Abstract
Habitat destruction and the resultant fragmentation of the remaining forest are a common phenomenon in the tropics. Most investigations emphasize the potential dangers of fragmentation in isolating patches of forest and exposing populations to loss of species diversity through founder effects, genetic drift, inbreeding, and restricted gene flow. However, a limited number of studies have shown that gene flow may be extensive in tropical trees, suggesting that it may occur between forest fragments and also “isolated” remnant trees. There is an urgent need to quantify pollen flow within and between forest fragments to test the veracity of such views and determine the genetic value of such fragments for in situ conservation. Microsatellite markers are used to genotype individuals of Swietenia humilis from a highly fragmented forest mosaic to directly quantify pollen-mediated gene flow. Distances of pollen flow more than 10 times greater than previously reported were detected. Our results show that some tropical angiosperm tree species may be much more adaptable and resilient to habitat destruction and fragmentation than previously considered. The description of many remnant trees as isolated or “living dead” may be more a conditioning of human perception than a true reflection of their potential conservation value.
Dramatic increases in deforestation have produced numerous small and fragmented forests throughout the tropics. The possible impacts of such disturbance and fragmentation on the biodiversity of tropical forests have been highlighted with respect to species loss and more recently to a reduction of within-species genetic diversity (1, 2). The impacts of forest disturbance on genetic structure and gene flow within such fragmented landscapes are, however, poorly understood. Fragmentation decreases the size and increases the spatial isolation of a population, and its effects on the dynamics of gene flow may directly influence genetic structure within a forest fragment. Genetic isolation, by means of a curtailment of gene flow between fragments, may ultimately have detrimental consequences on the evolutionary viability of a population, by way of increased levels of inbreeding and random genetic drift (3). Studies of the distribution of genetic diversity within and between tropical forest populations indirectly show the existence of extensive long-distance gene flow, with a greater partitioning of genetic variation within, rather than among, populations (4–6). Allozyme studies of the influence of fragmentation on genetic diversity, through direct estimates of pollen dispersal, have shown a consensus of extensive pollen flow into remnant fragments, which could mitigate the presumed effects of physical isolation (7–9).
However, integral to understanding the genetic impacts of fragmentation, descriptions of actual patterns of pollen flow and the degree of genetic, rather than physical, isolation have eluded researchers because of two main factors, namely (i) the resolution power of genetic markers to discriminate pollen donors and (ii) the initial sampling strategy and degree of fragmentation of the species under investigation. Of these limitations the former has been recognized, prompting the use of more variable genetic markers (10). Use of highly variable microsatellite markers has markedly increased the power to successfully resolve direct distances of pollen dispersal, by providing the highest paternity exclusion probabilities of any markers used in the study of plant populations (11–16). Equally important, however, is the maximum degree of separation between fragments sampled, which limits the detailed understanding of the impacts of fragmentation on the dynamics of pollen flow and the long-term genetic viability of populations.
Here, we report direct measures of the pattern and distances of pollen flow within a highly fragmented population of Swietenia humilis by using microsatellite markers. Populations over much of the range of S. humilis have been reduced and fragmented. Listed in 1973 on Appendix II of the Convention on International Trade in Endangered Species (17), S. humilis has become the focus of increasing conservation concern, and a strategy is required to define how remaining populations may be sustainably used and effectively conserved (18). Our assessment of genetic connectivity between the remnant stands of an intensively sampled highly fragmented forest mosaic provides the opportunity to address the specific question of whether fragmentation over distances of more than 1 km leads to the curtailment of pollen flow within a tropical tree species. Comparison with the patterns of pollen flow within a continuous “control” population will also determine whether forest fragmentation and reduction in population size alter the pattern of pollinator behavior, and ultimately pollen flow.
Materials and Methods
Species.
Swietenia humilis Zuccarini (Meliaceae) is a medium-sized deciduous tree and one of the three species commonly known as true or American mahogany (19). It grows within a very wide ecological range within its native Pacific watershed of Central America and Mexico (20). S. humilis is a monoecious species with unisexual flowers occurring in the same inflorescence (21), and under controlled pollination has been shown to be self-incompatible (D.H.B., unpublished data). The small white flowers are faintly sweetly scented and visited by small butterflies, bees, and other insects. The fruits are erect, strongly wooded, ovoid capsules containing seeds that are wind-dispersed (21).
