Abstract
Adult cardiomyocytes are irreversibly postmitotic but respond to a variety of stimuli by hypertrophic growth, which is associated with an increase in cell size and protein content, organization of sarcomeres, and activation of a fetal gene program. Recently, we described a novel cardiac helicase activated by MEF2 protein (CHAMP), which is expressed specifically in the heart throughout prenatal and postnatal development. Here we show that CHAMP acts as an inhibitor of cell proliferation and cardiomyocyte hypertrophy. Ectopic expression of CHAMP inhibits proliferation of HeLa cells and blocks cell cycle entry of serum-stimulated NIH 3T3 cells. Overexpression of CHAMP in primary neonatal cardiomyocytes blocks hypertrophic growth and the induction of fetal genes in response to stimulation by serum and phenylephrine but does not prevent sarcomere organization or early mitogenic signaling events including activation of extracellular signal-regulated kinases or up-regulation of c-fos. Inhibition of cardiomyocyte hypertrophy by CHAMP requires the conserved ATPase domain and is accompanied by up-regulation of the cyclin-dependent protein kinase inhibitor p21CIP1. These findings identify CHAMP as a cardiac-specific suppressor of cardiomyocyte hypertrophy and cell cycle progression and suggest that CHAMP may suppress these processes through the regulation of p21CIP1.
Cardiac myocytes proliferate rapidly during embryogenesis but lose their proliferative capacity soon after birth. However, adult cardiac myocytes retain the ability to respond to a variety of stimuli by hypertrophic growth, which is accompanied by an increase in cell size and protein synthesis, assembly of sarcomeres, and activation of a program of fetal gene expression. Cardiomyocyte hypertrophy occurs in response to numerous agonists, pressure or volume overload on the heart, cytoskeletal abnormalities, and intrinsic defects in contractility (reviewed in refs. 1–3). Although myocyte hypertrophy initially enhances cardiac output in response to stress, prolonged hypertrophy is associated with an increased risk for morbidity and mortality (4).
Numerous studies have demonstrated that the signaling pathways that control proliferation of nonmuscle cells also are involved in the withdrawal of cardiomyocytes from the cell cycle and cardiomyocyte hypertrophy (reviewed in ref. 5). Growth factor signaling pathways that induce hypertrophy are intimately interconnected with intracellular pathways that link changes in calcium homeostasis with the reprogramming of cardiac gene expression (6). The calcium, calmodulin-dependent protein phosphatase calcineurin serves as a point of convergence of these different intracellular pathways (7) and has been shown to be necessary and sufficient for hypertrophy in response to a wide range of signals in vivo and in vitro (ref. 8 and reviewed in refs. 9 and 10).
Mitogenic-signaling pathways drive cell cycle progression as a consequence of their effects on cyclins, which form complexes with cyclin-dependent kinases (CDKs) (reviewed in ref. 11). Growth factors up-regulate the activities of G1 cyclins, which activate CDKs, resulting in phosphorylation of tumor suppressor pocket proteins, such as the retinoblastoma protein, and S-phase entry. CDK inhibitors (CDKIs), such as p21CIP1 and p27KIP1, bind cyclin-CDK complexes and inhibit their growth-promoting activities (12). The progressive loss of proliferative potential of cardiomyocytes during postnatal development correlates with an increase in CDKI expression and a decrease in CDK activity (13, 14). Conversely, knockout mice lacking p27KIP1 show an increase in heart size and a prolonged duration of cardiac proliferation after birth (15–17).
CDKs and CDKIs also have been implicated in cardiac hypertrophy. Aortic constriction, a potent stimulus for hypertrophy, elicits a transient decrease in cardiac expression of p21CIP1 and p27KIP1 and a concomitant increase in CDK activity and expression (18). CDK activity also is stimulated in cardiomyocytes by hypertrophic agonists (19, 20), and the forced expression of CDKIs can prevent agonist-induced hypertrophy in vitro and pressure-overload hypertrophy in vivo (21). These findings are consistent with the notion that hypertrophic growth of cardiomyocytes involves the activation of cell cycle machinery and progression from quiescence to a restriction point in the G1 phase of the cell cycle. However, the blockade to cell cycle progression in cardiomyocytes prevents completion of cell division under most circumstances.
Recently, we described a cardiac-specific RNA helicase, cardiac helicase activated by MEF2 protein (CHAMP), in a screen for genes regulated by the MEF2C transcription factor in the developing heart (22). CHAMP is specifically expressed in the heart during development and adulthood. At embryonic day (E) 15.5, when ventricular cardiomyocytes form finger-like projections, known as trabeculae, CHAMP appears to be expressed preferentially in the trabecular region where the proliferative rate is diminished relative to the adjacent compact zone. Here, we show that ectopic expression of CHAMP inhibits proliferation of noncardiac cells and blocks hypertrophy of primary cardiomyocytes. The antihypertrophic activity of CHAMP requires the conserved ATPase motif that characterizes the RNA helicase superfamily and is associated with the up-regulation of the cell cycle inhibitor p21CIP1. These findings identify CHAMP as a cardiac-specific suppressor of cell proliferation and cardiac hypertrophy and suggest that these activities of CHAMP may be mediated by p21CIP1.
