Abstract
Species diversity is unevenly distributed across the globe, with terrestrial diversity concentrated in a few restricted biodiversity hotspots. These areas are associated with high losses of primary vegetation and increased human population density, resulting in growing numbers of threatened species. We show that conservation of these hotspots is critical because they harbor even greater amounts of evolutionary history than expected by species numbers alone. We used supertrees for carnivores and primates to estimate that nearly 70% of the total amount of evolutionary history represented in these groups is found in 25 biodiversity hotspots.
Biodiversity is distributed unequally across the globe—only a few, small areas hold most species (1). At least 44% of vascular plants and 35% of vertebrates are endemic to 25 biodiversity “hotspots” (2). Disconcertingly, people and, hence, threats to biodiversity are similarly distributed (3). Thus, none of the hotspots retain above 30% of their natural habitat, and together they represent only 1.4% of the planet's land (4). A hitherto unknown component of biodiversity hotspots is the evolutionary history of species residing within them—a more inclusive measure of biodiversity than species numbers (5). If this history is disproportionately extensive, we may face losses of phylogenetic diversity (PD) (6, 7) and/or evolutionarily ancient lineages (8) even more devastating than reflected by species losses alone. For example, the 103 endemic mammal species of Madagascar include no less than five endemic families and 14 endemic genera of primates (9, 10). Such areas may not only be important reservoirs for phylogenetic history, they may also be critical for preserving the future of evolutionary processes where biodiversity is created (11). Here we used complete phylogenies of two mammalian orders to show that significantly more primate and carnivore evolutionary history [343 million years (my)] is endemic to the hotspots than expected under a random model. Maybe even more serious, considering only threatened endemic species (12), hotspots also hold 163 my more evolutionary history than expected.
We use two measures of evolutionary history, or PD, that are derived by examining branch lengths in a phylogenetic tree (13). The first measure incorporates clade evolutionary history, which includes all of the branches within an included clade in a phylogeny (see Fig. 1), and hence takes higher-level diversity into account (6). For species corresponding to an area or set of areas, clade evolutionary history is equal to the amount of branch length uniquely represented by this set of species. It is the amount of phylogenetic diversity inevitably lost if those areas are lost. The other represents the species evolutionary history, measured as the branch length from the present to the time of last divergence (7, 14, 15). To obtain these measures, we used complete phylogenies (16) (with dated branch lengths) for all extant primate (17) and carnivore (18) species. For both orders, we measured the two PD parameters by using lists of species endemic to the hotspots. We then compared these values with those for 1,000 simulations of the same number of species removed at random (19) from the entire species lists for each order (9). We repeated this analysis by using lists of all primates and carnivores occurring within the hotspots.
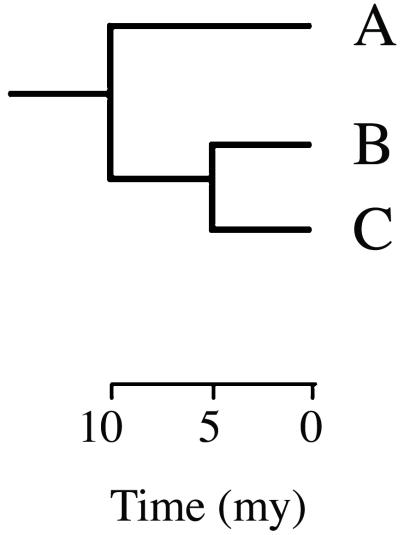
Figure 1.
A hypothetical phylogeny of three taxa. Measures of phylogenetic diversity are represented by branch lengths, with time (my, millions of years) measured across the horizontal axis. “Species evolutionary history” is calculated by the length of the branch to the most recent ancestral split. For example, for species C, this is equal to 5 my, whereas for species A the value is 10 my. “Clade evolutionary history” includes all branches in a set of taxa. For the clade containing species B and C this is equal to 15 my, because it includes all of the higher shared branches.
Methods
Species Range Designations to Hotspots.