Study Site.
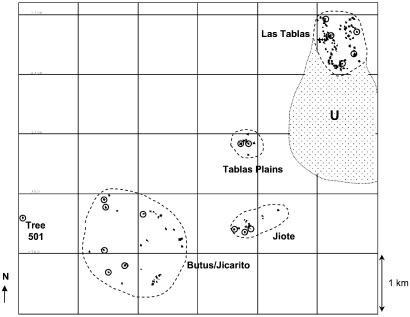
The fragmented population used for this study is located within the native range of S. humilis in the Punta Ratón region of the Honduran Pacific alluvial coastal plains. The study site is described in detail in G.M.W. et al. (6). It consists of two remnant stands of secondary dry deciduous forest principally confined to hillside areas [Jiote (n = 22 trees) and Butus/Jicarito (n = 44)] and one stand of remnant trees in pasture [Tablas Plains (n = 7)]. A plot of 97 trees within an approximately 68-hectare (ha) area of the Las Tablas Forest (total area ≈500 ha) was used as a control of continuous forest. The Las Tablas site lies within the largest area of dry forest in the Punta Ratón region and is predominantly secondary in nature. The relative locations of all of the mapped trees are shown in Fig. 1. To investigate the dynamics of pollen flow, 17 trees were selected for progeny analysis from across the population; 12 trees from the fragmented stands, including an isolated tree (no. 501) located at the most westerly point of S. humilis distribution within the area, and 5 trees from within the control subpopulation Las Tablas (the selected trees are encircled in Fig. 1).
Figure 1.
The relative locations of S. humilis trees (each represented by a dot) sampled in Punta Ratón, Choluteca, Honduras. Each fragment is enclosed with a hatched line and labeled accordingly. The location of the unsampled trees (U) adjacent to the Las Tablas study site is shown by the spotted encased area. The 17 trees selected for the progeny analysis are circled. All trees and progeny were genotyped at four microsatellite loci, and a paternity exclusion analysis was performed to identify the subsets of fathers for each progeny, the fractional likelihood of paternity, and distance from the tree.
Sample Collection and Microsatellite Analysis.
Bulked progeny arrays were collected from each tree in the field during 1994. DNA was extracted directly from 30 seeds per tree, using a modification of the extraction method of Doyle and Doyle (22). The starting material was 10–50 mg of seed tissue and a phenol:chloroform wash step was incorporated into the protocol after suspension of the DNA in 500 μl of TE buffer (10 mM Tris/1 mM EDTA, pH 7.4). Four microsatellite loci, mac38 (26 alleles), mac45 (11 alleles), mac49 (21 alleles), and mac58 (13 alleles) were used in this study; selected from a larger pool of loci based on their detection of high levels of allelic polymorphism, with a relatively high frequency of rare alleles throughout the study area (6). PCR conditions and product resolution were as described by G.M.W. and W.P. (23). Allelic products were scored according to their size in base pairs and showed Mendelian segregation patterns based on analysis of control pollinated full-sib progeny arrays. All of the trees within the fragmented stands and the control had been genotyped at each of the microsatellite loci (6).
Data Analysis.
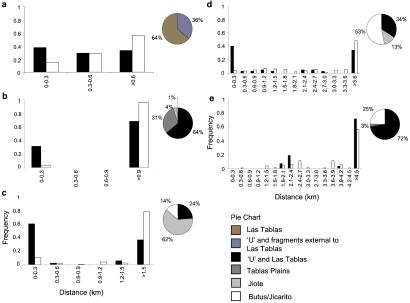
The dynamics of pollen flow were estimated by using a fractional paternity program (24) to analyze the progeny array data from each tree. With all parental genotypes known, the exclusion analysis identified subsets of fathers for each progeny, giving each pollen donor a fractional likelihood of paternity and its distance from the maternal tree. A constraint within the study population was an area of unsampled trees adjacent to the Las Tablas site, introducing an area of ungenotyped potential pollen donors between Las Tablas and the fragmented stands of trees (Fig. 1; area “U”). Specification of pollen donor distances could therefore be accurately characterized only up to this unmapped area. The frequency of progeny arising from pollen donors potentially within this area U were summed for each fragment and plotted as originating from a distance equal to or greater than the maximum specified limit. Distance limits for pollen flow characterization were 0.6 km for Las Tablas, 0.9 km for Tablas Plains, 1.5 km for Jiote, 3.6 km for Butus/Jicarito, and 4.5 km for Tree 501. Parent–offspring matches with a likelihood of 1.0 indicated a 100% certainty of paternal assignment, providing exact distances of pollen flow to the maternal trees. The frequency of the definite paternal donors was determined for 0.3-km distance categories from the maternal tree, which was calculated as the number of definites against the number of definites plus the number of progeny in that array for which the pollen source could not have been the father. The data were plotted per fragment, based on the cumulative frequencies per distance class within each fragment, together with the frequency of trees within each distance category, generating direct pollen flow-distance-frequency histograms (Fig. 2). All distance-frequency histograms were plotted as a proportion of 1.0, allowing comparisons of pollen flow among the trees and fragments, irrespective of the number of progeny assessed.
