Abstract
In response to starvation, an unstructured population of identical Myxococcus xanthus cells rearranges into an asymmetric, stable pattern of multicellular fruiting bodies. Central to this pattern formation process are changes in organized cell movements from swarming to aggregation. Aggregation is induced by the cell surface-associated C-signal. To understand how aggregation is accomplished, we have analyzed how C-signal modulates cell behavior. We show that C-signal induces a motility response that includes increases in transient gliding speeds and in the duration of gliding intervals and decreases in stop and reversal frequencies. This response results in a switch in cell behavior from an oscillatory to a unidirectional type of behavior in which the net-distance traveled by a cell per minute is increased. We propose that the C-signal-dependent regulation of the reversal frequency is essential for aggregation and that the remaining C-signal-dependent changes in motility parameters contribute to aggregation by increasing the net-distance traveled by starving cells per minute. In our model for symmetry-breaking and aggregation, C-signal transmission is a local event involving direct contacts between cells that results in a global organization of cells. This pattern formation mechanism does not require a diffusible substance or other actions at a distance. Rather it depends on contact-induced changes in motility behavior to direct cells appropriately
Formation of spatial patterns of cells from a mass of initially identical cells is a recurring theme in developmental biology. The dynamics that direct pattern formation in biological systems often involve morphogenetic cell movements (1–3). An example is fruiting body formation in the gliding bacterium Myxococcus xanthus in which an unstructured population of identical cells rearranges into an asymmetric, stable pattern of multicellular fruiting bodies in response to starvation (4).
M. xanthus cells are rod shaped and move by gliding, a process whereby a bacterial cell moves in the direction of its long axis on a solid surface (5). Fruiting body morphogenesis absolutely depends on starvation of cells at a high cell density on a solid surface (4). It represents a true de novo pattern formation process as it starts from a homogeneous and symmetric population of starving cells, and occurs without the contribution of external cues. In the presence of nutrients, M. xanthus cells form cooperatively spreading swarms. In response to starvation, swarming behavior is constrained, and, after 6 h of starvation, small aggregates are evident (6). Some of these aggregates enlarge into hemispheres as a consequence of continued accumulation of cells, and, after 24 h, haystack-shaped fruiting bodies have formed, each containing ≈100,000 densely packed cells. Within the nascent fruiting bodies, the motile, rod-shaped cells differentiate into non-motile spores. Aggregates that do not mature into fruiting bodies dissipate as their cells migrate to other aggregation centers. Before the appearance of aggregation centers, cells become organized in streams, in which the cells are arranged end-to-end and with their long axes roughly in parallel with each other (7), and cells move toward the aggregation centers organized in these streams (7–10).
Aggregation is induced by the cell surface-associated C-signal (11–16), the latest acting of several extracellular signals required for fruiting body morphogenesis (17, 18). The C-signal is a cell surface-associated protein encoded by the csgA gene (11, 12). Cells that carry mutations in the csgA gene are conditionally defective in aggregation and sporulation (19, 20). The developmental defects in csgA cells are rescued by codevelopment with wild-type (wt) cells (extracellular complementation; ref. 20). Extracellular complementation is based on wt cells providing the missing C-signal to the csgA cells, thus enabling them to complete their development (12, 20). The level of C-signaling increases during development because of accumulation of the C-signal (15, 16). The C-signal induces aggregation and sporulation in a concentration-dependent manner, i.e., intermediate level aggregation is induced and at high levels sporulation is induced (13–15). C-signal transmission occurs by a contact-dependent mechanism (12) that involves end-to-end contacts between cells and fails when cells are unable to make these even though they are able to make side-by-side contacts (21–24).
We have previously shown that C-signal in the absence of other cell–cell interactions induces cells to move with increased transient gliding speeds, in longer gliding intervals and with a reduced stop frequency (25). The present work was undertaken to determine which motility parameters are controlled by the C-signal at the high cell density required for fruiting body formation and to further the understanding of how C-signal induces aggregation. We show that, at the high cell density, C-signal controls specific motility parameters. The outcome of the C-signal-dependent changes in motility parameters is a switch from an oscillatory to a unidirectional type of motility behavior in which the net-distance traveled by a cell per minute is increased.
Materials and Methods
Bacterial Strains and Growth.
M. xanthus strains used in this work are all derivatives of the fully motile wt DK1622 (26). DK1622GFP (pilAp-gfp; ref. 27), DK5208GFP (csgA∷Tn5-132ΩLS205, pilAp-gfp) was constructed by generalized transduction into DK5208 (csgA∷Tn5-132ΩLS205; ref. 28) by using the transducing phage Mx8clp2ts3 propagated on DK1622GFP. Cells were grown in liquid CTT medium (10 g/liter Casitone/8 mM MgSO4/10 mM Tris⋅HCl/1 mM potassium phosphate, pH 7.6) or maintained on CTT agar plates (29) containing kanamycin at 40 μg/ml or oxytetracycline at 10 μg/ml.
Preparation of Cells for Video Microscopy.