Allocation of species to hotspots was made in two ways. Carnivore and primate species were either listed as being strictly endemic to a hotspot or as occurring in a hotspot. Species that were endemic to multiple hotspots were classified as hotspot endemics. Most primate species are endemic to them (127 species from a total 233; only 29 species also occur outside of hotspots). By contrast, fewer terrestrial carnivores are endemic to hotspots (only 51 of 234 species); however, no less than 208 carnivore species occur in at least one hotspot. We give full primary references used to assess primate and carnivore distributions on the web at http://www.faculty.virginia.edu/gittleman.
Tests of PD.
Phylogenetic information was based on complete trees of the primates (17) and carnivores (excluding pinnipeds) (18). Branch lengths are derived from a combination of absolute (fossil and point molecular estimates) and relative molecular dates. Date estimates were available for a majority of nodes in both trees. Nodes without times of divergence were dated by interpolation using a pure birth model, whereby a clade's age is proportional to the logarithm of the number of species it contains (17). Based on the numbers of species and their associated branch lengths in hotspots, 1,000 simulations were run on the same numbers of species removed at random (19); the phylogenies and computer programs used to conduct the simulations are available at http://www.bio.ic.ac.uk/evolve/.
Results and Discussion
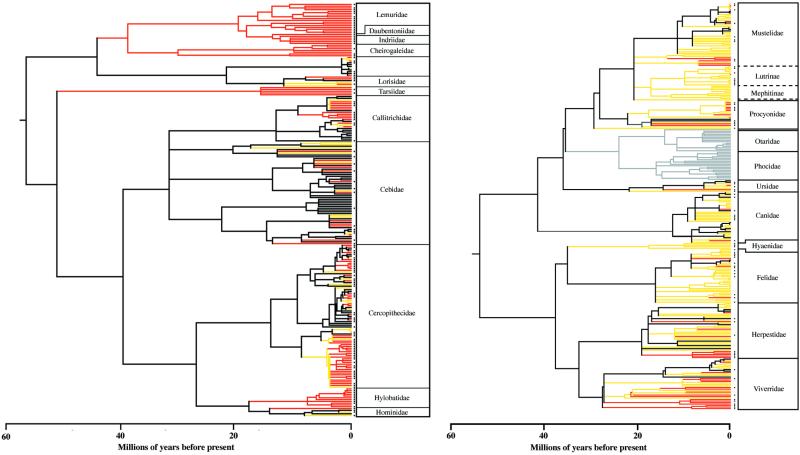
Approximately 55% of the world's primates and 22% of carnivores are endemic to hotspots. Entire primate lineages (Fig. 2a) are found in Madagascar (Cheirogaleidae, Megaladapidae, Lemuridae, Indriidae, and Daubentoniidae), Southeast Asia (Tarsiidae and Hylobatidae), and the Atlantic Forest of Brazil (Leontopithecus and Brachyteles). Comparatively few carnivores are endemic to hotspots (Fig. 2b), although notable endemic clades include the Malagasy viverrids and the majority of New World procyonids.
Figure 2.
Phylogenetic patterns of primates (Left) and carnivores (Right) residing in biodiversity hotspots. Species hotspot designation is used to color branches where extant PD exists. Red branches represent PD found only in hotspots (endemic), yellow branches represent PD occurring in hotspots, and black branches represent PD not occurring in hotspots. Threatened species are indicated with an asterisk. Across carnivores, the pinnipeds are represented in gray to denote that they were excluded from analyses because of their aquatic geographic range distributions.
With around 59% of species evolutionary history and 71% of clade evolutionary history, both measures of PD for hotspot endemic primates are significantly greater than expected under the random model (Table 1). Branch lengths with dates of divergence allow analysis of whether lineages within hotspots are on average younger or older than those outside of hotspots. There are no overall differences with respect to mean (log-transformed) ages of species endemic to hotspots compared with non-hotspot species, although there are some patterns within continents. Primates within the Indo–Burma hotspot are older than Asian non-hotspot species (t = 2.007, P < 0.05, df = 18). Conversely, species endemic to the West African Guinea Forests and the Tanzanian Eastern Arc show a younger mean age than African non-hotspot species (t = 2.144, P < 0.05, df = 60). No consistent patterns were found in South American hotspot regions.