Figure 2.
Distance-frequency histograms of direct pollen flow to selected S. humilis trees in each forest fragment (Punta Ratón, Choluteca, Honduras). The distance of pollen flow to the 17 maternal trees was determined by using a fractional paternity program. With all parental genotypes known, the paternity analysis identified parental donors, their fractional likelihood of paternity, and distance from the mother tree. The frequency of definite pollen donors, paternal likelihood of 100%, was calculated for 0.3-km distance classes from the maternal tree (solid bars) for each fragment [(a) Las Tablas, (b) Tablas Plains, (c) Jiote, (d) Butus/Jicarito, and (e) tree 501] and plotted together with the frequency of trees within each distance class (empty bars). The pie charts show the proportion and source of the pollen flow to the trees within each fragment.
Results
The high exclusion powers of the microsatellite markers (combined expected exclusion probability of 0.983; mac38 0.781, mac45 0.496, mac49 0.577, and mac58 0.642) resulted in unique paternal assignments for 45% of the parent–offspring matches, giving exact distances of pollen flow to the maternal trees. The distance frequency histograms of pollen flow per fragment are shown in Fig. 2. Common to each fragment (Fig. 2 a–d) was a high frequency of pollen flow from within the first 0.3 km of the maternal tree, indicating a predominance of near-neighbor gene exchange. In the Las Tablas control population (Fig. 2a), the source of the majority of pollen flow (64%) was from within the first 600 m. In Tablas Plains (Fig. 2b), just under a third of the pollen flow (31%) was from inside the fragment, within 300 m of the maternal trees. Pollen flow was not evident from between 300–900 m, coinciding with the intervening space (with no S. humilis trees) between Tablas Plains and neighboring fragments. The remaining 69% was from distances greater than 900 m, 4% of which was from Jiote and 1% from Butus/Jicarito, but the highest proportion (64%) was from Las Tablas or the unsampled area. In both the Jiote and Butus/Jicarito fragments, the highest proportion of pollen again came from within the fragment. Jiote (Fig. 2c) had a small proportion (4%) of the fathers in Butus/Jicarito, 1.1 km away, but 24% came from distances greater than 1.5 km. A similar pattern was evident in Butus/Jicarito (Fig. 2d) with 13% of the pollen from Jiote, but 34% from distances greater than 3.6 km, in the Las Tablas area.
The isolated tree, no. 501 (Fig. 2e), enabled characterization of pollen flow from up to 4.5 km. Pollen flow was evident from across this range, with the pattern depending, to some degree, on the frequency of trees rather than the distance. The neighboring fragment, Butus/Jicarito, provides the nearest pollen source (26%). Tablas Plains also contributes a small proportion (3%) but the majority of the pollen (71%) originated from either Las Tablas or the unsampled area, at a distance of over 4.5 km away. Tree 500, the nearest neighbor to 501, located only 402 m away, was not found to be a pollen donor, presumably as it is a much smaller tree and only recently reached flowering maturity. Tree 501 was also a potential father to progeny of tree 510 in Butus/Jicarito, trees 533 and 536 in Jiote, and tree 609 in Las Tablas.
The number of trees in a fragment is shown to influence patterns of pollen flow within and between the remnant stands (Table 1), with the mean percentage of pollen flow into fragments generally increasing with a reduction in subpopulation size. The smallest “fragment,” tree 501, had 100% external pollen sources in the pollen distribution curve, indicating that all of the progeny under study resulted from outcrossed mating. The largest mean distance of pollen flow detected for each fragment exceeded that to the nearest fragment, showing an extensive network of gene exchange between the neighboring fragments.