Cells were grown at 32°C in liquid CTT medium to a density of 5 × 108 cells/ml, harvested, and resuspended in A50-buffer (10 mM Mops, pH 7.2/10 mM CaCl2/10 mM MgCl2/50 mM NaCl; ref. 12) to a calculated density of 5 × 109 cells/ml. Two microliters of concentrated cell mixtures was spotted on 1.5-mm-thick A50–1.5% agarose prepared on a standard glass-microscope slide. Cells were incubated in a humid chamber at 32°C to induce development. Fruiting body formation occurs with normal timing on A50–1.5% agarose (25).
Time-Lapse Fluorescence Video Microscopy.
Green fluorescent protein (GFP)-expressing cells were observed by using a Nikon Eclipse E800 microscope equipped with a Plan Fluor ×40 objective (n.a. 0.75) and a Chroma FITC-HYQ filter. The microscope images were captured with an intensified charge-coupled device (CCD) camera (VideoScope, Dallas; model ICCD-1000F). The output channel of the camera was connected to a Hamamatsu (Middlesex, NJ) Argus 10 image processor, which performed real-time averaging of eight frames (240 ms). The resulting image was stored as a single image on an optical disk recorder (Panasonic TQ-FH224). Frames were captured every 15 s during a recording period of 900 s (60 frames). To reduce the exposure of the cells to light, the intensity of the 100-W mercury arc lamp was reduced 8-fold by using a neutral density filter. The camera was adjusted to maximal sensitivity, and the microscope was equipped with a computer-controlled shutter. Observations were performed at 22°C. Cells expressing GFP developed normally, and the exposure of cells to UV-light for 900 s did not interfere with development (data not shown).
To distinguish active cell movement from background noise, wt cells expressing GFP (DK1622GFP) were mixed with non-fluorescent wt cells (DK1622). This mixture was fixed with 1.0% glutaraldehyde, spotted on an A50-agarose, and then recorded immediately after the droplets had dried (2–3 min). The transient speed of the fixed GFP-containing cells was 0.39 ± 0.20 μm/min; thus, only gliding speeds > 0.59 μm/min were taken to indicate active cell movement.
Computerized Motion Analysis of Cells.
The captured images were digitized and transferred to a personal computer by using NIH image (developed at the National Institutes of Health; available at http://rsb.info.nih.gov/nih-image/). Images were calibrated by using an image of a slide micrometer. The x,y coordinates of a cell in a frame were defined as the position of a specific pole of that cell. The quantitative analysis of cell movements was performed as described (25). Briefly, transient gliding speeds were calculated as follows. The displacement of a cell was determined as the geometrical distance between the position of the specific pole of that cell in two subsequent frames. The displacement divided by 15 s (the time interval between two frames) then gives the transient gliding speed. The transient gliding speed for each cell in each 15-s time interval was calculated. From these transient gliding speeds, speed profiles were prepared for each individual cell. The transient gliding speeds in the first recording interval (0–15 s) were not significantly different from the transient gliding speeds in the last recording interval (885–900 s) according to a Kruskal-Wallis test (30). Therefore, an experiment in which 20 cells were followed for 900 s contributed 1,180 independent transient gliding speed measurements, and distributions of transient gliding speeds in the populations could be calculated. To test for the statistical difference between transient gliding speed distributions, the data sets underlying the distributions were subjected to a Kruskal-Wallis test. Mean gliding speed was calculated by using only transient gliding speeds > 0.59 μm/min, the detection limit for active movement. A gliding interval is the interval between a start and a stop; a stop interval is the interval between a stop and a start. A stop is a decrease in transient speed to <0.59 μm/min; a start is an increase in transient gliding speed to >0.59 μm/min. Stop frequency was calculated as the number of stops per minute of gliding with transient speeds > 0.59 μm/min. Reversal frequency was calculated as the number of reversals per minute of gliding with transient speeds > 0.59 μm/min. Net gliding distance was calculated as the geometrical distance between the position of a cell at t = 0 s and at t = 900 s. At each time point, three independent experiments were performed, and motion analyses were performed on at least 20 cells.
Results
To analyze cell movements during aggregation, we used a motility assay in which the behavior of individual cells could be monitored in the context of the high cell density required for fruiting body formation (Materials and Methods). In brief, cells that constitutively express GFP were mixed at a ratio of 1:400 with non-fluorescent cells and then exposed to starvation at a high cell density on a solid surface. At different time-points during development, the behavior of individual GFP-expressing cells was recorded by using fluorescence time-lapse video microscopy, and cell behavior was quantified. The detection limit for active cell movement in this assay was 0.59 μm/min, and only gliding speeds > 0.59 μm/min were taken to indicate active cell movement. The behavior of the GFP-expressing cells was analyzed in terms of transient gliding speeds, mean gliding speed, duration of mean gliding, and stop intervals and mean stop and reversal frequencies (25) and net gliding distance (cf. Materials and Methods for how these parameters were defined and calculated).
