C) Quantitation of CD4 solubility by TX100 in CEM and HL60 cells treated or not treated with Cyto. D. Data are expressed as the ratio of fluorescence associated with cells after TX100 addition to fluorescence associated with cells before TX100 addition. Results are means ± SE from three to four independent experiments.
These results indicate that in unstimulated p56Lck-positive cells, CD4 is associated to a high extent with cytoskeletal elements and suggest that p56Lck may act as a bridge between CD4 and the cytoskeleton.
Lateral Mobility of CD4 Is Restricted in p56Lck-Expressing Cells.
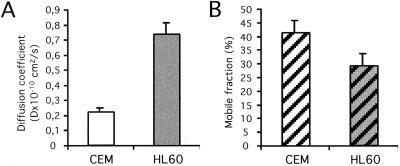
To further support a role of p56Lck in anchoring CD4 to particular domains/structures of the plasma membrane, the lateral mobility of CD4 in the plane of the plasma membrane was measured by FRAP analysis. As shown in Fig. 4, in CEM cells, the lateral diffusion coefficient of CD4 at 37°C is 3- to 4-fold lower than in HL60 cells, suggesting that p56Lck restricts CD4 mobility in the plasma membrane. The mobile fraction, in contrast, was lower in HL60 cells than in CEM cells, which could be related to the segregation of a large fraction of CD4 in clathrin-coated pits under these conditions.
Figure 4.
Lateral mobility of CD4 is restricted in p56Lck-expressing cells. The lateral diffusion of CD4 was measured at 37°C in individual, cultured CEM cells and HL60 cells by using FRAP analysis. (A) Mean diffusion constant. (B) Mobile fraction. Data are means ± SE of 35–59 cells analyzed for each condition.
These experiments demonstrating that p56Lck expression restricts CD4 surface mobility provide additional evidences supporting a role of p56Lck as a CD4 anchor to the cytoskeleton and/or particular membrane domains.
p56Lck Mediates CD4 Association with DRMs.
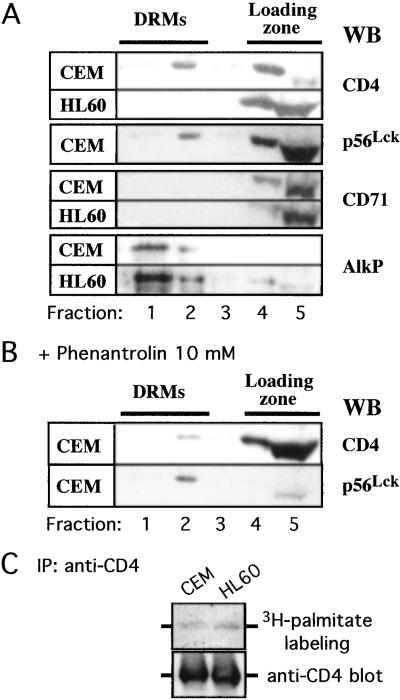
CD4 insolubility by TX100 at low temperature also might reflect its association with specialized lipid microdomains termed DRMs (27). CD4 and p56Lck previously have been reported to be concentrated in DRMs in T lymphocytes (28), but no information is available as to whether CD4 association with DRMs is p56Lck-dependent. To answer this question, we isolated biochemically DRMs in CEM and HL60 cells and analyzed the distribution of CD4 and p56Lck within these microdomains. Alkaline phosphatase (AlkP), a glycosylphosphatidylinositol-anchored protein specifically associated with DRMs, and the transferrin receptor (CD71), a transmembrane molecule excluded from DRMs, were used as markers of the fractionation procedure. As shown in Fig. 5A, CD4 and p56Lck are present in DRMs in CEM cells. However, in HL60 cells, CD4 was undetectable in DRMs, suggesting that the localization of this receptor in DRMs depended on p56Lck. In support of these data, disruption of the CD4-p56Lck complex in CEM cells by 1,10-O-phenanthroline substantially shifted CD4 out of DRMs whereas p56Lck remains associated with these microdomains (Fig. 5B).
Figure 5.
p56Lck triggers CD4 segregation in DRMs. DRMs were prepared from CEM and HL60 cells as described in Materials and Methods and fractions were analyzed by SDS/PAGE and Western blotting. (A) Typical distribution of CD4, p56Lck, CD71, and AlkP is shown. Data are representative of three independent experiments. (B) Disruption of the CD4-p56Lck complex by 1,10-O-phenanthroline redistributes CD4, but not p56Lck, out of DRMs. CEM cells were treated for 30 min at 37°C with 1,10-O-phenanthroline before starting the fractionation procedure, and distribution of CD4 and p56Lck was analyzed. Data are representative of three independent experiments. (C) CD4 is palmitoylated in both CEM and HL60 cells. Cells were metabolically labeled with [3H]palmitate, and CD4 palmitoylation was revealed as described in Materials and Methods. Data are representative of three independent experiments.