Table 1.
Numbers of endemic carnivore and primate species with associated amount of PD residing in hotspots
| Clade | No. of species | No. of endemic hotspot species | Phylogenetic diversity, my
|
|||
|---|---|---|---|---|---|---|
| Clade | Random | Species | Random | |||
| Primates | 233 | 127 | 838.9** | 613.0 | 617.6* | 564.2 |
| Threatened primates | 128 | 98 | 549.6** | 460.6 | 468.0 | 434.0 |
| Carnivores | 234 | 51 | 412.7** | 295.4 | 337.0* | 283.8 |
| Threatened carnivores | 75 | 31 | 249.3** | 175.3 | 211.7 | 171.6 |
PD is measured as clade evolutionary history (all branch lengths across clades) and species evolutionary history (branch length for each species from the most recent node) relative to the mean expected amounts from 1,000 random simulations for each measure. Similar results were obtained when data deficient species were counted as threatened (see text).
, P ≤ 0.05;
, P ≤ 0.01.
Carnivore species have unusually large distributions worldwide (20, 21), and few species have restricted ranges. Thus, the amount of evolutionary history endemic to hotspots is less than in primates. Nevertheless, with either measure of PD, hotspots do contain a greater amount of carnivore PD than under a random model (Table 1). The ages of hotspot endemic carnivores are not significantly different from non-hotspot endemic species. Taken together, the ages of primate and carnivore species are not consistently older or younger in hotspots; a similar pattern is observed in other taxa whereby there is no consistency in the ages of lineages living in particular eco–climatic zones (22, 23). Hotspots contain endemic primates and carnivores that, according to times of divergences in the phylogenies, represent over 343 my more evolutionary history than expected from the random model. The reason for hotspots containing a great amount of PD is that, with exceptions such as rapid radiations in Old World monkeys (24) and some canids (18), a clade's evolutionary history within a given geographic region is generally variable with respect to ages of divergence.
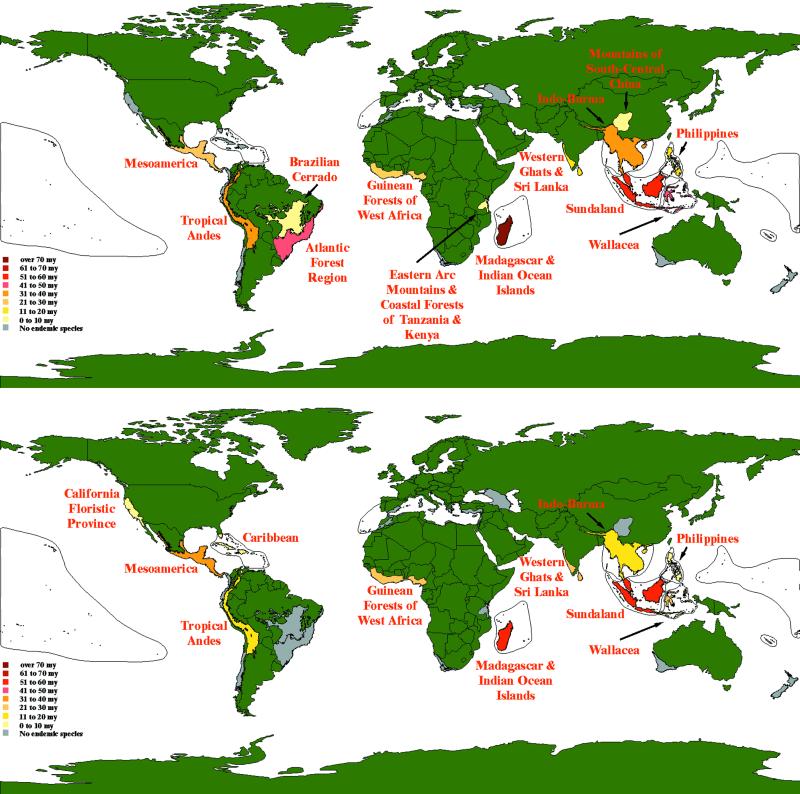
A rank ordering of the top five hotspots according to endemic evolutionary history reveals taxonomic differences in the amount of unique evolutionary history (in millions of years) residing in them (see Fig. 3). In primates, they are Madagascar (257), Sundaland (65), Wallacea (50), Brazil's Atlantic forest (44), and Indo-Burma (40). For carnivores, they are Sundaland (53), Madagascar (51), Mesoamerica (35), Western Ghats and Sri Lanka (26), and the Guinean Forests of West Africa (22). Madagascar and Sundaland are clearly outstanding for both groups.