Behavior of Wild-Type Cells in the Context of Other Wild-Type Cells.
The behavior of wt cells during development was analyzed by mixing GFP-expressing wt cells (DK1622GFP) with non-fluorescent wt cells (DK1622; Table 1). We first analyzed the behavior of wt cells starved for 3 h in details. Gliding speed profiles of individual cells showed that cells exhibited a range of speeds (cf. Fig. 1a for a representative profile). The variation in transient gliding speeds displayed by each individual cell is also reflected in the broad distribution of transient gliding speeds observed for 23 cells recorded at 3 h of starvation (Fig. 1c). All cells experienced periods of no active movement with the duration of the mean gliding interval being 1.31 ± 0.34 min and the duration of the mean stop interval being 1.13 ± 0.33 min (Table 1). The stop frequency was 10-fold higher than the reversal frequency, and cells on the average stopped their movement once per minute and reversed their direction every 10 min (Table 1). The mean net-gliding distance traveled by these cells during the 900-s recording period was 5.65 ± 0.34 μm.
Table 1.
Motility parameters of wt and csgA cells developed in the context of other wt and csgA cells, respectively
| Motility parameter | Cell type | Hrs of starvation
|
|||||
|---|---|---|---|---|---|---|---|
| 0 | 3 | 6 | 9 | 12 | 15 | ||
| Gliding speed, μm/min* | wt | 1.65 ± 0.01 | 1.72 ± 0.09 | 2.23 ± 0.10 | 2.42 ± 0.12 | 2.49 ± 0.22 | 2.66 ± 0.05 |
| csgA | 1.59 ± 0.10 | 1.68 ± 0.09 | 2.01 ± 0.04 | 2.17 ± 0.07 | 2.23 ± 0.10 | 2.19 ± 0.09 | |
| Duration of gliding interval, min† | wt | 0.88 ± 0.06 | 1.31 ± 0.34 | 2.69 ± 0.24 | 4.43 ± 0.04 | 3.70 ± 0.58 | 5.00 ± 0.91 |
| csgA | 0.99 ± 0.21 | 1.24 ± 0.15 | 1.79 ± 0.18 | 1.93 ± 0.33 | 2.11 ± 0.19 | 2.42 ± 0.05 | |
| Duration of stop interval, min‡ | wt | 1.12 ± 0.24 | 1.13 ± 0.33 | 0.48 ± 0.04 | 0.43 ± 0.17 | 0.35 ± 0.04 | 0.44 ± 0.14 |
| csgA | 1.04 ± 0.13 | 0.99 ± 0.18 | 0.81 ± 0.09 | 0.51 ± 0.05 | 0.50 ± 0.04 | 0.54 ± 0.02 | |
| Stop frequency, stop/min§ | wt | 1.25 ± 0.09 | 0.95 ± 0.15 | 0.51 ± 0.03 | 0.26 ± 0.05 | 0.25 ± 0.02 | 0.22 ± 0.04 |
| csgA | 1.24 ± 0.17 | 1.10 ± 0.09 | 0.79 ± 0.10 | 0.65 ± 0.07 | 0.59 ± 0.02 | 0.56 ± 0.03 | |
| Reversal frequency, reversal/min¶ | wt | 0.104 ± 0.011 | 0.095 ± 0.008 | 0.077 ± 0.002 | 0.054 ± 0.008 | 0.042 ± 0.005 | 0.045 ± 0.002 |
| csgA | 0.099 ± 0.007 | 0.101 ± 0.002 | 0.103 ± 0.010 | 0.109 ± 0.006 | 0.103 ± 0.008 | 0.113 ± 0.010 | |
| Net gliding distance, μm‖ | wt | 4.01 ± 0.68 | 5.65 ± 0.34 | 14.22 ± 2.52 | 19.37 ± 1.33 | 22.39 ± 4.96 | 24.39 ± 1.73 |
| csgA | 4.21 ± 0.43 | 5.13 ± 0.99 | 8.76 ± 1.85 | 12.63 ± 1.64 | 10.04 ± 2.52 | 11.34 ± 3.04 | |
Mean values and SEM are shown for three independent experiments. Strains used: in experiments with wt cells, DK1622 and DK1622GFP; in experiments with csgA cells, DK5208 and DK5208GFP.
Mean gliding speed: calculated using transient gliding speeds > 0.59 μm/min.
Gliding interval: interval between a start and a stop. A start: increase in transient gliding speed to >0.59 μm/min. A stop: decrease in transient gliding speed to <0.59 μm/min.
Stop interval: interval between a stop and a start.
Stop frequency: calculated as the number of stops per minute of gliding with transient speeds > 0.59 μm/min.
Reversal frequency: calculated as the number of reversals per minute of gliding with transient speeds > 0.59 μm/min. A reversal: change in transient gliding speeds from a value > 0.59 μm/min to a value < −0.59 μm/min, or vice versa.
Net gliding distance: calculated as the geometrical distance between the position of a cell at t = 0 s and at t = 900 s.
Figure 1.