Materials and Methods
Materials.
Phospho-p44/p42 mitogen-activated protein kinase (MAPK) Abs were purchased from Cell Signaling Technology (Beverly, MA). Anti-p21CIP1 Ab was purchased from PharMingen. Rabbit anti-atrial natriuretic factor (ANF) Ab was purchased from Peninsula Laboratories. Anti-α-actinin mAb and anti-tubulin Ab were purchased from Sigma. Rabbit anti-calsarcin Ab and anti-CHAMP Ab have been described (22, 23). All other Abs were purchased from Santa Cruz Biotechnology.
Construction of Adenovirus and Expression Vectors.
A cDNA clone encoding full-length CHAMP with an amino-terminal FLAG epitope tag was cloned into the pcDNA expression vector, as described (22). This cDNA fragment also was used to construct a recombinant adenovirus by using the Adeno-X Tet-off system according to manufacturer's protocols (CLONTECH). Target cells were coinfected with Adeno-X Tet-off virus (adTet-off) and adenovirus encoding FLAG-tagged CHAMP (adCHAMP). Cells were infected with a 1:2 ratio of adCHAMP to adTet-off virus at the multiplicities of infection (MOI) specified in the text. The expression level of CHAMP was controlled by the amount of doxycycline added to the medium with maximum expression being achieved in the absence of doxycycline. Because the basal level of CHAMP expression in the presence of doxycycline (1 μg/ml) had significant effects on HeLa cell proliferation and cardiomyocyte growth, no attempt was made to correlate the levels of exogenous CHAMP expression with its antiproliferative effect on cell growth and no doxycycline was used in the studies reported here. As a control, we routinely infected cells with adenovirus that constitutively expressed β-galactosidase (adβ-gal) at a similar MOI.
A mutant form of CHAMP in which the conserved ATPase domain (DEAGQ) was mutated to GGAAG was generated by using the QuickChange site-directed mutagenesis kit from Stratagene. The pcDNA-FLAG-CHAMP expression vector was used as the parental plasmid for mutagenesis.
Cell Proliferation Assay.
Cell proliferation assays were performed in 96-well microtiter plates by using cell proliferation ELISA, BrdUrd (chemiluminescence) kit (Roche Molecular Biochemicals). HeLa cells were seeded at a density of 0.5 × 104 cells/well in a volume of 100 μl of medium/well and cultivated in DMEM supplemented with 10% FBS. After 24 h, cells were infected with adenovirus at an MOI of 40 overnight at 37°C. The medium was replaced with fresh medium after infection, and cells were incubated for another 48 h. At the end of the incubation, BrdUrd was added to the medium, and cells were incubated for 2 h. At the end of the labeling period, cells were fixed and peroxidase-conjugated anti-BrdUrd Ab was added. Immune complexes were detected by addition of substrate and subsequent quantitation of luminescence by using a microplate luminometer.
Primary Neonatal Rat Cardiomyocyte Cell Culture.
Primary cultures of neonatal rat ventricular cardiomyocytes were prepared as described (8). Twenty-four hours after seeding, infection with adenovirus was carried out in plating medium for 2 h at an MOI of 2. After infection, the culture medium was changed to serum-free medium, and 24 h later hypertrophic stimuli [20 μg/ml phenylephrine (PE), 10% FBS, or 10 μM isopreterenol] were added. Cells were harvested at various time points after hypertrophic stimulation. RNA and protein were isolated for RNA dot blot and Western blot analysis. Only cultures containing >90% cardiomyocytes were used. At an MOI of 2, >90% of cardiomyocytes were infected by adCHAMP.
Measurements of Cell Size.
For cell size measurements, ≈100 cells from each condition were randomly chosen and photographed at ×40. Myocyte cross-sectional areas were measured by using a computerized morphometric system (scion image, National Institutes of Health).
Extracellular Signal-Regulated Kinase (ERK) Activity Assay.
MAPK activities were assayed by using phospho-p42/p44 MAPK (ERK1/2) Abs. Stimulated cardiomyocytes were harvested in SDS sample buffer at various time points. Approximately 20 μg of protein was separated on 10% SDS/PAGE and blotted to nitrocellulose membranes. Two identical blots were incubated with Ab specific for the dually phosphorylated, activated forms of ERK1 and ERK2 (Cell Signaling Technology), and an Ab specific for ERK2 that is independent of its phosphorylation state (Santa Cruz Biotechnology). Signals were detected by using horseradish peroxidase-conjugated secondary Ab and enhanced chemiluminescence (Amersham Pharmacia).
RNA Analysis.
Total RNA was isolated from cultured cardiomyocytes by using Trizol reagent (GIBCO/BRL) according to the manufacturer's instructions. RNA dot blotting was performed with 1 μg of total RNA dotted on nitrocellulose membrane and hybridized against a panel of oligonucleotide probes as described (24). Northern blot analysis with CHAMP and p21CIP1 cDNA probes and reverse transcription–PCR were performed following previously described procedures (22).
Western Blot Analysis.