Table 1.
Pollen flow into fragments of S. humilis, Punta Ratón, Choluteca, Honduras
| Fragment | Sample size | Separation distance from next fragment, km | Mean % “into fragment” pollen flow | Mean largest minimum pollen flow distance, km |
|---|---|---|---|---|
| Las Tablas | 97 | — | 36.0 | —* |
| Butus/Jicarito | 44 | 1.1 | 47.0 | 3.1 |
| Jiote | 22 | 1.1 | 38.3 | 1.7 |
| Tablas plains | 7 | 1.2 | 68.4 | 1.6 |
| Tree 501† | 1 | 1.4 | 100.0 | >4.5 |
The sample size of each fragment, the separation distance from the nearest fragment, the mean percentage of external pollen flow, and largest minimum distances of pollen flow are shown per fragment.
Las Tablas is a portion of continuous forest and is adjacent to unsampled trees.
Tree 501 is an isolated tree.
Discussion
The impact of forest fragmentation on pollen movement can be inferred by comparing pollen flow within and between the fragments to that within Las Tablas. Despite the high level of fragmentation and small size of the subpopulations there was an extensive network of gene exchange over the spatial scale of the study site. However, the pattern of pollen movement to the sampled trees depended on the spatial structure of the particular fragment, with subpopulation size (number of trees) a critical factor, directly influencing the proportion of immigrant pollen. Although the trend of near-neighbor mating was maintained within the fragments, reductions in subpopulation size were paralleled by an increase in the proportion of long-distance pollen flow, from outside the fragment. The isolated tree 501 proves an interesting situation. Separated from the nearest fragment of flowering trees by 1.1 km, the direct pollen flow measures supersede this distance, with over 70% coming from the maximum distance category (>4.5 km). Totally outcrossing with a wide array of pollen donors, it contrasts with the predictions of Murawski and Hamrick (25), who suggested that spatially isolated trees are more likely to deviate from random mating and presumably receive pollen from fewer donors. In fact, the increase in spatial isolation has promoted long-distance gene exchange between tree 501 and all of the fragments. Together with its potential role as a father to progeny in other fragments, tree 501 proves to be an integral part of the population and challenges the living dead title given to the conservation value of such remnant trees (26).
Direct distances of gene flow have been determined in only a few studies of tropical tree species. Boshier et al. (27) found pollen movement in Cordia alliodora by the detection of rare isozyme alleles as far as 280 m. Microsatellite analysis of pollen movement in Pithecellobium elegans (13) and Gliricidia sepium (14) detected maximum distances of 350 and 275 m, respectively. Long distances of pollen movement over the same scale as that detected in this study have been inferred only for species of Ficus, where estimates of effective population size, utilizing isozyme markers, suggested typical pollen dispersal distances of 6–14 km (7). The lack of variability detected by isozymes, together with the scale over which sampling of trees occurred, has constrained the power of resolution of gene flow. In this study, sampling a fragmented population over a much greater spatial scale compared with most studies (9), combined with the use of hypervariable microsatellite markers, provided the opportunity to detect extensive pollen movement in S. humilis and the furthest direct distance of pollen flow detected to date in tropical trees (>4.5 km). Furthermore, the pollen flow distances identified may be an underestimation of the true distances within the population owing to the constraints of the unsampled area of forest between the fragments and the control subpopulation Las Tablas. External gametes that mimic local ones may also be misidentified, such that progeny identified as possible with a given set of fathers may in fact have been sired from outside that same sample (12, 28). Correction for such cryptic pollen flow gave a ratio of total to apparent gene flow of 1.8:1. However, the spatial layout of the fragments and the unsampled area, mean that any cryptic pollen flow may well come from the same distances and would not alter the rank order of the pollen flow measures, nor the conclusions of the study. Pollen flow from the unsampled area would also account for the relatively low percentage of progeny assigned unique paternity, despite the high combined exclusion probability of the markers.