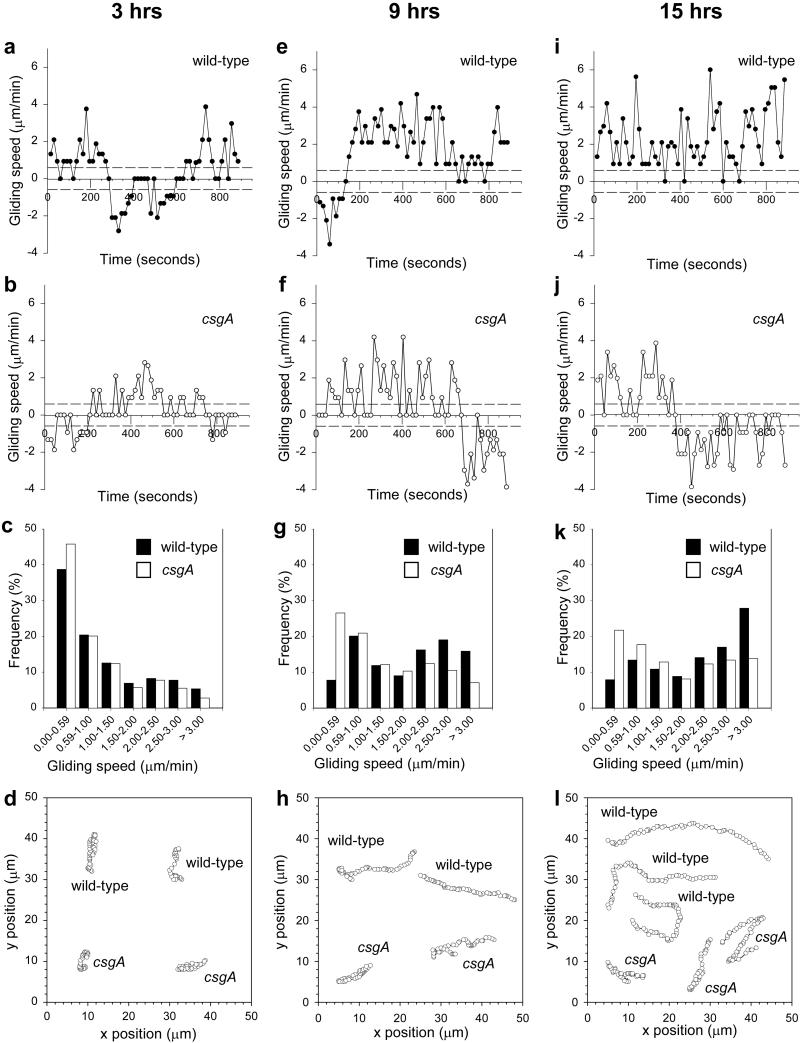
Motility parameters of wt and csgA cells during development. The data in the first, second, and third columns were collected at 3, 9, and 15 h, respectively. (a, e, and i) Gliding speed profiles of representative wt cells developed in the context of other wt cells. Each panel shows the speed profile of a single representative cell. The dotted lines indicate the detection limit for active movement. A reversal is a change in speed from a positive to a negative value, or vice versa. (b, f, and j) Gliding speed profiles of representative csgA cells developed in the context of other csgA cells. Each panel shows the speed profile of a single representative cell. The dotted lines indicate the detection limit for active movement. A reversal is a change in speed from a positive to a negative value, or vice versa. (c, g, and k) Distributions of transient gliding speeds in wt cells developed in the context of other wt cells and csgA cells developed in the context of other csgA cells. Each distribution was prepared from at least 1,180 transient gliding speeds calculated from at least 20 cells. (d, h, and i) The x,y positions of representative wt cells developed in the context of other wt cells and csgA cells developed in the context of other csgA cells. Each trajectory represents the x,y positions of a representative cell as recorded every 15 s during a recording period of 900 s.
Next, the motility behavior of wt cells was analyzed systematically at different time points during development (Table 1 and Fig. 1). Speed profiles of individual cells showed that, at all time points, cells displayed a range of transient gliding speeds and all cells experienced periods of no active movement (Fig. 1 a, e, and i for representative profiles). Consistently, the distributions of transient gliding speeds were broad at all time-points (Fig. 1 c, g, and k). The distribution changed significantly during development. At 3 h, only 62% of the transient gliding speeds were above the detection limit for active cell movement whereas, at 9 h and 15 h, 93% of the transient gliding speeds were above this limit. Also, more cells were gliding with higher transient gliding speeds, the longer the cells had been starved. The mean gliding speed increased 1.5-fold between 0 and 15 h of starvation. The duration of the mean gliding interval increased 5-fold from 0 and 9 h and then remained constant. Between 0 and 6 h the duration of the mean stop interval decreased 2- to 3-fold and then remained constant. From 0 to 9 h the stop frequency decreased 4- to 5-fold and then remained constant. From 0 to 9 h, the reversal frequency decreased 2-fold and then remained constant. The mean net-gliding distance traveled by cells during the 900 s recording period increased 5- to 6-fold from 0 to 12 h and then remained constant (Fig. 1 d, h, and l for representative trajectories).