Sorting of a protein into DRMs correlates in some cases with posttranscriptional modifications such as palmitoylation (29). Because CD4 is palmitoylated on two cysteines proximal to the transmembrane domain (30), we checked whether a difference in CD4 palmitoylation might explain the difference in CD4 segregation within DRMs recorded for CEM and HL60 cells. Cells were labeled metabolically with [3H]palmitic acid, and CD4 was immunoprecipitated. Fluorographic analysis of the immunoprecipitates revealed that CD4 is palmitoylated to the same extent in both CEM and HL60 cells (Fig. 5C), thus excluding the possibility raised above.
In summary, these data demonstrate that CD4 targeting to DRMs is mediated by its association with p56Lck and is not dependent on posttranscriptional CD4 acylation. Moreover, because the CD4 associates concomitantly with DRMs and microvilli, these observations suggest that CD4 segregates in DRMs associated with microvilli via p56Lck.
Discussion
In this study, we demonstrate that the tyrosine kinase p56Lck plays a crucial role in CD4 localization and trafficking at the plasma membrane. Specifically, we reached the following conclusions: (i) p56Lck prevents CD4 internalization by maintaining this receptor on microvilli; (ii) CD4 binding to p56Lck as well as the inhibition of CD4 internalization by p56Lck are independent of p56Lck activity; (iii) p56Lck mediates CD4 association with cytoskeletal elements, CD4 localization on microvilli, and CD4 segregation within DRMs, strongly suggesting that CD4 is associated with DRMs located on microvilli; and (iv) CD4 association with DRMs/microvilli in p56Lck-expressing cells correlates with a restricted lateral mobility of CD4 in the plasma membrane.
Compartmentalization of specific signaling or adhesion molecules on microvilli is now well established. Receptor segregation on microvilli might play a role in cell–cell and cell–substratum adhesion (31, 32) as well as in membrane fusion processes (33). Why localization of adhesion/fusion molecules on microvilli is functionally relevant is not clearly understood, but the morphology and dynamic of microvilli may provide a scaffold for the presentation of these molecules and potentially could be instrumental in targeting adhesion/fusion molecules where functionally appropriate. In addition, the cylindrical shape and narrow diameter of microvilli may provide the low radius of curvature required to overcome the inherent repulsive interactions between cell surfaces during cell adhesion processes. The functional relevance of signaling receptors' association with microvilli is also unclear. We have described previously that the insulin and epidermal growth factor receptors preferentially segregate on microvillar domains in various cell types (11). In addition, G-coupled seven-transmembrane proteins such as CCR5 and CXCR4 also associate predominantly with microvilli on macrophages and lymphocytes (34). On the basis of these observations, it is tempting to speculate that such localization might favor the binding of soluble, circulating ligands and, therefore, might represent a particular surface domain involved in receptor signaling.
The localization of receptors on microvilli might reflect their tight coupling to the cytoskeleton. Indeed, microvilli are actin-filled cell extensions enriched in a panel of actin-associated cytoskeletal proteins (26). In this regard, cytoskeletal proteins such as talin and α-actinin have been proposed to mediate localization of adhesion molecules on microvilli (31, 32). Other candidates are the members of the ERM family (35). Unfortunately, except for the epidermal growth factor receptor, which has been shown to bind actin (36), little information is available on potential cytoskeletal partners anchoring signaling receptors to microvilli. Present observations showing that the CD4 TX100 solubility is cytochalasin D-sensitive support the concept of such an association of signaling receptors with cytoskeletal elements on microvilli, but further studies are required to identified the exact molecules involved.
CD4 is not only sequestered on microvilli but also concentrated in DRMs when p56Lck is expressed. Based on the observations that (i) CD4 associates with microvilli and DRMs in the same experimental conditions (time and temperature) and (ii) CD4 segregation on microvilli and in DRMs is p56Lck-dependent, we propose that CD4 associates with DRMs located on microvilli. In support of these conclusions are the recent observations that cholesterol-enriched microdomains may be linked to cytoskeletal elements through protein of the annexin family (37) and that prominin, a pentaspan membrane protein, is retained on microvilli concomitantly with a segregation in cholesterol-based microdomains (38). Thus, microvilli, possibly through their high concentration in cytoskeletal elements, could play a key role in the sorting of proteins specifically segregated in DRMs.