The increase in pollen flow seen here, as a consequence of fragmentation, is contrary to the traditional view of the genetic effects of physically isolating populations, where an increase in spatial isolation and reduction in population sizes have been considered (29, 30) and shown in some species to reduce interfragment gene flow (31–34). Conversely, this study indicates that for S. humilis, fragmentation does not impose a genetic barrier between stands but instead promotes increased levels of interfragment gene flow over longer distances. The promotion of increased external pollen flow, coupled with a reduction in population sizes, has also been shown for another tropical tree, Spondias mombin (8). Allozyme markers indicated higher rates of pollen flow into small “island” populations, at distances up to 1 km, than into continuous and large island populations. While in three fragmentation studies (35–37) of the temperate tree Acer saccharum, reductions in allozyme genetic differentiation among patches compared with continuous forest indirectly suggested that fragmentation may facilitate increased gene flow by means of wind dispersal of pollen.
Inter-fragment pollen flow is facilitated by the ability of a vector to move pollen between spatially isolated fragments of trees. The capability of insect pollinators to move long distances has been shown (38), although the potential to move among stands depends on their behavioral response to habitat fragmentation. Changes in pollinator assemblages in fragments could strongly affect patterns of gene flow and genetic variation within remnant tree populations. Mark and recapture experiments on euglossine bees in remnant stands of Brazilian wet forest showed the bees were able to traverse distances up to 4 km among patches of isolated trees (39). Nason et al. (40) implied that, in contrast to most species, the genus Ficus with its species-specific wasp pollinator could apparently form extensive metapopulations in fragmented landscapes. The small generalist insect pollinators, including bees, of S. humilis are shown in this study to be able to fly across the large interfragment distances, providing genetic connectivity among stands. The distance of pollinator movement cannot, however, be specifically inferred, as it is probable that “carry-over” is also a contributing factor to long-distance pollen flow (41–43). With a range of nonspecialist pollinators, S. humilis is probably much less susceptible to the effects of habitat disturbance on the dynamics of pollen flow, compared with tree species with more specialist pollinator relationships. However, it is likely that all pollinators have a distance threshold beyond which they will not move. This threshold being pollinator-specific implies that the extent to which fragmentation affects trees may be highly variable among species.
Different patterns of pollinator foraging, as detected by variable patterns of pollen flow, depend on the spatial structure of the subpopulation. Stacey et al. (44) found that populations that had a lot of clumping of reproductive trees had a predominance of near-neighbor interactions, with an increase in pollinator distance mirrored by a decrease in density of the conspecific flowering trees. Also, Ellstrand et al. (45) found outcrossing in populations of Helianthus annuus, which are primarily pollinated by bees, to be negatively correlated with plant density. If pollinator flight distances are density- or number-dependent in S. humilis, the reduction in the number of flowering trees in the fragments may promote interfragment pollinator flight, and hence pollen flow among the smaller stands. Pollinator changes in fragmented landscapes may affect patterns of gene flow and reproduction in remnant tree populations. A decline in pollinator populations in such altered agroecosystems may eventually limit tree reproduction and therefore monitoring of pollinator numbers, as well as evidence of pollinator limitation (46), is needed.
The practical implications of our findings are also important. Policy makers more accustomed to traditional in situ approaches to conservation need to be aware of the importance and complementary role that remnant forest patches and trees play in providing connectivity and enhancing population variability. Similarly, such forests and trees should not be ignored as a source of germplasm for ex situ conservation or breeding programs. Self-compatible species, which normally show mixed mating (25), are likely to exhibit increased levels of inbreeding at much shorter distances of separation than self-incompatible species. Establishment of the presence or absence of self-incompatibility mechanisms (47) will provide a cheap and effective indication of a species susceptibility to increases in inbreeding after fragmentation and provide criteria for the inclusion of remnant trees in seed collection for ex situ conservation.
It is evident from this study that forest fragmentation does effect the dynamics of pollen flow of the tropical tree S. humilis, but not by imposing a genetic barrier between fragments. Fragmentation and spatial isolation over the scale studied here have promoted an increase in the frequency of long-distance pollen movement. The fragmented landscape persists as a metapopulation, with the remnant stands fragmented at a finer scale than that of the breeding associations. The fragments examined in this study did show reduced genetic diversity through the loss of lower frequency alleles (6) in agreement with results from previous studies of trees and other plants (33, 34). However, the altered patterns of interfragment gene flow over time will promote increased genetic mixing across this spatial scale and potentially restore, maintain, or even augment the levels of genetic variation of S. humilis within this modified ecosystem. Although the genetic impacts of fragmentation are undoubtedly complex and will vary among species, this study has clearly demonstrated that remnant fragmented stands and isolated trees of S. humilis may provide a buffer to the deleterious genetic consequences of habitat destruction and may be vital to the future long-term viability of the species.