Figure 3.
Map of the top 25 biodiversity hotspots (from ref. 4) containing endemic primates (Upper) and carnivores (Lower). Hotspots with endemic species are labeled by name and colored corresponding to the scale with the amount of species evolutionary history in my. Species endemic to multiple hotspots were not used to calculate these amounts (for clarity), and hence the measure of endemic clade evolutionary history has not been included.
The same analyses were performed by using all species occurring in the hotspots. The ranges of over 89% of carnivore species overlapped with at least one hotspot; for primates, the percentage is 67%. For both orders, the clade phylogenetic diversity occurring in hotspots was significantly greater than predicted by the random model (Table 2). The amount of species evolutionary history occurring in hotspots was also significantly greater for primates, but not for carnivores.
Table 2.
Numbers of occurring carnivore and primate species with associated amount of PD residing in hotspots
| Clade | No. of species | No. of occurring hotspot species | Clade, my | Phylogeneric diversity, my
|
||
|---|---|---|---|---|---|---|
| Random | Species | Random | ||||
| Primates | 233 | 156 | 985.5** | 778.9 | 735.5* | 691.0 |
| Threatened primates | 128 | 108 | 582.1* | 509.1 | 500.5 | 480.2 |
| Carnivores | 234 | 208 | 1,625.5** | 1,543.4 | 1,181.7 | 1,157.4 |
| Threatened carnivores | 75 | 69 | 500.0** | 404.9 | 443.1 | 411.1 |
Taxonomic revision can certainly affect the results of interspecific studies. Species level systematics are far from fully resolved even for groups as well known as primates and carnivores. For example, recent advances in primate taxonomy suggest that 110 Neotropical species exist (25), compared with the 84 traditionally listed (9). With the revised taxonomy, however, there would be only three additional species endemic to hotspots, and four more occurring in hotspots. Further, recent phylogenetic analyses (26) identifying up to seven mouse lemur species (genus Microcebus) would make the value of PD even greater in Madagascar. Carnivore taxonomy is rather more stable (18, 27, 28), and so changes within this order will have even smaller effects. Further analyses should allow the incorporation of more finely resolved systematic levels, such as subspecies or phylogenetic species (29), or even character diversity (30). Nonetheless, we doubt that these will alter our fundamental conclusion that evolutionary history is disproportionately concentrated in small—and highly threatened—areas. Generalizing these results, it is important to recognize that our analysis is restricted to primates and carnivores, two orders that comprise only 11% of all mammalian biodiversity (9). Although these taxa do represent both comparatively narrow (primates) and broad (carnivores) distributions globally (31), further mammalian and terrestrial vertebrate taxa should be examined across hotspots to evaluate differences in amounts of phylogenetic diversity living within them.
If hotspot lineages include many species listed on the International Union for the Conservation of Nature and Natural Resources Red List (12) as having a high probability of extinction in the medium-term future, impending losses of evolutionary history could be even more severe than the above results suggest (19, 32). We used the Red List to examine the patterns in threatened species (species classified as LRcd and above; data deficient, extinct in the wild, and extinct species excluded) (12). The majority of the species most likely to become extinct are endemic to the hotspots: 77% of threatened primates and 60% of threatened carnivores. For all primate species endemic to hotspots, 77% are threatened, whereas only 28% of species outside of hotspots are threatened; for carnivores, the numbers are 61% and 24%, respectively. We repeated the previous PD analyses by using only the threatened species. In terms of endemic clade evolutionary history, hotspots contain 63% of threatened primate PD and 37% of threatened carnivore PD; in both cases these figures are significantly higher than under the random model (see Table 1). When considering only threatened endemic species, hotspots also hold 163 my more evolutionary history than expected. When endemic species evolutionary history is analyzed, 82% of threatened primate PD and 45% of threatened carnivore PD is found only in the hotspots (Table 1). The results are qualitatively similar considering all primates and carnivores occurring within hotspots (Table 2). Not only are hotspots critical for a large portion of the diversity of primates and carnivores overall, but the evolutionary history of extinction-prone species is disproportionately clustered in these areas.