Behavior of csgA Cells in the Context of Other csgA Cells.
The behavior of csgA cells during development was determined by codeveloping GFP-expressing csgA cells (DK5208GFP) with non-fluorescent csgA cells (DK5208; Table 1 and Fig. 1). After 0 and 3 h of starvation, these csgA cells displayed a behavior similar to that of wt cells developed in the context of other wt cells (cf. Fig. 1 a and b; Fig. 1 c and d). At later time points, the behavior of these csgA cells deviated from that of these wt cells. The mean gliding speed of csgA cells was similar to that of wt cells at 6, 9 and 12 h and slightly lower at 15 h. Speed profiles (cf. Fig. 1 e and i and Fig. 1 f and j) and distributions of transient gliding speeds (Fig. 1 g and k) reveal that csgA cells displayed more frequent periods of no active movement than wt cells at 9 and 15 h. Moreover, csgA cells displayed significantly fewer periods of high transient gliding speeds than wt cells at 15 h (Fig. 1 g and k). From 0 to 15 h, the mean gliding interval increased only 2-fold in csgA cells. The duration of the mean stop interval decreased to the same extent in csgA cells as in wt cells. The stop frequency decreased only 2-fold between 0 and 15 h in csgA cells compared with the 4- to 5-fold decrease in wt cells. The reversal frequency remained unchanged in csgA cells, contrasting the 2-fold decrease observed in wt cells. The mean net-gliding distance traveled by csgA cells during the 900-s recording period increased 2- to 3-fold compared with the 5- to 6-fold increase in wt cells (Fig. 1 d, h, and l for representative trajectories).
C-Signaling Induces Changes in Motility Behavior.
To determine whether the differences in behavior, between wt codeveloped with other wt cells and csgA cells codeveloped with other csgA cells, were C-signal dependent, two experiments were performed in which wt cells and csgA cells were mixed. In the first experiment, GFP-expressing csgA cells (DK5208GFP) were mixed at a ratio of 1:400 with non-fluorescent wt cells (DK1622), and then the behavior of the GFP-expressing csgA cells was monitored after 9 h of starvation. In this experiment, wt cells rescue the development of csgA cells by extracellular complementation, i.e., the wt cells provide the csgA cells with the missing C-signal (12, 20). In this experiment, the behavior of the csgA cells was indistinguishable from that of wt cells starved for 9 h in the context of other wt cells (Table 2; cf. Table 1). Thus, the behavioral defects of csgA cells were corrected by codevelopment with wt cells.
Table 2.
Motility parameters of csgA and wt cells developed in the context of wt and csgA cells, respectively
| Motility parameter | Cell types mixed** | |
|---|---|---|
| Gliding speed, μm/min* | csgAGFP + wt | 2.59 ± 0.17 |
| wtGFP + csgA | 1.91 ± 0.21 | |
| Duration of gliding interval, min† | csgAGFP + wt | 4.02 ± 0.24 |
| wtGFP + csgA | 1.87 ± 0.19 | |
| Duration of stop interval, min‡ | csgAGFP + wt | 0.52 ± 0.12 |
| wtGFP + csgA | 0.59 ± 0.09 | |
| Stop frequency, stop/min§ | csgAGFP + wt | 0.22 ± 0.05 |
| wtGFP + csgA | 0.71 ± 0.04 | |
| Reversal frequency, reversal/min¶ | csgAGFP + wt | 0.037 ± 0.008 |
| wtGFP + csgA | 0.095 ± 0.002 | |
| Net gliding distance, μm∥ | csgAGFP + wt | 23.15 ± 2.47 |
| wtGFP + csgA | 11.78 ± 1.94 |
Mean values and SEM are shown for three independent experiments. Cells were monitored after starvation for 9 hr. Strains used: wt, DK1622; wtGFP, DK1622GFP; csgA, DK5208; csgAGFP, DK5208GFP.
†P^§¶∥ See Table 1.
Cells expressing GFP were mixed at a ratio of 1:400 with nonfluorescent cells.
In the reciprocal experiment, GFP-expressing wt cells (DK1622GFP) were mixed at a ratio of 1:400 with non-fluorescent csgA cells (DK5208). In this mixture, wt cells do not form fruiting bodies and are unable to rescue the development of csgA cells by extracellular complementation (data not shown), presumably because only 1 cell per 400 is wt. In the context of csgA cells, GFP-expressing wt cells after 9 h of starvation displayed a behavior similar to that of csgA cells developed in the context of other csgA cells (Table 2; cf. Table 1). Thus, these wt cells display behavioral defects similar to that of csgA cells developed in a csgA context. From these experiments we conclude that it is not sufficient for a cell to be csgA+ to display the full set of behavioral changes during development. Rather, cells need to be in a C-signaling context to display the full set of behavioral changes.