CD4 association with DRMs and cytoskeletal elements in p56Lck-expressing cells is corroborated further by our FRAP analysis. Indeed, interactions with the cytoskeleton and the membrane cholesterol/phospholipid ratio play a crucial role in the mobility of cell membrane glycoprotein (39, 40). Thus, the net decrease in CD4 mobility measured for cells expressing p56Lck could reflect dynamic interactions of the CD4-p56Lck complex with the cytoskeleton and/or lipids.
The concomitant localization of CD4 and p56Lck in DRMs and microvilli is consistent with previously described mechanisms leading to T cell activation. Indeed, DRMs concentrate the bulk of the signaling machinery necessary to activate T cells in response to antigen stimulation, including CD4 and p56Lck (41). Upon TCR activation, a coalescence of lipid rafts containing relevant molecules for the immune response as well as a capping of CD4 are observed (42, 43). The role of the cytoskeleton in the aggregation of other components of the TCR-signaling machinery is still unclear; however, in the case of CD4, this coreceptor is within lipid rafts and capping of this molecule depends on the cytoskeleton reorganization (18), thus establishing a link between lipid rafts as signaling platforms and the cytoskeleton. These observations raise the interesting hypothesis that plasma membrane domains highly concentrating cytoskeletal elements such as microvilli could represent potential cellular sites mediating these signaling processes.
How does p56Lck direct CD4 to DRMs/microvilli? p56Lck interacts with CD4 early in the secretory pathway, and both reach the plasma membrane together (44). Consistent with our data suggesting that p56Lck mediates CD4 segregation on microvilli/DRMs, p56Lck targeting to the plasma membrane and association with DRMs occur independently of CD4 expression, indicating that p56Lck contains the determinants necessary to localize in DRMs (7, 44). One of these determinants is likely to be posttranscriptional modifications such as myristoylation or palmitoylation (45, 46). Indeed, p56Lck palmitoylation has been shown to be crucial to allow p56Lck association with CD4 and membranes (44, 47). However, in the case of CD4, palmitoylation of the molecule does not correlate with segregation within DRMs in p56Lck-negative cells, and, thus, palmitoylation does not appear to be sufficient to trigger association with DRMs. These observations could appear in contradiction with those of Arcaro et al. (29), who suggested that CD8 palmitoylation is necessary to target CD8 to DRMs and to mediate the interaction with p56Lck. However, in light of our results, it is possible that unpalmitoylated CD8 could not associate with DRMs because it cannot interact with p56Lck. Thus, taken together, the present and previous studies support a model in which palmitoylation of CD4 might be crucial to allow interaction with p56Lck along the secretory pathway and traveling of the complex to the plasma membrane. However, targeting of these complexes to DRMs likely is due only to p56Lck potentially through the presence of other determinants in p56Lck such as myristoylated moieties.
In p56Lck-positive cells, CD4 has to translocate out of DRMs and slide out of microvilli to be internalized by clathrin-coated pits (48). Physiologically, CD4 internalization in T lymphocytes occurs in response to antigenic stimulation, a process initiated by p56Lck activation subsequently to CD4 and TCR binding to MHC II (24). By analogy with tyrosine kinase receptors (49), we hypothesized that activation of p56Lck could trigger the dissociation of the CD4-p56Lck complex, leading to CD4 translocation and internalization. However, we observed that p56Lck activation does not result in the p56Lck-CD4 complex dissociation and CD4 internalization is not induced. In addition, the stability of the p56Lck-CD4 complex is not dependent on the lipid raft or cytoskeleton integrity because neither cytochalasin D nor cyclodextrin affects the complex stability (data not shown). Thus, it is likely that p56Lck-independent signaling is responsible for inducing CD4-p56Lck dissociation and CD4 internalization. An interesting candidate is protein kinase C-α, which, in response to phorbol 12-myristate 13-acetate, induces CD4 internalization by dissociating the CD4-p56Lck complex in DRMs and promoting CD4 (but not p56Lck) translocation out of these domains (50).
The physiologic role of CD4 in p56Lck-negative cells remains an open question. Signaling through CD4 independently of p56Lck has been described, but information is still fragmentary and no consensus function for CD4 has been reached (3). However, our data and others (51), indicating a different CD4 localization and trafficking in p56Lck-negative cells as compared with T lymphocytes, presume a distinct function for CD4 and raise exciting questions for specialists working on hematopoiesis and macrophage function.
Acknowledgments
We thank G. Porcheron-Berthet and C. Giroud for technical assistance as well as Dr. K.-E. Magnusson for valuable discussions. This work was supported by Fonds National Suisse pour la Recherche Scientifique Grants 31-55170.98, 31-53686.98, and 31-65392.01; the Swedish Medical Research Council (Project 6251); and by the Swedish Research Council for Engineering Sciences (Project 230-99-392).