Extracts from cardiomyocytes or adult mouse hearts containing 20 μg of protein were subjected to SDS/PAGE. Protein was transferred to poly(vinylidene difluoride) membrane and subjected to Western blot analysis with anti-fos Ab, anti-tubulin Ab, and anti-CHAMP Ab as described (22).
Immunofluorescence.
The immunofluoresecence staining of cardiomyocytes was performed as described (22).
Results
Inhibition of Cell Proliferation by CHAMP.
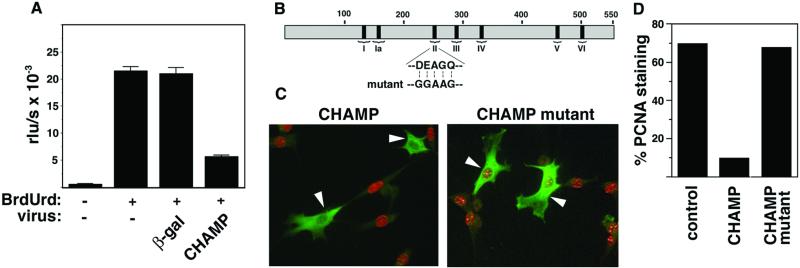
In light of the preferential expression of CHAMP in the trabecular region of the developing heart (22), in which the proliferative rate of cardiomyocytes is reduced relative to the adjacent compact zone (25), we investigated whether CHAMP might suppress cell proliferation. To test this possibility, we expressed CHAMP ectopically in HeLa cells by using an adenoviral expression vehicle and examined the effect on cell proliferation as measured by incorporation of BrdUrd into newly synthesized DNA. As shown in Fig. 1A, BrdUrd incorporation was inhibited by ≈75% in HeLa cells infected with adCHAMP compared to cells expressing adβ-gal as a control (Fig. 1A).
Figure 1.
Inhibition of cell proliferation by CHAMP. (A) HeLa cells grown in 96-well microtiter plates in the presence of 10% FBS were infected with adβ-gal or adCHAMP, and BrdUrd incorporation into newly synthesized DNA in proliferating cells was quantitated by using the cell proliferation ELISA, BrdUrd (chemiluminescence) kit as described. BrdUrd incorporation is expressed as relative light units/s (rlu/s). Values represent the averages ± SD of three experiments. (B) Schematic diagram of CHAMP showing the positions of the six conserved helicase domains (I–VI). The conserved ATPase domain II was mutated as shown to residues predicted to eliminate helicase activity. (C) NIH 3T3 cells were incubated for 24 h in 0.5% FBS before transfection with expression vectors encoding wild-type and mutant CHAMP, as described in Materials and Methods. Then 24 h later, medium containing 10% FBS was added to initiate synchronized cell cycle progression. After an additional 24 h, cells were fixed and double-stained with anti-CHAMP Ab (green) and anti-PCNA Ab (red), which are specific for cells in S phase. Representative fields are shown. Cells expressing CHAMP (Left) or the CHAMP mutant (Right) are indicated by white arrowheads. (D) Quantitation of cells labeled with anti-CHAMP and anti-PCNA. Approximately 70% of untransfected cells or cells transfected with mutant-CHAMP showed PCNA staining. In contrast, 10% of cells transfected with wild-type CHAMP showed PCNA staining.
Because HeLa cells are highly transformed and do not undergo complete cell cycle arrest in response to growth restriction, we further examined whether CHAMP could prevent the transition of NIH 3T3 cells from quiescence to S phase in response to serum stimulation. As a control, we also generated a mutant form of CHAMP in which the ATPase domain (domain II), which is conserved in members of the helicase superfamily, was mutated from DEAGQ to GGAAG (Fig. 1B). The wild-type and mutant forms of CHAMP were expressed at comparable levels in the cytoplasm of transfected cells (Fig. 1C).
NIH 3T3 cells maintained in 0.5% FBS for 24 h were transfected with an expression vector encoding wild-type and mutant CHAMP. Twenty-four hours later, fresh medium supplemented with 10% FBS was added to induce synchronous reentry into the cell cycle, and proliferative activity was assayed by staining for proliferating cellular nuclear antigen (PCNA) after an additional 24 h. As shown in Fig. 1 C and D, only 10% of cells that expressed CHAMP were PCNA-positive, compared to 70% of untransfected cells. In contrast, 68% of cells expressing the mutant form of CHAMP were able to enter the cell cycle and show positive PCNA staining. Based on cell morphology and Hoechst staining of nuclei, there was no evidence for apoptosis of CHAMP-expressing cells. These results demonstrate that CHAMP blocks cell proliferation and suggest that the ATPase activity of the conserved helicase motif is required for its antiproliferative effects.
Inhibition of Cardiomyocyte Hypertrophy by CHAMP.