Discussion
To identify the motility parameters controlled by C-signal during fruiting body formation and to further our understanding of how aggregation is accomplished, we carried out a systematic, quantitative analysis of the behavior of M. xanthus cells during the aggregation stage of fruiting body formation. The motility behavior of individual cells was monitored in the context of the high cell density required for fruiting body formation. The experiments show that wt cells developed in the context of other wt cells and csgA cells developed in the context of other csgA cells behave similarly until 3 h. From 6 h on, these wt and csgA cells behaved differently, and csgA cells only displayed a subset of the behavioral changes displayed by wt cells. The behavioral defects in csgA cells were corrected by developing csgA cells in the context of a 400-fold excess of wt cells. And vice versa, wt cells developed in a context of a 400-fold excess of csgA cells behaved in a manner similar to that of csgA cells developed in the context of other csgA cells. From these observations, we conclude that cell behavior is independent of the csgA genotype. Rather, cell behavior is context dependent, and cells display the full set of behavioral changes only when they are developed in a C-signaling context. These observations strongly support a model in which cell behavior is regulated by the C-signal from 6 h. From a comparison of the behavior of cells in C-signaling and non-C-signaling contexts, we conclude that the C-signal induces a motility response, which includes increases in transient gliding speeds and in the duration of gliding intervals and decreases in the stop and reversal frequencies. In addition, starvation per se induces a motility response, which includes increases in transient gliding speeds and in the duration of the gliding intervals as well as decreases in the duration of stop intervals and in the stop frequency.
The C-signal also induces a motility response in isolated cells (25) that is similar to that observed at the high cell density as reported here except that, in isolated cells, C-signal does not cause a decrease in the reversal frequency. Thus, the C-signal induces two types of motility responses: a cell density-independent response, which includes increases in transient gliding speeds and in the duration of the mean gliding interval and a decrease in the stop frequency, and a cell density-dependent motility response, which includes a decrease in the reversal frequency. The C-signal excites the Frz signal transduction system and causes methylation of the FrzCD protein (31), a cytoplasmic methyl-accepting chemotaxis protein (32). Increased methylation of FrzCD correlates with a decrease in the reversal frequency (33, 34). These observations are consistent with the finding reported here that C-signal induces a decrease in the reversal frequency.
Two genetic systems govern motility in M. xanthus (35). The social-motility system controls gliding of groups of cells, and the adventurous-motility system controls gliding of single cells. Cells starved for 24 to 48 h display a low reversal frequency contingent on an intact social-motility system and on a high cell density (8, 9). We, therefore, suggest that cell–cell interactions involved in social motility are required in order for the C-signal to manifest its effect on the reversal frequency.
The effect of the C-signal-dependent changes in motility parameters is a switch from an oscillatory to a unidirectional type of motility behavior in which cells travel longer net-distances per minute (net-distance calculated as the geometrical distance between the position of a cell at t = 0 s and at t = 900 s). Wt cells starved for 15 h in a context of other wt cells are predicted to travel ≈15-fold longer than similar cells starved for 0 h (1.5-fold increase in mean gliding speed, 5-fold increase in gliding interval, and a 2-fold decrease in reversal frequency). However, as cells move in meandering trajectories rather than in straight lines (cf. Fig. 1 d, h, and l), the mean net-distance traveled by wt cells is observed to increase only ≈6-fold from 0 to 15 h. How, then, are the C-signal-dependent changes in behavior coupled to the directional movements that lead to aggregation?
In order for bacterial cells to display directional movements, they need to be able to regulate their direction-changing frequency appropriately. In swimming bacteria, the direction-changing event, which is regulated during chemotaxis, is either a reversal, a stop, or a tumble in combination with buffeting by Brownian motion (36). Of the motility parameters controlled by the C-signal, only a reversal results in a change in the direction of movement. A stop does not change the direction of movement because M. xanthus cells adhere to each other and to the surface and are not buffeted by Brownian motion. We propose that the C-signal-dependent regulation of the reversal frequency is essential for aggregation and that the remaining C-signal-dependent changes in motility parameters contribute to aggregation by increasing the net-distance traveled by starving cells per minute. Consistently, the motility response induced by starvation per se, i.e., in the absence of C-signaling, is not sufficient to induce aggregation. This motility response represents a subset of the response observed in C-signaling cells and does not include a decrease in the reversal frequency.