Acknowledgments
Many organizations and people helped with the field and laboratory work related to this study. We thank Conservation and Silviculture of Honduran Dry Forest Species/Honduran Corporation of Forest Development for facilitating the fieldwork in Honduras, Sea Farms, S.A.; and Srs. Cornelio and Hernan Corrales, Sr. Hernan Videl, Sr. Enrique Weddle for access to their land. We thank José Dimas Rodriguez and Modesto Castillo for their help in seed and leaf collection, and Allan Booth for his assistance in both the DNA extractions and running the gels. This publication is an output from research projects funded by the United Kingdom Department for International Development (DFID) Forestry Research Program Grants R5729, R6080, and R6516. W.P. is funded by the Scottish Executive Environment and Rural Affairs Department.
References
- 1.Bawa K S. In: Proceedings on International Symposium on Genetic Conservation and Production of Tropical Forest Tree Seeds. Drysdale R M, John S E T, Yappa A C, editors. Muak-lek, Thailand: Assoc. South East Asian Nat.–Canada; 1994. pp. 10–16. [Google Scholar]
- 2.Laurance W F, Bierregaard R O, editors. Tropical Forest Remnants, Ecology, Management and Conservation of Fragmented Communities. Chicago: University of Chicago Press; 1997. [Google Scholar]
- 3.Young A G, Boyle T, Brown T. Trends Ecol Evol. 1996;11:413–418. doi: 10.1016/0169-5347(96)10045-8. [DOI] [PubMed] [Google Scholar]
- 4.Hamrick J L, Loveless M D. In: The Evolutionary Ecology of Plants. Bock J H, Linhart Y B, editors. Boulder, CO: Westview Press; 1989. pp. 129–146. [Google Scholar]
- 5.Hamrick J L, Murawski D A. J Tropical Ecol. 1991;7:395–399. [Google Scholar]
- 6.White G M, Boshier D H, Powell W. Mol Ecol. 1999;8:1809–1909. doi: 10.1046/j.1365-294x.1999.00790.x. [DOI] [PubMed] [Google Scholar]
- 7.Nason J D, Herre E A, Hamrick J L. J Biogeogr. 1996;23:501–512. [Google Scholar]
- 8.Nason J D, Hamrick J L. J Hered. 1997;88:264–276. [Google Scholar]
- 9.Kaufman S R, Smouse P E, Alvarez-Buylla E R. Heredity. 1998;81:164–173. [Google Scholar]
- 10.Chase M R, Kesseli R, Bawa K. Am J Bot. 1996;83:51–57. [Google Scholar]
- 11.Dow B D, Ashley M V, Howe H F. Theor Appl Genet. 1995;91:137–141. doi: 10.1007/BF00220870. [DOI] [PubMed] [Google Scholar]
- 12.Dow B D, Ashley M V. Mol Ecol. 1996;5:615–627. [Google Scholar]
- 13.Chase M R, Moller C, Kesseli R, Bawa K S. Nature (London) 1996;383:398–399. [Google Scholar]
- 14.Dawson I K, Waugh R, Simons A J, Powell W. Mol Ecol. 1997;6:179–183. [Google Scholar]
- 15.Aldrich P R, Hamrick J L, Chavarriaga P, Kochert G. Mol Ecol. 1998;7:933–944. doi: 10.1046/j.1365-294x.1998.00396.x. [DOI] [PubMed] [Google Scholar]
- 16.Aldrich P R, Hamrick J L. Science. 1998;281:103–105. doi: 10.1126/science.281.5373.103. [DOI] [PubMed] [Google Scholar]
- 17.CITES, Proposal to the Convention on International Trade in Endangered Species to Include Swietenia macrophylla on Appendix II, March 3, 1973, Washington, DC.