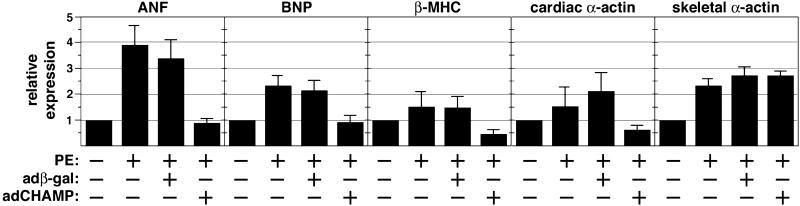
Hypertrophic growth of cardiac myocytes in response to extracellular agonists is controlled by many of the same signal transduction pathways that control proliferation of nonmuscle cells. In light of the ability of CHAMP to block cell proliferation, we tested whether it also could interfere with agonist-dependent hypertrophy of cardiomyocytes. Hypertrophy was assayed by expression of fetal genes after stimulation by the α-adrenergic agonist PE. As shown in Fig. 2, PE stimulated the expression of ANF, brain natriuretic factor, β-myosin heavy chain (MHC), and skeletal and cardiac α-actin to varying levels. In the presence of adCHAMP, the up-regulation of ANF, brain natriuretic factor, β-MHC, and cardiac α-actin by PE was blocked. In contrast, adCHAMP had no effect on expression of skeletal α-actin or glyceraldehyde-3-phosphate dehydrogenase, which is expressed ubiquitously. The suppression of hypertrophic gene expression was a specific response to adCHAMP and was not observed with adβ-gal. A similar inhibitory effect of adCHAMP on induction of hypertrophic marker genes was observed in cardiomyocytes stimulated with serum and isoproterenol (data not shown).
Figure 2.
Effect of CHAMP on fetal gene expression in neonatal cardiomyocytes. Primary neonatal rat cardiomyocytes were infected with adβ-gal or adCHAMP for 2 h. After infection, the culture medium was replaced with serum-free medium. Cells were incubated for a further 24 h at which time PE (20 μg/ml) was added as indicated. Total RNA was isolated 48 h later, and RNA dot blots were probed with oligonucleotide probes for the indicated genes. Expression of each transcript was quantified by phosphoimaging and is expressed as the level of expression relative to untreated cultures. Expression levels were normalized to the level of expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was used as an internal control for RNA loading (not shown). The adβ-gal had no effect on the up-regulation of hypertrophic marker genes by PE. The results are the average ± SD of three independent experiments.
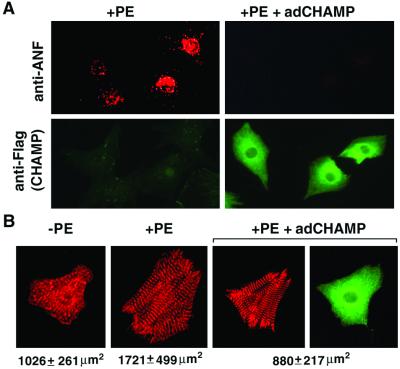
We also examined the effect of adCHAMP on hypertrophic responsiveness by immunostaining of cardiomyocytes with anti-ANF Ab. Cardiomyocytes were identified by immunostaining for α-actinin and CHAMP expression was confirmed by staining with a polyclonal anti-CHAMP Ab. As shown in Fig. 3A, ANF shows a perinuclear staining pattern in cardiomyocytes stimulated with PE. In adCHAMP-infected cells stimulated with PE, ANF staining was undetectable (Fig. 3A). PE also stimulates sarcomere organization, as shown by α-actinin staining, and induces an increase in cell size (Fig. 3B). adCHAMP completely inhibited the PE-induced increase in cell size, but it did not appear to prevent the organization of sarcomeres (Fig. 3B). Cells that expressed ectopic CHAMP appeared healthy, despite their inability to mount a hypertrophic response. There also was no increase in apoptosis in CHAMP-expressing cells, as determined by terminal deoxynucleotidyltransferase-mediated UTP end labeling staining (data not shown). The antihypertrophic effect of CHAMP on cardiomyocytes was observed over a wide range of adCHAMP expression (from 3- to 100-fold compared to the endogenous level, data not shown).
Figure 3.
Inhibition of ANF expression and cardiomyocyte growth by CHAMP. (A) Primary neonatal rat cardiomyocytes were infected with adCHAMP (Right) or were uninfected (Left). Serum-free medium was added to cells 2 h after infection, and 24 h later they were stimulated with PE (20 μg/ml) for 48 h. Cells were then double-stained with anti-ANF Ab (red) and anti-FLAG Ab (green), which recognizes CHAMP. (B) Serum-starved neonatal cardiomyocytes were incubated with or without PE for 2 days. After fixation, cells were stained with anti-α-actinin (red) and anti-CHAMP (green) Abs. Approximately 100 cells from each condition were photographed, and the cell area was measured as described in Material and Methods. Representative cells are shown. Cardiomyocytes grown in the absence of PE showed fragmented sarcomeres as visualized by α-actinin staining (Left). After 2 days of stimulation with PE, myocytes increased in cell size by ≈68% (P < 0.001 vs. no PE treatment) and sarcomeres became organized. adCHAMP completely blocked the increase of myocyte size induced by PE (P < 0.001 vs. +PE). However, sarcomere organization appeared to be unaffected by adCHAMP (Right). The level of expression of endogenous CHAMP in the left two panels is below the level of detection with anti-CHAMP Ab.
CHAMP Does Not Affect Early Mitogenic Responses.