On the basis of the results reported here and the suggestion that C-signal transmission occurs by a contact-dependent mechanism involving specific end-to-end contacts between cells, we propose the following model for symmetry-breaking and aggregation. The basic event in this model is an end-to-end contact between two cells with C-signal transmission. Cells engaged in end-to-end contacts with C-signal transmission acquire the ability to travel longer net-distances for as long as they maintain the end-to-end contact with C-signal transmission, i.e., as long as they move in the same direction. These end-to-end contacts are predicted to occur frequently at the high cell density required for fruiting body formation. Sequential end-to-end contacts with C-signal transmission between cells in a field of starving cells are predicted to result in the formation of chains of cells. Cells in a chain are moving with the same speed and in the same direction. The direction of movement of a chain is determined by the cell at the leading end of the chain and is conveyed from cell to cell by the end-to-end contacts. At the high cell density, cells tend to stay in side-by-side contact with each other (21, 23, 24). Cell movement in chains may create alignment of neighboring cells, thus giving rise to the formation of secondary chains, which are associated with the initiating chains by the lateral side-by-side contacts. Together, an initiating chain and the associated secondary chains are predicted to result in the formation of the streams. With the formation of chains and streams, symmetry is broken. Aggregation centers are predicted to be established by the coalescence of streams or by streams turning on themselves in spiral movements, thus, essentially trapping cells in circular tracks (37). Once the leading end of a stream is trapped in an aggregation center, the remaining cells in that stream follow the leading cell into that aggregation center. As cells accrete in aggregation centers, they continue to move with a low reversal frequency in circular tracks (8). Gradually, aggregates enlarge into hemispheres. As the high level of C-signaling required for induction of sporulation is attained by cells inside the aggregates, differentiation of the rod-shaped cells to non-motile spores brings an end to active cell movements and stabilizes the aggregates. As long as cells are motile, the aggregates are unstable because a change from a circular to a more straight trajectory of a stream trapped in an aggregation center is predicted to drain that center of cells and, thus, result in the dissipation of the aggregation center.
This model accounts for several experimental observations, including the cellular organization of streams, stream movement before the appearance of aggregation centers, and why cells enter aggregation centers organized in streams, as well as why some aggregation centers dissipate (7–10). In the model, C-signal transmission is a local event between two cell ends that occurs without reference to the global pattern, and the result is a global organization of cells in a field. Therefore, C-signal-induced aggregation is a self-organizing process.
Formally, C-signal-induced aggregation in M. xanthus is analogous to chemotaxis toward an attractant in Escherichia coli (36). C-signal regulates the direction-changing event as does an attractant. Direction changing in M. xanthus, i.e., a reversal, is followed by a long gliding period (intermittently interrupted by stops, which do not have an effect on the direction of movement). In E. coli, direction changing is followed by a run when cells are moving up a gradient of attractant. Moreover, both responses involve methylation of a methyl-accepting chemotaxis protein. The important difference is that, in E. coli, as in other swimming bacteria, the attractant is highly diffusible whereas C-signal is cell surface associated and, thus, nondiffusible. Why has M. xanthus adopted this strategy rather than using a highly diffusible aggregation signal? E. coli swims with a speed in the order of 20 μm/s whereas M. xanthus is slow-moving, with the highest mean gliding speed reaching 2.66 ± 0.05 μm/min after 15 h (cf. Table 1). Therefore, it has been argued that the directive properties of a highly diffusible aggregation signal might disperse before M. xanthus cells could reorient in a gradient of such a signal (38). The specific quality of a cell surface-associated signal is that it moves with the same speed as cells and then the direction of movement is relayed from cell to cell by the end-to-end cell contacts. In conclusion, the model described here represents a pattern formation mechanism that is distinct from other models of pattern formation such as chemical prepattern models (39, 40) or models based on chemotaxis in response to a diffusible molecule, as in Dictyostelium discoideum fruiting body formation (41) and chemotaxis in swimming bacteria (36), in that the organizing signal is cell surface associated and the direction of cell migration is relayed from cell to cell by the direct cell contacts.
Interestingly, C-signal is also involved in a type of organized cell movements called rippling (19, 22, 27, 42). Rippling occurs only under certain starvation conditions. During rippling, cells organize in a series of equidistant ridge-like structures that move in a pattern resembling ripples on a water surface. Rippling is induced at lower levels of C-signaling than aggregation (14, 15). Kaiser and coworkers (22, 27) have provided evidence that C-signal transmission stimulates cell reversals during rippling. These observations taken together with the observations reported here suggest that the C-signal may induce two types of motility responses depending on signaling levels. At the macroscopic level, the outcomes of these two responses are rippling at low levels of C-signaling and aggregation at higher levels of C-signaling. In this model, C-signal is a bi-modal morphogen, and the pattern-formation properties of the C-signal depend on signaling levels.
Acknowledgments
We thank Roy Welch and Dale Kaiser for stimulating discussions and for providing us with the strain containing the pilAp-gfp fusion. We thank Dale Kaiser for use of equipment and Mandy Ward and Raymond Cox for many helpful comments on the manuscript. This work was supported by the Danish Natural Science Research Council, the Female Researchers in Joint Action program, and the Carlsberg Foundation.