- 18.Newton A C, Cornelius J P, Baker P, Gillies A C M, Hernández M, Ramnarine S, Mesén J F, Watt A D. Bot J Linn Soc. 1996;122:61–73. [Google Scholar]
- 19.Styles B T. In: Flora Neotropica. Pennington T D, Styles B T, Taylor D A H, editors. New York: New York Botanical Garden; 1981. , No. 28, pp. 359–418. [Google Scholar]
- 20.Lamb F B. Mahogany of Tropical America: Its Ecology and Management. Ann Arbor, MI: University of Michigan Press; 1966. [Google Scholar]
- 21.Styles B T. Silvae Genetica. 1972;21:175–182. [Google Scholar]
- 22.Doyle J J, Doyle J L. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 23.White G, Powell W. Mol Ecol. 1997;6:851–860. [Google Scholar]
- 24.Devlin B, Roeder K, Ellstrand N C. Theor Appl Genet. 1988;74:369–380. doi: 10.1007/BF00265336. [DOI] [PubMed] [Google Scholar]
- 25.Murawski D A, Hamrick J L. Biotropica. 1992;24:99–101. [Google Scholar]
- 26.Janzen D H. Oikos. 1986;47:1–2. [Google Scholar]
- 27.Boshier D H, Chase M R, Bawa K S. Am J Bot. 1995;82:484–490. [Google Scholar]
- 28.Devlin B, Ellstrand N C. Evolution (Lawrence, Kans) 1990;44:248–259. doi: 10.1111/j.1558-5646.1990.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 29.Ehrlich P R, Raven P H. Science. 1969;165:1228–1232. doi: 10.1126/science.165.3899.1228. [DOI] [PubMed] [Google Scholar]
- 30.Hamrick J L. In: Resolving Tropical Forest Resource Concerns Through Tree Improvement, Gene Conservation and Domestication of New Species, Proceedings of the International Union of Forestry Research Organization Conference, Cartagena-Cali, Colombia, October, 1992. Lambeth C C, Dvorak W, editors. North Carolina State Univ., Raleigh, NC: Central America and Mexico Coniferous Resources; 1992. pp. 74–82. [Google Scholar]
- 31.Wilcove D S, McLellan C H, Dobson A P. In: Conservation Biology: The Science of Scarcity and Diversity. Soule M E, editor. Sunderland, MA: Sinauer; 1987. pp. 237–256. [Google Scholar]
- 32.Templeton A R, Shaw K, Routman E, Davis S K. Ann Mo Bot Gard. 1990;77:13–27. [Google Scholar]
- 33.Billington H L. Conserv Biol. 1991;5:115–119. [Google Scholar]
- 34.Saunders D A, Hobbs R J, Margules C R. Conserv Biol. 1991;5:18–32. [Google Scholar]
- 35.Fore S A, Hickey R J, Vankat J L, Guttman S I, Schaefer RL. Can J Bot. 1992;70:1659–1668. [Google Scholar]
- 36.Young A G, Merriam H G, Warwick S I. Heredity. 1993;71:227–289. [Google Scholar]
- 37.Ballal S R, Fore S A, Guttman S I. Can J Bot. 1994;72:1311–1315. [Google Scholar]
- 38.Webb C J, Bawa K S. Evolution (Lawrence, Kans) 1983;37:1258–1270. doi: 10.1111/j.1558-5646.1983.tb00242.x. [DOI] [PubMed] [Google Scholar]
- 39.Raw A. Revta Bras Ent. 1989;33:103–107. [Google Scholar]
- 40.Nason J D, Herre E A, Hamrick J L. Nature (London) 1998;391:685–687. [Google Scholar]
- 41.Woodell S R J. The Pollination of Flowers by Insects. 1978. , Linn. Soc. Symp. Ser. 6, ed. Richards, A. J. (Academic, London), pp. 31–39. [Google Scholar]
- 42.Schaal B A. Nature (London) 1980;284:450–451. [Google Scholar]
- 43.Adams W T. New Forests. 1992;6:217–240. [Google Scholar]
- 44.Stacey E A, Hamrick J L, Nason J D, Hubbell S P, Foster R B, Condit R. Am Nat. 1996;48:275–288. [Google Scholar]
- 45.Ellstrand N C, Torres A M, Levin D A. Syst Bot. 1978;3:403–407. [Google Scholar]
- 46.Allen-Wardell G, Bernhardt P, Bitner R, Burquez A, Buchmann S, Cane J, Cox P A, Dalton V, Feinsinger P, Inouye D, et al. Conserv Biol. 1998;12:8–17. [Google Scholar]
- 47.Bawa K S. Evolution (Lawrence, Kans) 1974;28:85–92. doi: 10.1111/j.1558-5646.1974.tb00729.x. [DOI] [PubMed] [Google Scholar]