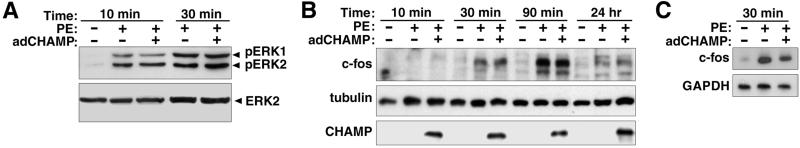
PE-induced cardiomyocyte hypertrophy involves activation of cascades of MAPKs, especially p44 (ERK1) and p42 (ERK2) (26). To determine the effect of CHAMP on PE-stimulated ERK1/2 activities, cardiomyocytes were harvested at multiple time points after PE stimulation and MAPK assays were performed by immunoblotting with Abs specific for activated phospho-ERKs (Fig. 4A). As reported previously, PE stimulation of cultured cardiomyocytes led to a pronounced increase in ERK1 and ERK2 phosphorylation. Cardiomyocytes infected with adCHAMP showed comparable activation of ERKs (Fig. 4A).
Figure 4.
Lack of an effect of CHAMP on early mitogenic responses of cardiomyocytes. (A) Primary neonatal rat cardiomyocytes were grown, infected with adCHAMP, and treated with PE as described in Fig 2 legend. Cells were harvested at various time points after addition of PE, and activities of ERKs1 and 2 were determined by immunoblot analysis by using an Ab specific for the dually phosphorylated, activated forms of the kinases. To control for equal protein loading, identical blots were probed with an Ab that recognizes the ERK2 protein independent of the phosphorylation state. CHAMP did not inhibit PE-induced activation of ERK1/2 kinase activities. (B and C) Cardiomyocytes were stimulated with PE for the indicated times and expression of c-fos protein or mRNA was determined by Western blot (B) or reverse transcription–PCR (C) analysis, respectively, as described in Materials and Methods. CHAMP did not inhibit PE-induced transient up-regulation of immediate-early gene c-fos.
Expression of c-fos is a sensitive marker of early mitogenic signaling events. Up-regulation of c-fos expression by PE, as measured by reverse transcription–PCR and Western blot, was unaffected by adCHAMP (Fig. 4 B and C). Thus, the inhibition of hypertrophic signaling by CHAMP does not appear to be attributable to a disruption of early mitogenic-signaling events.
CHAMP Up-Regulates p21CIP1.
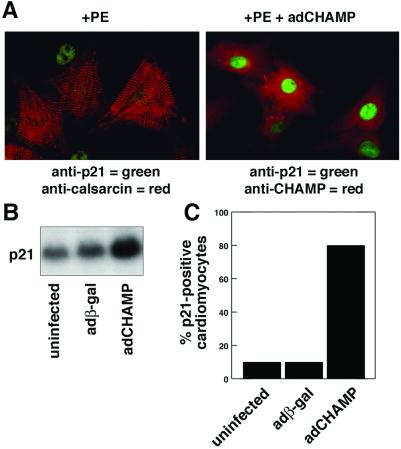
The CDK inhibitor p21CIP1 acts as a suppressor of cell proliferation and has been implicated as a negative regulator of cardiomyocyte hypertrophy (18, 27). To further investigate the basis for the antihypertrophic activity of CHAMP, we analyzed the expression of p21CIP1 by immunofluorescence staining of PE-stimulated cardiomyocytes in the presence and absence of adCHAMP (Fig. 5). Cardiomyocytes were distinguished from contaminating fibroblasts by staining with an Ab for calsarcin, a muscle-specific component of the Z-band (23). Only a small fraction of neonatal cardiomyocytes (< 10%) showed p21CIP1-positive staining in the absence of adCHAMP. In contrast, >80% of adCHAMP-infected cardiomyocytes showed strong p21CIP1 staining (Fig. 5).
Figure 5.
Up-regulation of p21CIP1 expression by CHAMP. (A) Primary neonatal rat cardiomyocytes were infected with adCHAMP or were uninfected and then incubated in serum-free medium containing PE (20 μg/ml) for 2 d. Uninfected cultures (Left) were stained with anti-calsarcin Ab to mark cardiomyocytes (red) and with anti-p21CIP1 Ab (green). adCHAMP-infected myocytes (Right) were stained with anti-CHAMP (red) and anti-p21CIP1 (green) Ab. The green p21CIP1 nuclear staining (Left) is from a fibroblast cell that does not have calsarcin staining. (B) p21CIP1 mRNA expression was measured by Northern blot analysis of RNA isolated from uninfected cultures and cultures infected with adβ-gal and adCHAMP. p21CIP1 mRNA expression was unaffected by adβ-gal, but was up-regulated by adCHAMP. (C) The percentage of cells in cardiomyocyte cultures that stained for p21CIP1 expression was determined. Weak p21CIP1 staining was seen in <10% of uninfected or adβ-gal-infected cultures. In contrast, >80% cells in adCHAMP-infected cultures showed strong p21CIP1 staining.
Down-Regulation of CHAMP Expression in Hypertrophic Hearts from α-MHC-Calcineurin Transgenic Mice.