Abbreviations
- DRMs
detergent-resistant membranes
- TX100
Triton X-100
- EM
electron microscopy
- FRAP
fluorescence recovery after photobleaching
References
- 1.Maddon P J, Molineaux S M, Maddon D E, Zimmerman K A, Godfrey M, Alt F W, Chess L, Axel R. Proc Natl Acad Sci USA. 1987;84:9155–9159. doi: 10.1073/pnas.84.24.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss A, Littman D R. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 3.Foti M, Lew D P, Carpentier J L, Krause K H. J Lab Clin Med. 1995;126:233–239. [PubMed] [Google Scholar]
- 4.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. Nature (London) 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 5.Veillette A, Bookman M A, Horak E M, Bolen J B. Cell. 1988;55:301–308. doi: 10.1016/0092-8674(88)90053-0. [DOI] [PubMed] [Google Scholar]
- 6.Huse M, Eck M J, Harrison S C. J Biol Chem. 1998;273:18729–18733. doi: 10.1074/jbc.273.30.18729. [DOI] [PubMed] [Google Scholar]
- 7.Bijlmakers M J, Isobe-Nakamura M, Ruddock L J, Marsh M. J Cell Biol. 1997;137:1029–1040. doi: 10.1083/jcb.137.5.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsh M, Pelchen-Matthews A. Curr Top Microbiol Immunol. 1996;205:107–135. doi: 10.1007/978-3-642-79798-9_6. [DOI] [PubMed] [Google Scholar]
- 9.Lama J, Mangasarian A, Trono D. Curr Biol. 1999;9:622–631. doi: 10.1016/s0960-9822(99)80284-x. [DOI] [PubMed] [Google Scholar]
- 10.Pelchen-Matthews A, Boulet I, Littman D R, Fagard R, Marsh M. J Cell Biol. 1992;117:279–290. doi: 10.1083/jcb.117.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpentier J L. Histochemistry. 1993;100:169–184. doi: 10.1007/BF00269090. [DOI] [PubMed] [Google Scholar]
- 12.Amrein K E, Sefton B M. Proc Natl Acad Sci USA. 1988;85:4247–4251. doi: 10.1073/pnas.85.12.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin S D, Anderson S J, Forbush K A, Perlmutter R M. EMBO J. 1993;12:1671–1680. doi: 10.1002/j.1460-2075.1993.tb05812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umesono K, Murakami K K, Thompson C C, Evans R M. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ausubel F, Brent R, Kingstone R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 16.Foti M, Mangasarian A, Piguet V, Lew D P, Krause K H, Trono D, Carpentier J L. J Cell Biol. 1997;139:37–47. doi: 10.1083/jcb.139.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piguet V, Gu F, Foti M, Demaurex N, Gruenberg J, Carpentier J L, Trono D. Cell. 1999;97:63–73. doi: 10.1016/s0092-8674(00)80715-1. [DOI] [PubMed] [Google Scholar]
- 18.Geppert T D, Lipsky P E. J Immunol. 1991;146:3298–3305. [PubMed] [Google Scholar]
- 19.Johansson B, Sundqvist T, Magnusson K E. Cell Biophys. 1987;10:233–244. doi: 10.1007/BF02797343. [DOI] [PubMed] [Google Scholar]
- 20.Axelrod D, Koppel D E, Schlessinger J, Elson E, Webb W W. Biophys J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobson K, Derzko Z, Wu E S, Hou Y, Poste G. J Supramol Struct. 1976;5:565–576. doi: 10.1002/jss.400050411. [DOI] [PubMed] [Google Scholar]
- 22.Ilangumaran S, Arni S, van Echten-Deckert G, Borisch B, Hoessli D C. Mol Biol Cell. 1999;10:891–905. doi: 10.1091/mbc.10.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Trible R P, Samelson L E. Immunity. 1998;9:239–246. doi: 10.1016/s1074-7613(00)80606-8. [DOI] [PubMed] [Google Scholar]
- 24.Weyand C M, Goronzy J, Fathman C G. J Immunol. 1987;138:1351–1354. [PubMed] [Google Scholar]
- 25.Lin R S, Rodriguez C, Veillette A, Lodish H F. J Biol Chem. 1998;273:32878–32882. doi: 10.1074/jbc.273.49.32878. [DOI] [PubMed] [Google Scholar]
- 26.Louvard D. Curr Opin Cell Biol. 1989;1:51–57. doi: 10.1016/s0955-0674(89)80036-5. [DOI] [PubMed] [Google Scholar]