Studies of past and present extinction rates have repeatedly shown taxonomically nonrandom patterns of threat to species (33–36). Further, the distributions of both biodiversity (4) and of people, and hence threats to this biodiversity (3), are also nonrandom across geographic space (37). We expand these findings to show co-occurrence between the phylogenetic and the geographic clumping of both biodiversity and threats. A third of the evolutionary history of all primates and carnivores is completely encompassed within the hotspots, and cannot be saved unless the hotspots are conserved. Conversely, however, if we can save the hotspots, we can represent over 2.6 billion years of primate and carnivore evolutionary history, almost 70% of their total. Hotspots are not only vital areas of species-level endemism, but are also highly significant reservoirs of unique and threatened evolutionary history.
Acknowledgments
We thank Georgina Mace, Paul-Michael Agapow, Janis Antonovics, Kaycie Billmark, Dan Faith, Kate Jones, Norman Myers, Charlie Nunn, John Pilgrim, Cindy Sechrest, Paul Williams, Michael Rosenzweig, and an anonymous reviewer for comments and suggestions on the analysis and manuscript. This work was partially supported by the Center for Applied Biodiversity Science, Conservation International, and the National Center for Ecological Analysis and Synthesis through a National Science Foundation Grant DEB-94-21535 (to J.L.G.).
Abbreviations
- PD
phylogenetic diversity
- my
million years
References
- 1.Gaston K J. Nature (London) 2000;405:220–227. doi: 10.1038/35012228. [DOI] [PubMed] [Google Scholar]
- 2.Mittermeier R A, Myers N, Gil P R, Mittermeier C G. Hotspots. Mexico City: Cemex; 2000. [Google Scholar]
- 3.Cincotta R P, Wisnewski J, Engelman R. Nature (London) 2000;404:990–992. doi: 10.1038/35010105. [DOI] [PubMed] [Google Scholar]
- 4.Myers N, Mittermeier R A, Mittermeier C G, da Fonseca G A, Kent J. Nature (London) 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 5.Purvis A, Hector A. Nature (London) 2000;405:212–219. doi: 10.1038/35012221. [DOI] [PubMed] [Google Scholar]
- 6.Vane-Wright R I, Humphries C J, Williams P H. Biol Conserv. 1991;55:235–254. [Google Scholar]
- 7.Faith D P. In: Systematics and Conservation Evaluation. Forey P L, Humphries C J, Vane-Wright R I, editors. Oxford: Clarendon; 1994. pp. 251–268. [Google Scholar]
- 8.May R M. Nature (London) 1990;347:129–130. [Google Scholar]
- 9.Wilson D E, Reeder D M, editors. Mammal Species of the World. Washington, DC: Smithsonian Institution Press; 1993. [Google Scholar]
- 10.Harcourt A H. Biol Conserv. 2000;93:163–175. [Google Scholar]
- 11.Myers N, Knoll A H. Proc Natl Acad Sci USA. 2001;98:5389–5392. doi: 10.1073/pnas.091092498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilton-Taylor C. 2000 IUCN Red List of Threatened Animals. Gland, Switzerland: Int. Union for Conservation of Nature and Natural Resources; 2000. [Google Scholar]
- 13.Faith D P. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 14.Erwin T L. Science. 1991;253:750–752. doi: 10.1126/science.253.5021.750. [DOI] [PubMed] [Google Scholar]
- 15.Altschul S F, Lipman D J. Nature (London) 1991;348:493–494. doi: 10.1038/348493c0. [DOI] [PubMed] [Google Scholar]