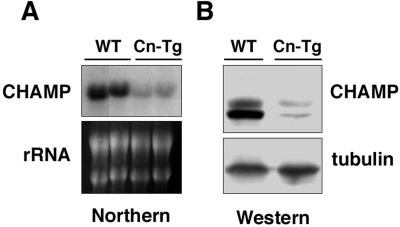
Based on the ability of CHAMP to block cardiomyocyte hypertrophy in vitro, we investigated whether CHAMP might be down-regulated in response to hypertrophic stimuli in vivo, thereby facilitating a hypertrophic growth response. The possible regulation of CHAMP expression during hypertrophy was examined by using transgenic mice that expressed a constitutively activated form of the calcineurin phosphatase under control of the α-MHC promoter. These mice develop severe cardiac hypertrophy by 4 wk of age, which progresses to dilated cardiomyopathy and heart failure (8). As shown in Fig. 6, CHAMP mRNA and protein were down-regulated 5-fold in hypertrophic hearts from α-MHC-calcineurin transgenic mice at 8 wk of age.
Figure 6.
Down-regulation of CHAMP expression in hypertrophic hearts from α-MHC-calcineurin transgenic mice. (A) Total RNA was isolated from two hearts of wild-type (WT) and α-MHC-calcineurin transgenic (Cn-Tg) littermates at 8 wk of age. CHAMP mRNA was detected by Northern blot analysis, and 18S and 28S RNA were detected by staining with ethidium bromide. (B) Protein extracts were prepared from hearts of wild-type (WT) and α-MHC-calcineurin (Cn-Tg) transgenic littermates at 8 wk of age. CHAMP protein and tubulin protein as a control for protein loading were detected by Western blot analysis.
Discussion
In a screen for downstream target genes of the MEF2C transcription factor in the developing mouse heart, we identified CHAMP, a RNA helicase that is down-regulated at the looping heart tube stage of mef2c mutant embryos (22). CHAMP expression is initiated in the linear heart tube at E8.0–8.5, immediately after the expression of MEF2C. At E15.5, CHAMP expression appears to be graded within the ventricular chambers, with highest expression in the trabeculae, where the proliferation rate of cardiomyocytes is diminished, and lowest in the adjacent compact zone, where cardiomyocytes are highly proliferative (22). The results of the present study demonstrate that CHAMP can act as an inhibitor of cell proliferation, as well as cardiomyocyte hypertrophy, which are governed by many of the same signaling pathways.
Suppression of Cell Proliferation and Hypertrophy by CHAMP.
Ectopic expression of CHAMP in HeLa and NIH 3T3 cells inhibits proliferation. These findings demonstrate that CHAMP acts through general cell cycle machinery, rather than through a cardiac-specific mechanism, to inhibit cell cycle progression. Given that many of the signaling pathways that control proliferation of nonmuscle cells also mediate hypertrophic growth of postnatal cardiomyocytes (5), it seems likely that the antiproliferative and antihypertrophic effects of CHAMP are mediated by the same mechanism and involve the same targets.
The block to hypertrophy imposed by CHAMP is accompanied by up-regulation of the cell cycle inhibitor p21CIP1, which inhibits cyclin-CDK activity. Although the present results do not allow us to distinguish whether the up-regulation of p21CIP1 is a causal component or a secondary consequence of the antiproliferative activity of CHAMP, the dramatic up-regulation of p21CIP1 coupled with the fact that p21CIP1 expression does not necessarily accompany cell cycle arrest in all cell types, suggests that p21CIP1 is an important mediator of CHAMP activity.
Developmental Regulation of CHAMP Expression and Activity.
CHAMP is expressed throughout the linear and looping heart tube when cardiomyocytes are able to proliferate. This raises the question as to why it does not block proliferation during development of the heart. We can imagine several possible explanations. (i) The antiproliferative activity of CHAMP may be over-ridden by other mechanisms during early development. (ii) The level of CHAMP expression during early embryogenesis may be inadequate to inhibit cardiomyocyte proliferation. In this regard, CHAMP expression is up-regulated after birth as cardiomyocytes progressively lose the ability to proliferate (unpublished results). (iii) Although the detailed expression pattern of p21CIP1 during embryogenesis has not been described, like CHAMP, p21CIP1 expression increases during fetal to neonatal development, reaching a maximum at adulthood (29, 30). Therefore, if CHAMP requires p21CIP1 to inhibit growth, CHAMP would be unable to suppress proliferation of embryonic cardiomyocytes. It also is notable that during the trabeculation stage of mouse heart development, the expression pattern of CHAMP is similar to that of the CDKI p57KIP2, which shows a graded expression pattern with highest expression in the trabecular region and undetectable expression in the proliferative zone (22, 28).
Because CHAMP was originally discovered as a MEF2C-dependent gene in the developing heart, and MEF2 activity is up-regulated in response to hypertrophic stimuli (31, 32), the observation that CHAMP is down-regulated in hypertrophic hearts from α-MHC-calcineurin transgenic mice seems paradoxical. p21CIP1 is also down-regulated in pressure-overload hypertrophy (18). Because MEF2C is down-regulated in the heart during late fetal development (33, 34), it is possible that CHAMP expression in the postnatal heart is independent of MEF2C. Whether this down-regulation of CHAMP expression in vivo is an essential prerequisite for hypertrophy remains to be determined.