- 27.Hooper N M. Mol Membr Biol. 1999;16:145–156. doi: 10.1080/096876899294607. [DOI] [PubMed] [Google Scholar]
- 28.Parolini I, Sargiacomo M, Lisanti M P, Peschle C. Blood. 1996;87:3783–3794. [PubMed] [Google Scholar]
- 29.Arcaro A, Gregoire C, Boucheron N, Stotz S, Palmer E, Malissen B, Luescher I F. J Immunol. 2000;165:2068–2076. doi: 10.4049/jimmunol.165.4.2068. [DOI] [PubMed] [Google Scholar]
- 30.Crise B, Rose J K. J Biol Chem. 1992;267:13593–13597. [PubMed] [Google Scholar]
- 31.Pavalko F M, Walker D M, Graham L, Goheen M, Doerschuk C M, Kansas G S. J Cell Biol. 1995;129:1155–1164. doi: 10.1083/jcb.129.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abitorabi M A, Pachynski R K, Ferrando R E, Tidswell M, Erle D J. J Cell Biol. 1997;139:563–571. doi: 10.1083/jcb.139.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson N F, Snell W J. Trends Cell Biol. 1998;8:93–96. doi: 10.1016/s0962-8924(98)01234-3. [DOI] [PubMed] [Google Scholar]
- 34.Singer I I, Scott S, Kawka D W, Chin J, Daugherty B L, DeMartino J A, DiSalvo J, Gould S L, Lineberger J E, Malkowitz L, et al. J Virol. 2001;75:3779–3790. doi: 10.1128/JVI.75.8.3779-3790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsukita S, Yonemura S. Trends Biochem Sci. 1997;22:53–58. doi: 10.1016/s0968-0004(96)10071-2. [DOI] [PubMed] [Google Scholar]
- 36.den Hartigh J C, van Bergen en Henegouwen P M, Verkleij A J, Boonstra J. J Cell Biol. 1992;119:349–355. doi: 10.1083/jcb.119.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harder T, Kellner R, Parton R G, Gruenberg J. Mol Biol Cell. 1997;8:533–545. doi: 10.1091/mbc.8.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roper K, Corbeil D, Huttner W B. Nat Cell Biol. 2000;2:582–592. doi: 10.1038/35023524. [DOI] [PubMed] [Google Scholar]
- 39.Hoover R L, Dawidowicz E A, Robinson J M, Karnovsky M J. J Cell Biol. 1983;97:73–80. doi: 10.1083/jcb.97.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braun J, Unanue E R. Semin Hematol. 1983;20:322–333. [PubMed] [Google Scholar]
- 41.Cherukuri A, Dykstra M, Pierce S K. Immunity. 2001;14:657–660. doi: 10.1016/s1074-7613(01)00156-x. [DOI] [PubMed] [Google Scholar]
- 42.Kupfer A, Singer S J, Janeway C A, Jr, Swain S L. Proc Natl Acad Sci USA. 1987;84:5888–5892. doi: 10.1073/pnas.84.16.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janes P W, Ley S C, Magee A I. J Cell Biol. 1999;147:447–461. doi: 10.1083/jcb.147.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bijlmakers M J, Marsh M. J Cell Biol. 1999;145:457–468. doi: 10.1083/jcb.145.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Resh M D. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 46.Paige L A, Nadler M J, Harrison M L, Cassady J M, Geahlen R L. J Biol Chem. 1993;268:8669–8674. [PubMed] [Google Scholar]
- 47.Kwong J, Lublin D M. Biochem Biophys Res Commun. 1995;207:868–876. doi: 10.1006/bbrc.1995.1266. [DOI] [PubMed] [Google Scholar]
- 48.Keller G A, Siegel M W, Caras I W. EMBO J. 1992;11:863–874. doi: 10.1002/j.1460-2075.1992.tb05124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carpentier J L, Paccaud J P, Gorden P, Rutter W J, Orci L. Proc Natl Acad Sci USA. 1992;89:162–166. doi: 10.1073/pnas.89.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parolini I, Topa S, Sorice M, Pace A, Ceddia P, Montesoro E, Pavan A, Lisanti M P, Peschle C, Sargiacomo M. J Biol Chem. 1999;274:14176–14187. doi: 10.1074/jbc.274.20.14176. [DOI] [PubMed] [Google Scholar]
- 51.Pelchen-Matthews A, da Silva R P, Bijlmakers M J, Signoret N, Gordon S, Marsh M. Eur J Immunol. 1998;28:3639–3647. doi: 10.1002/(SICI)1521-4141(199811)28:11<3639::AID-IMMU3639>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]