- 16.Sanderson M J, Purvis A, Henze C. Trends Ecol Evol. 1998;13:105–109. doi: 10.1016/S0169-5347(97)01242-1. [DOI] [PubMed] [Google Scholar]
- 17.Purvis A. Philos Trans Roy Soc London. 1995;348:405–421. doi: 10.1098/rstb.1995.0078. [DOI] [PubMed] [Google Scholar]
- 18.Bininda-Emonds O R P, Gittleman J L, Purvis A. Biol Rev. 1999;74:143–175. doi: 10.1017/s0006323199005307. [DOI] [PubMed] [Google Scholar]
- 19.Purvis A, Agapow P-M, Gittleman J L, Mace G M. Science. 2000;288:328–330. doi: 10.1126/science.288.5464.328. [DOI] [PubMed] [Google Scholar]
- 20.Hunt R M., Jr . In: Carnivore Behavior, Ecology, and Evolution. Gittleman J L, editor. Vol. 2. Ithaca, NY: Cornell Univ. Press; 1996. pp. 485–541. [Google Scholar]
- 21.Mills M G L, Freitag S, Van Jaarsveld A S. In: Carnivore Conservation. Gittleman J L, Funk S, Macdonald D W, Wayne R K, editors. Cambridge, U.K.: Cambridge Univ. Press; 2001. pp. 467–483. [Google Scholar]
- 22.Fjeldså J, Lovett J C. Biodiversity Conserv. 1997;6:325–346. [Google Scholar]
- 23.Chown S L, Gaston K J. Trends Ecol Evol. 2000;15:311–315. doi: 10.1016/s0169-5347(00)01910-8. [DOI] [PubMed] [Google Scholar]
- 24.Purvis A, Nee S, Harvey P H. Proc R Soc London B. 1995;260:329–333. doi: 10.1098/rspb.1995.0100. [DOI] [PubMed] [Google Scholar]
- 25.Rylands A B, Schneider H, Langguth A, Mittermeier R A, Groves C P, Rodriquez-Luna E. Neotropical Primates. 2000;8:61–93. [Google Scholar]
- 26.Yoder A D, Rosoloarison R M, Goodman S M, Irwin J A, Atsalis S, Ravosa M J, Ganzhorn J U. Proc Natl Acad Sci USA. 2000;97:11325–11330. doi: 10.1073/pnas.200121897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flynn J J, Neff N A, Tedford R H. In: The Phylogeny and Classification of the Tetrapods. Benton M J, editor. Oxford: Clarendon; 1988. pp. 73–115. [Google Scholar]
- 28.Wozencraft W C. In: Carnivore Behavior, Ecology, and Evolution. Gittleman J L, editor. Ithaca, NY: Cornell Univ. Press; 1989. pp. 569–593. [Google Scholar]
- 29.Wheeler Q D, Platnick N I. In: Species Concepts and Phylogenetic Theory. Wheeler Q D, Meier R, editors. New York: Columbia Univ. Press; 2000. pp. 55–69. [Google Scholar]
- 30.Williams P H, Humphries C H. In: Biodiversity: A Biology of Numbers and Difference. Gaston K J, editor. Oxford: Blackwell Science; 1996. pp. 54–76. [Google Scholar]
- 31.Anderson S, Jones J N., Jr . Orders and Families of Recent Mammals of the World. New York: Wiley; 1984. [Google Scholar]
- 32.Pimm S L, Russell G J, Gittleman J L, Brooks T M. Science. 1995;269:347–350. doi: 10.1126/science.269.5222.347. [DOI] [PubMed] [Google Scholar]
- 33.Purvis A, Gittleman J L, Cowlishaw G, Mace G M. Proc R Soc London. 2000;267:1947–1952. doi: 10.1098/rspb.2000.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKinney M L. Annu Rev Ecol Syst. 1997;28:495–516. [Google Scholar]
- 35.Russell G J, Brooks T M, McKinney M L, Anderson C G. Conserv Biol. 1998;12:1365–1376. [Google Scholar]
- 36.von Euler F. Proc R Soc London B. 2001;268:127–130. doi: 10.1098/rspb.2000.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pimm S L, Raven P. Nature (London) 2000;403:843–845. doi: 10.1038/35002708. [DOI] [PubMed] [Google Scholar]