Potential Mechanisms of Action of CHAMP.
The molecular mechanism whereby CHAMP inhibits proliferation and hypertrophy and up-regulates p21CIP1 expression remains to be determined. CHAMP contains seven conserved RNA helicase domains. Our results demonstrate that the conserved ATPase domain is required for the growth-inhibitory activity of CHAMP.
Members of the RNA helicase superfamily play diverse roles in RNA metabolism that include regulation of transcription, ribosome biogenesis, pre-mRNA editing and splicing, RNA export to the cytoplasm, translation initiation and termination, and RNA degradation (35). CHAMP shares highest homology with members of RNA helicase subfamily I, which includes yeast Upf1 and its mammalian homologs, which are involved in translation termination and mRNA turnover (36). The signature motif of this group of helicases is the DEAGQ sequence in conserved region II. Our results show that this motif is required for the antiproliferative function of CHAMP.
With regard to the possible functions of CHAMP as a regulator of RNA processing or stability, it is interesting to note that p21CIP1 mRNA can be stabilized by the binding of an AU-rich element-binding protein to the 3′ untranslated region (37, 38). Thus, CHAMP could up-regulate p21CIP1 mRNA by stabilizing the transcript through its putative RNA-binding activity. Alternatively, CHAMP could function as an ATPase-driven RNA helicase and regulate p21CIP1 RNA processing.
The functions of CHAMP in vivo remain to be explored. It will be especially interesting to determine whether targeted deletion of the gene results in enhanced proliferation of cardiomyocytes or supersensitivity to hypertrophic stimuli. Whether pharmacologic inhibitors of the helicase activity of CHAMP might permit cardiomyocyte proliferation after myocardial infarction or apoptosis also warrants consideration as a strategy for restoring cardiac mass. The results of the present study also predict that elevated expression of CHAMP in the heart through adenoviral delivery, for example, would diminish the responsiveness of cardiomyocytes to hypertrophic stimuli. Because hypertrophy of the myocardium is correlated with poor clinical prognosis and frequently progresses to heart failure and sudden death, strategies to elevate CHAMP expression could be of therapeutic value.
Acknowledgments
We thank Norbert Frey for anti-calsarcin Ab, Robert D. Gerard for assistance with adenoviruses, and A. Tizenor for assistance with graphics. We are grateful to Rhonda Bassel-Duby for critical review of the manuscript. E.N.O. was supported by grants from the National Institutes of Health, the D. W. Reynolds Cardiovascular Clinical Research Center, the Robert A. Welch Foundation, the Texas Advanced Technology Program, and the W. G. McGowan Charitable Fund.
Abbreviations
- ANF
atrial natriuretic factor
- CDK
cyclin-dependent kinase
- CDKI
cyclin-dependent kinase inhibitor
- CHAMP
cardiac helicase activated by MEF2 protein
- adCHAMP
adenovirus encoding FLAG-tagged CHAMP
- E
embryonic day
- ERK
extracellular signal-regulated kinase
- MAPK
mitogen-activated protein kinase
- MHC
myosin heavy chain
- MOI
multiplicity of infection
- PCNA
proliferating cellular nuclear antigen
- PE
phenylephrine
- adβ-gal
adenovirus that constitutively expresses β-galactosidase
References
- 1.Sadoshima J, Izumo S. Annu Rev Physiol. 1997;59:551–571. doi: 10.1146/annurev.physiol.59.1.551. [DOI] [PubMed] [Google Scholar]
- 2.McKinsey T A, Olson E N. Curr Opin Genet Dev. 1999;9:267–274. doi: 10.1016/s0959-437x(99)80040-9. [DOI] [PubMed] [Google Scholar]
- 3.Hunter J J, Chien K R. N Engl J Med. 1999;341:1276–1283. doi: 10.1056/NEJM199910213411706. [DOI] [PubMed] [Google Scholar]
- 4.Benjamin E J, Levy D. Am J Med Sci. 1999;317:168–175. doi: 10.1097/00000441-199903000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Brooks G, Poolman R A, Li J M. Cardiovasc Res. 1998;39:301–311. doi: 10.1016/s0008-6363(98)00125-4. [DOI] [PubMed] [Google Scholar]
- 6.Lim H W, New L, Han J, Molkentin J D. J Biol Chem. 2001;276:15913–15919. doi: 10.1074/jbc.M100452200. [DOI] [PubMed] [Google Scholar]
- 7.Frey N, McKinsey T A, Olson E N. Nat Med. 2000;6:1221–1227. doi: 10.1038/81321. [DOI] [PubMed] [Google Scholar]