Abbreviations
- wt
wild type
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Le Douarin N M. Cell. 1984;38:353–360. doi: 10.1016/0092-8674(84)90490-2. [DOI] [PubMed] [Google Scholar]
- 2.De Felici M, Dolci S, Pesce M. Int J Dev Biol. 1992;36:205–213. [PubMed] [Google Scholar]
- 3.Melchers F, Rolink A G, Schaniel C. Cell. 1999;99:351–354. doi: 10.1016/s0092-8674(00)81521-4. [DOI] [PubMed] [Google Scholar]
- 4.Dworkin M. Microbiol Rev. 1996;60:70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spormann A M. Microbiol Mol Biol Rev. 1999;63:621–641. doi: 10.1128/mmbr.63.3.621-641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuner J M, Kaiser D. J Bacteriol. 1982;151:458–461. doi: 10.1128/jb.151.1.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Connor K A, Zusman D R. J Bacteriol. 1989;171:6013–6024. doi: 10.1128/jb.171.11.6013-6024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sager B, Kaiser D. Proc Natl Acad Sci USA. 1993;90:3690–3694. doi: 10.1073/pnas.90.8.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi W, Ngok F K, Zusman D R. Proc Natl Acad Sci, USA. 1996;93:4142–4146. doi: 10.1073/pnas.93.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reichenbach H, Heunert H H, Kuczka H. Encyclopedia Cinematographica, Film C 893. Göttingen, Germany: Inst. Wissensch. Film; 1965. [Google Scholar]
- 11.Shimkets L J, Rafiee H. J Bacteriol. 1990;172:5299–5306. doi: 10.1128/jb.172.9.5299-5306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S K, Kaiser D. Cell. 1990;61:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- 13.Kim S K, Kaiser D. J Bacteriol. 1991;173:1722–1728. doi: 10.1128/jb.173.5.1722-1728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Lee B-U, Shimkets L J. Genes Dev. 1992;6:401–410. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- 15.Kruse T, Lobedanz L, Berthelsen N M S, Søgaard-Andersen L. Mol Microbiol. 2001;40:156–168. doi: 10.1046/j.1365-2958.2001.02365.x. [DOI] [PubMed] [Google Scholar]
- 16.Gronewold T M A, Kaiser D. Mol Microbiol. 2001;40:744–756. doi: 10.1046/j.1365-2958.2001.02428.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim S K, Kaiser D, Kuspa A. Annu Rev Microbiol. 1992;46:117–139. doi: 10.1146/annurev.mi.46.100192.001001. [DOI] [PubMed] [Google Scholar]
- 18.Downard J, Ramaswamy S V, Kil K S. J Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimkets L J, Gill R E, Kaiser D. Proc Natl Acad Sci USA. 1983;80:1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagen D C, Bretscher A P, Kaiser D. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 21.Kim S K, Kaiser D. Science. 1990;249:926–928. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- 22.Sager B, Kaiser D. Genes Dev. 1994;8:2793–2804. doi: 10.1101/gad.8.23.2793. [DOI] [PubMed] [Google Scholar]
- 23.Kroos L, Hartzell P, Stephens K, Kaiser D. Genes Dev. 1988;2:1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- 24.Julien B, Kaiser A D, Garza A. Proc Natl Acad Sci USA. 2000;97:9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jelsbak L, Søgaard-Andersen L. Proc Natl Acad Sci USA. 1999;96:5031–5036. doi: 10.1073/pnas.96.9.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser D. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welch R, Kaiser D. Proc Natl Acad Sci USA. 2001;98:14907–14912. doi: 10.1073/pnas.261574598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroos L, Kaiser D. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 29.Hodgkin J, Kaiser D. Proc Natl Acad Sci USA. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zar J H. Biostatistical Analysis. Englewood Cliffs, NJ: Prentice-Hall; 1996. [Google Scholar]
- 31.Søgaard-Andersen L, Kaiser D. Proc Natl Acad Sci USA. 1996;93:2675–2679. doi: 10.1073/pnas.93.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride M J, Weinberg R A, Zusman D R. Proc Natl Acad Sci USA. 1989;86:424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride M J, Köhler T, Zusman D R. J Bacteriol. 1992;174:4246–4257. doi: 10.1128/jb.174.13.4246-4257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi W, Köhler T, Zusman D R. Mol Microbiol. 1993;9:601–611. doi: 10.1111/j.1365-2958.1993.tb01720.x. [DOI] [PubMed] [Google Scholar]
- 35.Hodgkin J, Kaiser D. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 36.Armitage J P. Sci Progress. 1992;76:451–477. [PubMed] [Google Scholar]
- 37.White D. In: Myxospore and Fruiting Body Morphogenesis. Dworkin M, Kaiser D, editors. Washington, DC: Am. Soc. Microbiol.; 1993. pp. 307–332. [Google Scholar]
- 38.Kim S K. Trends Genet. 1991;7:361–365. doi: 10.1016/0168-9525(91)90256-p. [DOI] [PubMed] [Google Scholar]
- 39.Turing A M. Philos Trans R Soc London. 1952;237:37–72. [Google Scholar]
- 40.Wolpert L. J Theor Biol. 1969;25:1–47. doi: 10.1016/s0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- 41.Devreotes P N. Neuron. 1994;12:235–241. doi: 10.1016/0896-6273(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 42.Shimkets L J, Kaiser D. J Bacteriol. 1982;152:451–461. doi: 10.1128/jb.152.1.451-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]