- 8.Molkentin J D, Lu J R, Antos C L, Markham B, Richardson J, Robbins J, Grant S R, Olson E N. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leinwand L A. Proc Natl Acad Sci USA. 2001;98:2947–2949. doi: 10.1073/pnas.051033698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olson E N, Williams R S. Cell. 2000;101:689–692. doi: 10.1016/s0092-8674(00)80880-6. [DOI] [PubMed] [Google Scholar]
- 11.Morgan D O. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 12.Harper J W, Elledge S J. Curr Opin Genet Dev. 1996;6:56–64. doi: 10.1016/s0959-437x(96)90011-8. [DOI] [PubMed] [Google Scholar]
- 13.Flink I L, Oana S, Maitra N, Bahl J J, Morkin E. J Mol Cell Cardiol. 1998;30:563–578. doi: 10.1006/jmcc.1997.0620. [DOI] [PubMed] [Google Scholar]
- 14.Koh K N, Kang M J, Frith-Terhune A, Park S K, Kim I, Lee C O, Koh G Y. J Mol Cell Cardiol. 1998;30:463–474. doi: 10.1006/jmcc.1997.0611. [DOI] [PubMed] [Google Scholar]
- 15.Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh D Y, Nakayama K. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 16.Kiyokawa H, Kineman R D, Manova-Todorova K O, Soares V C, Hoffman E S, Ono M, Khanam D, Hayday A C, Frohman L A, Koff A. Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 17.Fero M L, Rivkin M, Tasch M, Porter P, Carow C E, Firpo E, Polyak K, Tsai L H, Broudy V, Perlmutter R M, et al. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 18.Li J M, Brooks G. Eur Heart J. 1999;20:406–420. doi: 10.1053/euhj.1998.1308. [DOI] [PubMed] [Google Scholar]
- 19.Sadoshima J, Aoki H, Izumo S. Circ Res. 1997;80:228–241. doi: 10.1161/01.res.80.2.228. [DOI] [PubMed] [Google Scholar]
- 20.Tamamori M, Ito H, Hiroe M, Terada Y, Marumo F, Ikeda M A. Am J Physiol. 1998;275:H2036–H2040. doi: 10.1152/ajpheart.1998.275.6.H2036. [DOI] [PubMed] [Google Scholar]
- 21.Nozato T, Ito H, Watanabe M, Ono Y, Adachi S, Tanaka H, Hiroe M, Sunamori M, Marum F. J Mol Cell Cardiol. 2000;33:1493–1504. doi: 10.1006/jmcc.2001.1412. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z P, Nakagawa O, Nakagawa M, Yanagisawa H, Passier R, Richardson J A, Srivastava D, Olson E N. Dev Biol. 2001;234:497–509. doi: 10.1006/dbio.2001.0277. [DOI] [PubMed] [Google Scholar]
- 23.Frey N, Richardson J A, Olson E N. Proc Natl Acad Sci USA. 2000;97:14632–14637. doi: 10.1073/pnas.260501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicol R L, Frey N, Pearson G, Cobb M, Richardson J, Olson E N. EMBO J. 2001;20:2757–2767. doi: 10.1093/emboj/20.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sedmera D, Pexieder T, Vuillemin M, Thompson R P, Anderson R H. Anat Rec. 2000;258:319–337. doi: 10.1002/(SICI)1097-0185(20000401)258:4<319::AID-AR1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 26.Clerk A, Sugden P H. Am J Cardiol. 1999;83:64H–69H. doi: 10.1016/s0002-9149(99)00261-1. [DOI] [PubMed] [Google Scholar]
- 27.von Harsdorf R, Hauck L, Mehrhof F, Wegenka U, Cardoso M C, Dietz R. Circ Res. 1999;85:128–136. doi: 10.1161/01.res.85.2.128. [DOI] [PubMed] [Google Scholar]
- 28.Kochilas L K, Li J, Jin F, Buck C A, Epstein J A. Pediatr Res. 1999;45:635–642. doi: 10.1203/00006450-199905010-00004. [DOI] [PubMed] [Google Scholar]
- 29.Burton P B, Yacoub M H, Barton P J. Eur Heart J. 1999;20:604–611. doi: 10.1053/euhj.1998.1231. [DOI] [PubMed] [Google Scholar]
- 30.Poolman R A, Brooks G. J Mol Cell Cardiol. 1998;30:2121–2135. doi: 10.1006/jmcc.1998.0808. [DOI] [PubMed] [Google Scholar]
- 31.Kolodziejczyk S M, Wang L, Balazsi K, DeRepentigny Y, Kothary R, Megeney L A. Curr Biol. 1999;9:1203–1206. doi: 10.1016/S0960-9822(00)80027-5. [DOI] [PubMed] [Google Scholar]
- 32.Passier R, Zeng H, Frey N, McKinsey T A, Overbeek P, Richardson J A, Grant S R, Olson E N. J Clin Invest. 2000;105:1395–1406. doi: 10.1172/JCI8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edmondson D G, Lyons G E, Martin J F, Olson E N. Development (Cambridge, UK) 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- 34.Martin J F, Schwarz J J, Olson E N. Proc Natl Acad Sci USA. 1993;90:5282–5286. doi: 10.1073/pnas.90.11.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanner N K, Linder P. Mol Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 36.Weng Y, Czaplinski K, Peltz S W. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joseph B, Orlian M, Furneaux H. J Biol Chem. 1998;273:20511–20516. doi: 10.1074/jbc.273.32.20511. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Shen X, Nguyen V A, Kunos G, Gao B. J Biol Chem. 2000;275:11846–11851. doi: 10.1074/jbc.275.16.11846. [DOI] [PubMed] [Google Scholar]